Abstract
The reversible ubiquitylation of histone H2B has long been known to regulate gene transcription, and is now understood to modulate DNA replication as well. In this review, we describe how recent, genome-wide analyses have demonstrated that this histone mark has further reaching effects on transcription and replication than once thought. We also consider the ongoing efforts to elucidate the molecular mechanisms by which H2B ubiquitylation affects processes on the DNA template, and outline the various hypothetical scenarios.
Keywords: chromatin, DNA replication, H2B ubiquitylation, histone modification transcription
Abbreviations
- ARS
autonomous replication site
- H2Bub
monoubiquitylated H2B
- HU
hydroxyurea
- RNAP II
RNA Polymerase II
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- ssDNA
single-stranded DNA
Introduction
The eukaryotic genome exists in the form of chromatin, a tight complex of DNA, histones, and other proteins.1 Chromatin restricts the accessibility of eukaryotic DNA to the cellular machinery, enabling careful regulation of processes that occur on the DNA template. Such accessibility can be regulated through various mechanisms, including the posttranslational modification of histones. One such modification involves the addition of an ubiquitin monomer to lysine 120 of human histone H2B (or lysine 123 of yeast H2B), which regulates both transcriptional initiation and elongation, in part through mediating methylation of lysines 4 and 79 of histone H3.2,3 Both the addition and removal of ubiquitin are critical for the transcriptional process, with deubiquitylation being performed by the USP22 protein (Ubp8 in yeast) of the SAGA co-activator complex.4,5 More recently, ubiquitylated H2B (H2Bub) has been found to be associated with the recruitment of chromatin remodeling complexes during DNA repair and transcription, underscoring the critical role of this modification.6,7 Two reports published in late 2014 showed the genome-wide effects of H2Bub, revealing that regulation of this modification has far wider effects on transcription and DNA replication than previously thought. Loss of H2Bub resulted in rapid progression of perturbed DNA replication throughout the yeast genome, while disruption of the SAGA complex affected RNA Polymerase II recruitment and transcription at all active genes (Figs. 1 and 2).8,9 However, despite the importance of H2Bub and related machinery in these integral cellular processes, the underlying molecular mechanisms remain elusive. This review will summarize recent findings on the effects of H2Bub on transcription and replication, and briefly outline possible directions for future research into H2Bub.
Figure 1.
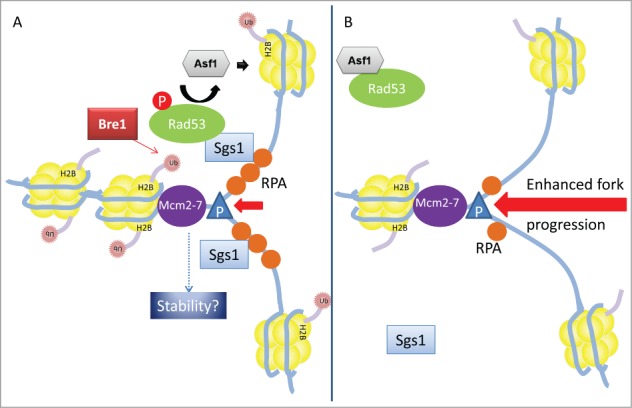
Model of the possible role of H2Bub during DNA replication stress. (A) Ubiquitylation (Ub; pink stars) of H2B by Bre1 (red rectangle) may facilitate chromatin reassembly under replication stress, thereby delaying progression of the replisome, which consists of DNA Polymerase I (blue triangle, P) and the Mcm2–7 helicase (purple oval), among other proteins. This serves to preserve the stability of the replication fork until it can be repaired. Increased assembly of nucleosomes (composed of 8 histones, shown as yellow circles) behind the replisome may also facilitate recruitment of the helicase Sgs1 (required to unwind DNA structures generated during DNA repair; shown as a blue rectangle) and phosphorylation of Rad53 (green oval), thereby further enhancing stability of the fork. Phosphorylated Rad53 dissociates from the histone chaperone Asf1 (gray hexagon), enabling the latter to deposit acetylated H3/H4 onto DNA (blue lines). (B) In cells lacking H2Bub, movement of the replication fork is less restricted, which results in the formation of shorter tracts of single-stranded DNA coated with RPA (orange circles). This reduces Sgs1 recruitment, and delays Rad53 phosphorylation.
Figure 2.
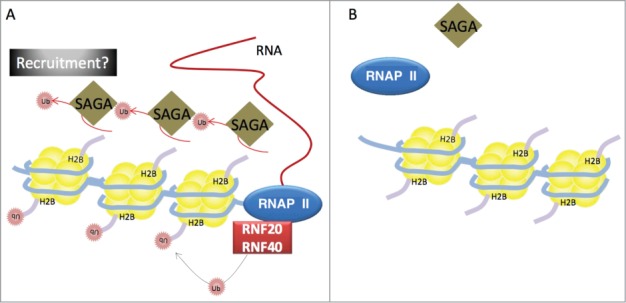
Model of the possible role of H2Bub in transcription. (A) RNA Polymerase II (RNAP II, blue oval) and associated proteins are recruited to the promoters of protein-coding genes; nucleosomes are subsequently dissociated to allow the transcription machinery to traverse the gene and synthesize mRNA (red line). Nucleosomes are reassembled behind RNAP II. Recent findings suggest that H2Bub may facilitate the recruitment of the SAGA complex (brown diamond) to protein-coding genes, in a process independent of the advancing RNAP II. SAGA-mediated deubiquitylation of H2B may enable nucleosome disassembly; the ubiquitin E3 ligase RNF20/RNF40 (red rectangle; mammalian homologues of yeast Bre1), which travels with the transcriptional machinery, subsequently ubiquitylates H2B, facilitating histone reassembly. (B) In the absence of H2Bub, RNAP II is not able to efficiently transcribe certain genes. The “adaptor model” suggests this is because H2Bub creates an epitope necessary for the recruitment and/or retention of the transcriptional machinery, while the “structural model” suggests that H2Bub induces changes in chromatin structure required for RNAP II to traverse the gene.
H2Bub Regulates Replication Fork Progression Under Stress
Accurate DNA replication is critical for faithful transmission of the genome and, as such, several cellular mechanisms have evolved to protect replicating DNA from intracellular and extracellular hazards (known collectively as “replicative stress”).10 One source of replicative stress is the chemical hydroxyurea (HU); the presence of HU results in the activation of the intra-S phase checkpoint, which serves to prevent the DNA replication fork from collapsing.11 However, our understanding of the mechanism underlying intra-S checkpoint activation remains incomplete; in one model, stress causes decoupling between DNA polymerase and the Mcm2-7 helicase (Fig. 1). The unconstrained helicase unwinds double-stranded DNA, causing the accumulation of single-stranded DNA (ssDNA); excess ssDNA results in the phosphorylation and activation of the Mec1/ATR kinase and its downstream effector, Rad53.12,13 Phosphorylation of Rad53 is facilitated by the Sgs1 helicase; Sgs1 also contributes to the re-initiation of stalled forks, by resolving DNA structures formed during the recombination repair pathway.14,15
Yeast cells lacking H2Bub are hypersensitive to HU, and H2Bub was thus hypothesized to relieve replication stress.16 While testing this hypothesis, Lin et al. made the unexpected finding that replication fork progression is faster in H2Bub-deficient mutants than in wild-type cells under HU stress, when observed at the genome-wide level.9 Importantly, the increase in fork progression appears to be independent of DNA damage-induced ribonucleotide production, suggesting that H2Bub may play a direct role in regulating fork progression.17 In addition, the genetic studies of Lin et al. suggest that H2Bub may act in parallel with the Mec1 and/or Sgs1 pathways to overcome replication stress.9 In support of this idea, the loss of H2Bub is able to partially rescue the HU hypersensitivity of a Rad53 mutant, indicating that H2Bub may act upstream of Rad53.9 Phosphorylation of Rad53 causes it to dissociate from the histone chaperone Asf1; Asf1 deposits acetylated H3/H4 dimers onto nascent DNA.18,19,20 Thus, the findings of Lin et al. suggest that H2Bub may regulate nucleosome assembly in response to replication stress by controlling the availability of Asf1.9 The authors also confirmed an earlier report that H3 occupancy at replication origins during S phase under HU is reduced in cells lacking H2B.9,16 Such a finding suggests defective assembly and/or stability of nucleosomes in the absence of H2Bub. Such H2Bub-mediated nucleosome assembly behind the replication fork may slow replication by (i) impeding the movement of the replication fork during the subsequent round of replication, or (ii) transmitting a signal to the fork during the current round of replication. The latter scenario is a formal possibility, but it is not clear how such a signal would be transmitted. However, it is apparent that H2Bub coordinates intra-S checkpoint activation and chromatin assembly during replication under stress, and such regulation may help to maintain genomic stability (Fig. 1).
SAGA Regulates Genome-wide Transcription
It is well established that H2Bub is required for transcription, with both ubiquitylation and deubiquitylation of H2B involved in the progression of RNA Polymerase II (RNAP II) along DNA.4,21 During transcription, H2Bub is deubiquitylated by USP22 (Ubp8 in yeast), a component of the co-activator SAGA complex. Genome-wide location studies of SAGA complex subunits in different organisms reached various conclusions as to the extent of SAGA binding; while some reported only a few hundred binding sites for SAGA subunits, mainly at a subset of promoters, others observed more wide-spread occupancy.22,23,24,25,26 Bonnet et al. recently confirmed that SAGA deubiquitylates H2B in the coding region of all expressed genes in yeast and human cells—whereas earlier studies reached their conclusions by inspecting the occupancy of SAGA subunits, Bonnet et al. examined deubiquitylase activity by measuring H2Bub distribution in the coding regions of active genes upon disruption of SAGA.8 Earlier studies that concluded that SAGA is associated with a subset of active genes may have detected only the most stable interactions of the examined SAGA subunit with DNA.
Confirmation that SAGA acts on more genes than previously believed raises the question of whether it has a more general effect on RNAP II-mediated transcriptional regulation than once thought. Disruption of the SAGA complex in yeast did not affect background levels of RNAP II, but did reduce RNAP II occupancy at active genes.8 Bonnet et al. proceeded to examine whether this effect on RNAP II occupancy was accompanied by altered transcription patterns. As cells compensate for global changes in transcription by regulating mRNA degradation (RNA buffering), Bonnet et al. uncoupled transcription from degradation; this revealed that SAGA disruption reduces transcription of active genes, reflecting the decrease in RNAP II occupancy (Fig. 2).8,27 Finally, both Bonnet et al. and Fuchs et al. observed that global H2Bub is lost rapidly following inhibition of transcriptional elongation, and this loss is abolished in the absence of SAGA.8,28 As such, it appears that SAGA does not require RNAP II to deubiquitylate H2Bub.
What's Happening at the Molecular Level?
Both DNA replication and transcription require that nucleosomes be disassembled to expose the underlying DNA; nucleosomes are subsequently reassembled following the passage of the transcriptional or replication complexes.29,30 This dynamic of histone assembly and disassembly is reminiscent of the mercurial nature of H2B ubiquitylation. Indeed, H2Bub appears to contribute to nucleosome assembly during both transcription and DNA replication; in yeast, histone occupancy at early origins of replication under HU was reduced by elimination of H2Bub, and histone occupancy at the coding region of the GAL1 gene during transcription is reduced in double mutants of H2Bub and the histone chaperone Spt16.9,31 In addition, H2Bub may be required for the recruitment of SAGA to nucleosomes and the retention of Sgs1 at autonomous replication sites (ARSs).8,9 Furthermore, the loss of SAGA function dramatically reduces the presence of RNAP II at active yeast genes, suggesting that H2Bub may prohibit RNAP II binding or recruitment.8 Does reversible H2Bub modulate nucleosome assembly by facilitating the recruitment of certain proteins, or vice versa? To answer this question, we need to delineate the molecular mechanism(s) by which H2Bub affect(s) chromatin. Here, we outline 3 formal possibilities:
It is possible that dynamic ubiquitylation of H2B creates (or destroys) a binding site for a protein that acts as a reader of this particular modification (“Adaptor model”). This reader may serve to recruit other proteins. There is tremendous precedent for such a hypothetical reader of H2Bub, as several proteins that recognize methylated, acetylated, and phosphorylated amino acids in histones have been identified.32 Recently, over 90 human proteins were reported to preferentially bind to chromatin containing H2Bub; some of these proteins are known to be involved in transcription, DNA replication, and/or chromatin remodeling.7 However, none of these proteins have known ubiquitin-binding sites. Instead, Shema-Yaacoby et al. propose that some of the bound proteins may (i) recognize a structure at the junction between H2B and ubiquitin, (ii) cooperatively bind a region encompassing ubiquitin and other histones, or (iii) recognize a particular conformational state of chromatin induced by ubiquitylation of H2B.7
Alternatively, ubiquitin may stabilize nucleosomes or affect higher-order chromatin structure, thereby affecting DNA accessibility (“Structural model”). H2B ubiquitylation has been reported to increase nucleosome stability in vivo, which is consistent with the finding that levels of H2Bub correlate with genome-wide nucleosome occupancy.33,34 Such enhancement of nucleosome stability may restrict access of transcriptional or replication machinery to the underlying DNA.34 However, H2Bub appears to disrupt higher-order chromatin structure in vitro.35 This effect on higher-order structure is congruent with the suggestion that human H2Bub may mediate homologous recombination via relaxation of chromatin structure.6 Consequently, H2Bub may play distinct roles at the levels of nucleosome and chromatin fiber.
Finally, it should be noted that the (i) ‘adaptor’ and (ii) ‘structural’ models need not be mutually exclusive; the chromatin remodeler Swf1 was among the H2Bub-bound proteins identified by Shema-Yaacoby and colleagues.7 This suggests that the presence of H2Bub may create a zone in which peripheral chromatin is subjected to remodeling.
We propose a model whereby cellular activities, such as transcription and DNA replication, induce reversible and self-sustaining ubiquitylation of H2B (Fig. 3). In the absence of H2Bub, chromatin may exist as a restrictive and static structure, which is not conducive to the progression of processes on the DNA template. Extra- or intracellular signals may induce the ubiquitylation of H2B on the nucleosomes at a given gene or replication origin. Such ubiquitylation is dynamic, and may be predicted to induce fluctuations between permissive and restrictive chromatin structure, through one of the mechanisms outlined above. A permissive nucleosome may be required to retain and/or recruit the transcriptional or replication machinery on DNA, while partial or total formation of (restrictive) nucleosomes may be required to allow such machinery to depart from recognition sequences. As mentioned above, blocking transcriptional elongation results in the loss of H2Bub from active genes, and global H2Bub is gradually lost under replication stress; these findings suggest that processes on the DNA template are required to maintain rhythmic H2B ubiquitylation, and thereby facilitate ongoing remodeling of the chromatin environment.8,9,28 Signals that terminate these processes may thus result in the loss of dynamic H2Bub, and the return of chromatin to a steady, static state. Through being coupled to transcription in this manner, H2Bub may be a self-perpetuating signal of active transcription; this signal may facilitate the engagement of RNAP II at active genes, obviating the need for stochastic recruitment processes that would delay transcription. A similar scenario may exist for DNA replication, with enriched H2Bub marking replication origins to guide DNA polymerase; future technologies for examining rhythmic changes in genome-wide chromatin structure (chromatin breathing) during cellular processes may help to test this hypothesis. It is also possible that dynamic H2B ubiquitylation enables the cell an extra level of control over the rate of transcription and DNA replication at active genes and replication origins, by controlling the pace of chromatin breathing.
Figure 3.
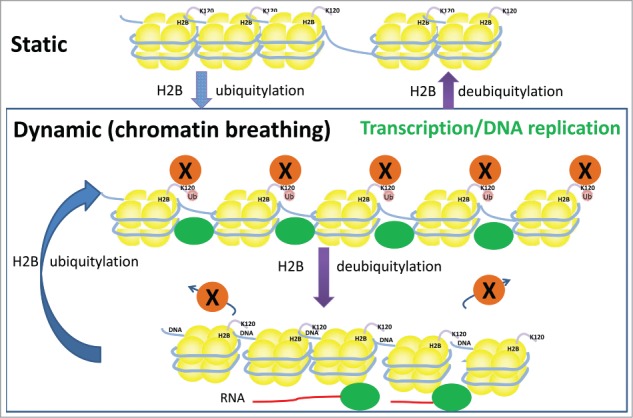
Model for how introduction of reversible H2B ubiquitylation may maintain a dynamic chromatin state. Chromatin in which H2B is not undergoing ubiquitylation and deubiquitylation may exist in a relatively static state (top panel), which is not amenable to the passage of transcriptional or replication machinery. A change in cellular conditions (metabolic state, extracellular signaling, detection of DNA damage, etc.) may induce the ubiquitylation of H2B at a given gene or replication origin. Such ubiquitylation is dynamic, and may be self-sustaining when coupled with ongoing transcription or DNA replication. Dynamic ubiquitylation is predicted to result in a fluctuating chromatin environment (lower panel; chromatin breathing), necessary for transcription/DNA replication. Ubiquitylation of H2B may enable the recruitment/retention of transcriptional/replication machinery (green ovals) at chromatin [possibly mediated by unknown protein(s) X (orange circles)], while deubiquitylation is required to enable the machinery to detach and advance along the DNA template. Blocking transcriptional elongation results in the loss of H2Bub from active genes, suggesting that transcription itself maintains the dynamic state of H2B ubiquitylation. Furthermore, replication stress results in the gradual loss of global H2Bub, and it is thus possible that DNA replication also maintains dynamic H2Bub.8,9,28 Cessation of these processes may thus result in the underlying chromatin returning to a static and silent state (top panel).
Future studies on the interactions of H2Bub in vivo are required to help determine how it influences DNA-based processes. Such studies are complicated by the transient nature of H2Bub, which makes it difficult to both purify H2Bub in association with interacting proteins, and to determine how dynamic H2Bub affects chromatin breathing. A further difficulty arises from the observation that ubiquitin is attached to H2B at locations other than K120, as well as to adjacent histones, complicating the nature of the interactions.36,37 These challenges may be overcome through the development of live imaging systems that allow visualization of vigorous nucleosome transformation and interactions, as well as by the generation of a biochemical system that enables the monitoring of long chromatin arrays with transiently-ubiquitylated H2B (possibly involving fluorescence resonance energy transfer).35 In conclusion, successive findings continue to emphasize the importance of the H2Bub mark, while the core mechanism remains, for the time being, a closed book. Opening it will no doubt lead to further insights into the roles of H2Bub and dynamic chromatin structure in the cell.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank 2 anonymous reviewers for their help in improving the article.
Funding
The authors thank the Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan, for funding.
References
- 1. Chen C, Dent SYR. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 2014; 9:749-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002; 418:104-8; PMID:12077605; http://dx.doi.org/ 10.1038/nature00883 [DOI] [PubMed] [Google Scholar]
- 3. Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 2002; 418:498; PMID:12152067; http://dx.doi.org/ 10.1038/nature00970 [DOI] [PubMed] [Google Scholar]
- 4. Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubquitylation, mediated by SAGA-associated Ubp8. Genes Dev 2003; 17:2648-63; PMID:14563679; http://dx.doi.org/ 10.1101/gad.1144003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 2004; 279:1867-71; PMID:14660634; http://dx.doi.org/ 10.1074/jbc.C300494200 [DOI] [PubMed] [Google Scholar]
- 6. Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 2011; 41:515-528; PMID:21362548; http://dx.doi.org/ 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 7. Shema-Yacoby E, Nikolov M, Haj-Yahya M, Siman P, Allemand E, Yamaguchi Y, Muchardt C, Urlaub H, Brik A, Oren M, et al. Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep 2013; 4:601-8; PMID:23933260; http://dx.doi.org/ 10.1016/j.celrep.2013.07.014 [DOI] [PubMed] [Google Scholar]
- 8. Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 2014; 28:1999-2012; PMID:25228644; http://dx.doi.org/ 10.1101/gad.250225.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin CY, Wu MY, Gay S, Marjavaara L, Lai MS, Hsiao WC, Hung SH, Tseng HY, Wright DE, Hsu GS, et al. H2B Mono-ubiquitylation facilitates fork stalling and recovery during replication stress by coordinating Rad53 activation and chromatin assembly. PLoS Genet 2014; 10:e1004667; http://dx.doi.org/ 10.1371/journal.pgen.1004667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014; 16:2-9; PMID:24366029; http://dx.doi.org/ 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 2010; 11:208-19; PMID:20177396; http://dx.doi.org/ 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- 12. Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009; 8:1038-46; PMID:19482564; http://dx.doi.org/ 10.1016/j.dnarep.2009.04.014 [DOI] [PubMed] [Google Scholar]
- 13. Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009; 8:1077-88; PMID:19505853; http://dx.doi.org/ 10.1016/j.dnarep.2009.04.023 [DOI] [PubMed] [Google Scholar]
- 14. Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J 2005; 24:405-17; PMID:15616582; http://dx.doi.org/ 10.1038/sj.emboj.7600511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ashton TM, Hickson ID. Yeast as a model system to study RecQ helicase function. DNA Repair (Amst) 2010; 9:303-14; PMID:20071248; http://dx.doi.org/ 10.1016/j.dnarep.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 16. Trujillo KM, Osley MA. A role for H2B ubiquitylation in DNA replication. Mol Cell 2012; 48:734-46; PMID:23103252; http://dx.doi.org/ 10.1016/j.molcel.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poli J, Tsaponina O, Crabbé L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 2012; 31:883-94; PMID:22234185; http://dx.doi.org/ 10.1038/emboj.2011.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emili A, Schieltz DM, Yates JR, 3rd, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell 2001; 7:13-20; PMID:11172707; http://dx.doi.org/ 10.1016/S1097-2765(01)00150-2 [DOI] [PubMed] [Google Scholar]
- 19. Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes Dev 2001; 15:1061-6; PMID:11331602; http://dx.doi.org/ 10.1101/gad.873201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell 2010; 140:183-95; PMID:20141833; http://dx.doi.org/ 10.1016/j.cell.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004; 18:184-95; PMID:14752010; http://dx.doi.org/ 10.1101/gad.1149604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010; 142:967-80; PMID:20850016; http://dx.doi.org/ 10.1016/j.cell.2010.08.020 [DOI] [PubMed] [Google Scholar]
- 23. Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 2011; 30:2829-42; PMID:21685874; http://dx.doi.org/ 10.1038/emboj.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone Acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-Independent enhancers. Mol Cell 2011; 44:410-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 2011; 41:480-92; PMID:21329885; http://dx.doi.org/ 10.1016/j.molcel.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev 2011; 25:1499-509; PMID:21764853; http://dx.doi.org/ 10.1101/gad.2046211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. Global levels of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell 2013; 52:52-62; PMID:24119399; http://dx.doi.org/ 10.1016/j.molcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- 28. Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res 2014; 24:1572-83; PMID:25049226; http://dx.doi.org/ 10.1101/gr.176487.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem 2011; 286:30504-12; PMID:21757688; http://dx.doi.org/ 10.1074/jbc.M111.264721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 2004; 14:65766; PMID:15175160; http://dx.doi.org/ 10.1016/j.molcel.2004.05.016 [DOI] [PubMed] [Google Scholar]
- 31. Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 2008; 31:57-66; PMID:18614047; http://dx.doi.org/ 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- 32. Musselman CA, Lalonde ME, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 2012; 19:1218-27; PMID:23211769; http://dx.doi.org/ 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009; 106:16686-91; PMID:19805358; http://dx.doi.org/ 10.1073/pnas.0907862106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011; 25:2254-65; PMID:22056671; http://dx.doi.org/ 10.1101/gad.177238.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher order chromatin compaction. Nat Chem Biol 2011; 7:113-9; PMID:21196936; http://dx.doi.org/ 10.1038/nchembio.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 2004; 16: 631-9; PMID:15546622; http://dx.doi.org/ 10.1016/j.molcel.2004.10.016 [DOI] [PubMed] [Google Scholar]
- 37. Wu L, Li L, Zhou B, Qin Z, Dou Y. H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol Cell 2014; 54:920-31; PMID:24837678; http://dx.doi.org/ 10.1016/j.molcel.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]


