Abstract
DNA methylation is one of the main epigenetic mechanisms that can regulate gene expression and is an important means for creating phenotypic variation. In the present study, we performed methylation profiling of 2 candidate genes for personality traits, namely DRD4 and SERT, in the great tit Parus major to ascertain whether personality traits and behavior within different habitats have evolved with the aid of epigenetic variation. We applied bisulphite PCR and strand-specific sequencing to determine the methylation profile of the CpG dinucleotides in the DRD4 and SERT promoters and also in the CpG island overlapping DRD4 exon 3. Furthermore, we performed pyrosequencing to quantify the total methylation levels at each CpG location. Our results indicated that methylation was ∼1–4% higher in urban than in forest birds, for all loci and tissues analyzed, suggesting that this epigenetic modification is influenced by environmental conditions. Screening of genomic DNA sequence revealed that the SERT promoter is CpG poor region. The methylation at a single CpG dinucleotide located 288 bp from the transcription start site was related to exploration score in urban birds. In addition, the genotypes of the SERT polymorphism SNP234 located within the minimal promoter were significantly correlated with novelty seeking behavior in captivity, with the allele increasing this behavior being more frequent in urban birds. As a conclusion, it seems that both genetic and methylation variability of the SERT gene have an important role in shaping personality traits in great tits, whereas genetic and methylation variation at the DRD4 gene is not strongly involved in behavior and personality traits.
Keywords: DRD4, methylation, novelty seeking, personality traits, SERT, urbanization
Introduction
The behavioral system of vertebrates is thought to be controlled via the dopaminergic and serotonergic neurogenic systems.1 Dopamine receptor D4 (DRD4) and serotonin transporter (SERT) are 2 prime candidate genes for personality traits. Single nucleotide polymorphisms (SNPs) within the third exon of the DRD4 gene are related to variation in novelty seeking behavior in humans and other mammals.2-9 In birds, the great tit (Parus major) is a classic model species to study avian personality, and an association between DRD4 SNP830 and exploratory behavior has also been described.10
However, the relationship between genes and behavior appears far more complex than previously described, as the relationship holds in some populations but not in others.11 This made Mueller et al. 12 suggest that the link between DRD4 gene and personality is context-dependent, probably mediated by certain characteristics of habitat at a local scale. In a recent study, we found that urban and forest great tits differed greatly in novelty seeking behavior, but we were unable to link this variation in behavior to DRD4 SNP830 (Riyahi & Senar personal observation). Heterogeneous association of DRD4 gene with exploratory behavior in different environments suggests a possible gene-gene or gene-environment interaction.11
In addition to DRD4, the SERT (also known as SLC6A4) gene is recognized as a candidate gene for anxiety, harm avoidance, and other behavioral syndromes in humans. A recent study on 12 paired urban and rural populations of the blackbird Turdus merula found a highly significant divergence at an exonic microsatellite of SERT gene between habitats.13 They concluded that the SERT gene can be considered as one of the candidate loci for local adaptation to novel conditions and urbanization. Hence, the possible gene-environment interaction has been suggested in both DRD4 and SERT genes. One of the ways to detect gene-environment interaction is investigating epigenetic modifications. DNA methylation is one of several epigenetic modifications that can directly affect gene expression. Any change in environmental condition can potentially modify the phenotype through epigenetic modification.14-16 In many cases, epigenetic marks are not inherited in a Mendelian fashion but different lines of evidence suggest that genetics can influence the epigenetic marks.17,18 Currently, several studies have shown the effect of the genotype on the epigenotype. Particularly, SNPs can influence DNA methylation, a phenomenon referred to as allele-specific methylation (ASM) or methylation quantitative trait loci (meQTL).19
Associations of DNA methylation in the SERT and DRD4 genes and personality have been described before in humans. In humans, prenatal exposure to maternal depression affects methylation patterns of the SERT promoter.20,21 Moreover, the methylation level of the SERT promoter was associated with abuse during childhood.22 In addition, the DNA methylation level of both DRD4 and SERT genes were negatively associated with attention deficit hyperactivity disorder symptom score.23 Furthermore, in genetically identical twins, methylation differences of SERT and DRD4 genes have been described, which are mostly attributable to environmental factors.24 As a result, methylation is a strong candidate mechanism to explain differences in personality between habitats. The urban habitat, due to rapid changes in diet as well as environmental pollution, is undoubtedly impacting not only the human epigenome, but also the evolution of many other species. Since urban habitats have independently been found to affect levels of DNA methylation through pollution effects25 and also personality traits,13 we hypothesize that differences in personality between urban and forest Parus major populations could be related to variation in methylation.
Since the promoter region of a gene primarily influences transcript expression, in this study we determine the methylation patterns of the DRD4 and SERT gene promoters in the great tit from 2 different environments, city and forest. In addition, we investigate the potential function of SNP830 of the DRD4 gene, which has previously been found to relate to exploratory behavior,10,11 by looking at the DNA methylation profile of the flanking interval. The great tit inhabiting Barcelona city is an ideal model species to test for that, since the urban population is genetically isolated from the surrounding forest populations26 and urban birds display higher levels of exploration and novelty seeking behavior compared to the nearby forest (Riyahi & Senar personal observations).27
This study aims to answer the following specific questions: (1) Are DRD4 and SERT promoters differently methylated in urban and forest great tits? (2) Is novelty seeking behavior linked to the level of DNA methylation in the promoter of these 2 genes? (3) Is methylation of SERT and DRD4 promoters associated with allelic states at nearby SNPs? In particular, does the synonymous substitution SNP830 in the DRD4 gene, associated with diversity in the exploratory behavior in the great tit,10,11 exert its effect by influencing methylation at nearby CpG sites?
Methods
DNA sample set
We obtained blood samples from 96 different birds that were captured from October to March in 2012 and 2013 for personality experiments in captivity. The sample was composed by 46 forest birds from Can Catà field station, located in Collserola National Park, close to Barcelona city (3 km). It is a mixed forest consisting mainly of pure oak (Quercus ilex and Quercus cerrioides) located at the bottom of the valleys and Aleppo pine (Pinus halepensis) forests in the upper hills. A further 50 city birds were captured at 2 different urban parks in Barcelona city (Ciutadella Park and Setmenat-Sarria Park). The blood samples were stored in pure ethanol at 4°C. Finally, we collected brain biopsies from 13 great tits (from city and forest) that died during fieldwork or in captivity. DNA was extracted using Ecogen MasterPure DNA Purification kit (MCD85201).
Gene sequencing
The genotyping for SNP830 of DRD4 in all the samples was performed following the protocol described by Fidler et al.10 Additional polymorphisms within the DRD4 and SERT loci were genotyped by standard PCR amplification and direct sequencing of the resulting amplicons. Approximately 200 ng genomic DNA was amplified in 25 µl reactions containing 1x NH4 reaction buffer, 25 ng of each oligonucleotide primer, 0.2 mM of each dNTP, 1.5 mM MgCl2 and 2 units DNA Taq DNA Polymerase (Bioline). PCR was performed for 35 cycles (see Table 5 for primer sequences). The resulting amplicons were purified using ethanol precipitation and subsequently sequenced in both directions using the BigDye Terminator reaction kits on an ABI 3730 DNA analyzer (PE Bio-systems). The sequence electropherograms were interrogated using Sequencherv4.6 (Gene Codes Corporation, MI, Ann Arbor, USA) to distinguish heterozygous and homozygous samples. The sequence of SERT gene and SNPs has been deposited in GenBank: accession number KP869099.
Table 5.
List of primers. The primers used for bisulphite conversion and pyrosequencing are marked in bold font
| DRD4 genotype promoter 1F | GGGAAGGACAGTGCTTGGATCTG |
| DRD4 genotype promoter 1R | AGTGCTCTAGATAAGTTGGCAAATGCA |
| DRD4 genotype promoter exon 1F | CTCGGAGTAGACGTAGAGGGGCAGGAC |
| DRD4 genotype promoter exon 1R | AGGCTCCTCCCGCGGCTCGGCGGGCA |
| DRD4promoter Bis F | AGTAGTTGGTGGTGGTTTTGA |
| DRD4pyro seq F | TGAGGAGGATGAGGAGGATGT |
| DRD4promoter Bis R | CTAACATTACTTAAAAAACAACCC |
| DRD4exon 3 Bis F | TGGGATAAGTTGGTATATTTTTTAT |
| DRD4pyro seq F | TTTTAGTTATTTTTGATAGTTATT |
| DRD4exon 3 Bis R | CAAAAAACTATATCACCCCCCAACC |
| SERT gentype F | CATCTTCTCCTTTGCTACAGCC |
| SERT genotype R | ACAGAGCCTCAGAAGTTAGTTGA |
| SERTpromoter Bis F | TTAGGGGTTTTGTTTTATTGTTTGTG |
| SERTpyro seq 1F | GAGATTTGTTTTGGTTAT |
| SERTpyro seq 2F | GGTTATTAATTATTATAGTA |
| SERTpromoter Bis R | CAAATTACCTACTCCATAATTC |
Bisulphite treatment and strand-specific methylation analysis
Approximately 1 μg DNA was subjected to sodium bisulphite treatment and purified using the EZ GOLD methylation kit (ZYMO, Orange, CA, USA) and was used for bisulphite PCR analysis. Bisulphite PCR primers for each region (see Table 5 for primer sequences) were used with Hotstar Taq polymerase (Qiagen, Crawley, UK) at 40 cycles and the resulting PCR product cloned into pGEM-T easy vector (Promega, Madrid, Spain).
Quantitative methylation pyrosequencing
Pyrosequencing was used as an accurate method of quantifying total methylation at CpG dinucleotides within a PCR amplicon. Standard bisulphite PCR was used to amplify across the region of interest with the exception that the reverse primers were biotinylated. The entire biotinylated PCR product (diluted to 40 μl) was mixed with 38 μl of binding buffer and 2 μl (10 mg/ml) streptavidin-coated polystyrene beads. Bead-amplicon complexes were captured on a vacuum prep tool (Qiagen, Crawley, UK) and the PCR products denatured using 0.2 M NaOH. The denatured DNA was resuspended in 40 pmol of sequencing primer dissolved in 12 μl water and primers annealing was achieved by heating the sample to 80°C for 2 min before cooling to room temperature. For sequencing, internal forward primers were designed to the complementary strand (see Table 5). The pyrosequencing reaction was carried out on a PyroMark Q96 instrument (Qiagen, Crawley, UK). The interrogated peak heights of C/T variants at CpG dinucleotides were determined using the pyrosequencing commercial software.
Phenotyping
Two different standard tests were performed. Each bird was tested alone in the whole sequence of tests and used only once. On the morning of the second day after capture, we performed a standard novel object tests inside the individual cages (1 × 1 × 1.5 m) using a pen light battery put on the feeder where we had previously introduced a few mealworms.28,29 We measured the latency to approach the feeder (in seconds) within a period of 10 min. Birds were then allowed to continue with their activities for one hour.
We then performed a standard novel environment test.29 Birds were first introduced into an individual cage (100 × 40 × 40 cm) within the experimental room. After 30 min, the individual cages were opened with a remote control string to allow the birds to fly into the observation room. The size of the room was 3 × 2 × 2 m and contained 5 artificial trees, as in Verbeek et al.29 We observed the birds from a one-way screen and we recorded movement of birds with a video camera. Number of flights and hops within the first 2 min after entering the room was used as exploration score.30 Exploration score was standardized by date (days from September 1) to account for within-season temporal trends.30,31
Statistical methods
Comparisons between genomic DNA of different species were performed using BLASTN (http://www.ncbi.nlm.nih.gov). Screening for CpG islands (>200 bp; obs/exp 0.6; GC content >50%) utilized the CpG island finder bioinformatics tool (http://dbcat.cgm.ntu.edu.tw). To normalize the data, the average value of DNA methylation percentage were arcsin transformed. The relation between exploration score and DNA methylation of SERT and DRD4 was tested using General Linear Model (GLM) in STATISTICA 8 software (StatSoft 2013). In relation to the novel object test, the response included censored observations (birds that did not approach the novel object). Applying standard statistical methods to censored data, or not taking them into account, can lead to biased estimates. Hence we applied a stratified Cox proportional hazards regression model, a specialized nonparametric regression survival analysis which deals with this problem,32 to check the latency to approach to the novel object according to DNA methylation. We included the methylation levels of SERT and DRD4 as independent variables and habitat (forest vs. city) as a grouping variable.
The association of each SNP with average CpG DNA methylation percentage of DRD4 promoter was tested using GLM in STATISTICA 8 software. In addition, we used the same method to reveal SNP associations within the SERT promoter and DNA methylation.
Haplotypes were reconstructed with the PHASE program.33 Population genetic statistics and neutrality tests such as Tajima's D were calculated using DNAsp5.1034 and Arlequin3.1135 software packages. Linkage disequilibrium was determined using Haploview.36 Chi-square test was performed to check whether genotype frequencies of each SNP in each population followed the Hardy-Weinberg equilibrium and to test whether the genotypes frequencies differed between habitats. We used a Cox proportional hazards analysis to assess the association between neophobia (novel object test) and DRD4 and SERT different SNPs one by one, and we estimated the association between exploration score in a novel environment and DRD4 and SERT SNPs with General Linear model (GLM). Cox and GLM analysis were done using STATISTICA 8 (StatSoft 2013) and R software (R Development Core Team 2011).
Results
Extensive genotyping of the DRD4 promoter interval in great tit
We designed 2 PCR assays which resulted in amplicons covering ∼1.5 kb of the DRD4 exon 1 and proximal promoter interval. Subsequent sequencing of the PCR products revealed 9 SNPs, 7 located in the promoter region (detailed in Table 1). Four of the SNP variants (SNP784, SNP830, SNP835, and SNP876) deviated from Hardy-Weinberg equilibrium in our urban population. Allele frequencies showed significant differences between the urban and the forest population only for SNP830 (10086), which is located in exon 3 (χ22 = 9.99, P < 0.01). Contrary to other northern Europe populations,11,12 the In/del ID15 was not observed in our populations. Two SNPs were observed in exon 1 of the DRD4 gene. One of these coding polymorphisms, SNP1884, resulted in an amino acid substitution, a change from alanine to valine in amino acid position 11. A valine at this position is conserved among passerine birds, while it is not found in other avian orders,37 which makes the functional significance of this amino acid change uncertain. However, the functional prediction returned by SIFT (http://sift.jcvi.org) was “likely benign.”
Table 1.
Allele names and minor allele frequency (MAF) of each SNP for the DRD4 gene in each population. Population sample size: 46 forest great tits and 50 urban great tits.
| Major/minor | MAF | MAF | HW P-value | |||
|---|---|---|---|---|---|---|
| DRD4 locus | allele(s) | location | (forest) | (urban) | (forest) | HW P-value (urban) |
| 1. SNP784 | G/A | promoter | 0 | 0.057 | 1 | <0.001 |
| 2. SNP795 | G/A | promoter | 0.026 | 0.029 | 1 | 1 |
| 3. SNP830g | G/T | promoter | 0 | 0.057 | 1 | <0.001 |
| 4. SNP835 | T/G | promoter | 0.079 | 0.057 | <0.001 | <0.001 |
| 5.SNP876 | T/G | promoter | 0 | 0.057 | 1 | <0.001 |
| 6. SNP1006 | G/A | promoter | 0.132 | 0.129 | 1 | 1 |
| 7. SNP1173 | C/G | promoter | 0.368 | 0.414 | 1 | 0.256 |
| 8. SNP1878 | C/T | exon1 | 0.042 | 0.014 | 1 | 1 |
| 9. SNP1884 | C/T | exon1 | 0 | 0.014 | 1 | 1 |
| 10.SNP830(10086) | C/T | exon3 | 0.180 | 0.150 | 0.97 | 0.98 |
Characterization of the SERT promoter in great tit
The promoter sequence of the SERT gene has not previously been reported for the great tit. As a prerequisite for identifying possible genetic variants at this interval, we performed sequence homology comparisons between additional bird species for which genomic sequence was available to enable us to identify the orthologous region in great tits. We performed BLAST2 sequence analysis comparing the genomic sequence of common blackbird (Turdus merula) for which the SERT promoter interval was already available (Genbank accession number KC584781.1). We subsequently designed PCR primers to conserved regions identified in multiple species [Collared flycatcher (Ficedula albicollis), Atlantic canary (Serinus canaria), American crow (Corvus brachyrhynchos), Rock dove (Columba livia)] and performed PCR on DNA derived from great tits. The resulting ∼850 bp PCR product was sequenced, revealing that the amplicon comprised SERT exon 1 and a 770 bp interval within the proximal promoter (Fig. 2a), which contained 11 SNP variants in our populations (Table 2). All of the SNPs were in the Hardy-Weinberg equilibrium except SNP65, in both populations. Allele frequencies were significantly different between urban and forest populations in SNP136 and SNP234 (χ22 = 6.45, P = 0.039; χ21 = 9.96, P < 0.001, respectively), so that AA genotype for SNP136 and TT genotype for SNP234 were more frequent in the urban population while GA (SNP136) and TA (SNP234) genotypes were more abundant in forest birds (Table 2).
Figure 1.
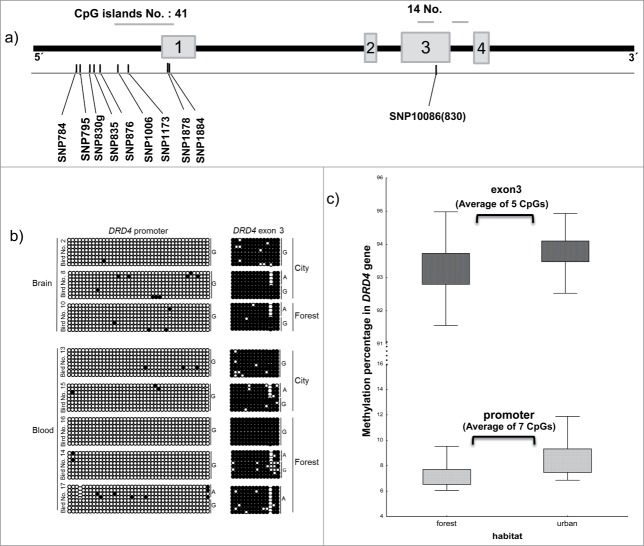
Structure and methylation profile of the DRD4 gene. (a) Schematic representation of the DRD4 gene. Horizontal bars show the number and location of CpG islands and gray boxes represent exons. The location of SNPs is shown by black bars. (b) Methylation status at DRD4 locus in brain and blood-derived DNA samples. The left panel shows the methylation profile of the promoter and the right panel shows the results for the CpG island within exon 3. Each circle represents a single CpG dinucleotide on a DNA strand. (•): methylated cytosine; (○): unmethylated cytosine. (c) Methylation percentage of DRD4 promoter and exon 3 regions comparing urban and forest populations. Differences are significant (see text).
Figure 2.
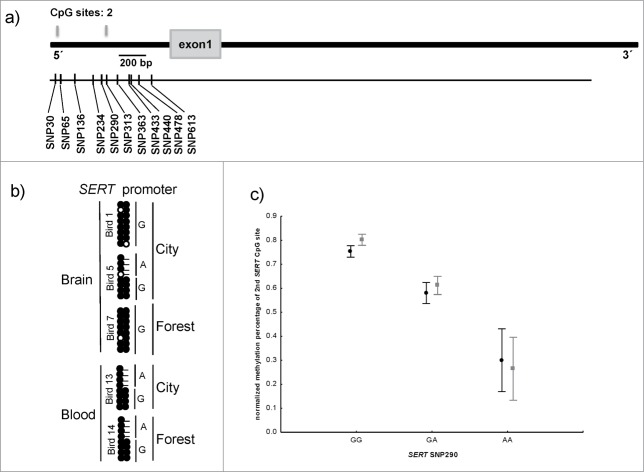
Structure and methylation profile of the SERT promoter. (a) The Schematic representation of SERT gene. Gray boxes showing the location of CpG sites and the gray box represents the exon. The location of SNPs is shown by black bars. (b) The methylation status at 2 CpG sites in the SERT promoter in brain and blood-derived samples. (c) Variation in methylation percentage of second CpG site in the SERT promoter, according to habitat and SNP290 genotype. The black data points represent forest birds and the gray urban birds. Methylation percentage was both affected by genotype and habitat, but the lack of significant interaction suggests that the difference between habitats was similar for the different genotypes (see text for tests).
Table 2.
Allele names and minor allele frequency (MAF) of each SNP for the SERT gene in each population. Population sample size: 46 forest great tits and 50 urban great tits.
| SERT locus | Major/minor allele(s) | location | MAF (forest) | MAF (urban) | HW P-value (forest) | HW P-value (urban) |
|---|---|---|---|---|---|---|
| 1. SNP30 | t/g | promoter | 0.088 | 0.022 | 1 | 1 |
| 2. SNP65 | c/t | promoter | 0.188 | 0.267 | 0.002 | 0.011 |
| 3. SNP136 | a/g | promoter | 0.238 | 0.111 | 1 | 0.144 |
| 4. SNP234 | t/a | promoter | 0.212 | 0.067 | 0.229 | 1 |
| 5. SNP290 | g/a | promoter | 0.138 | 0.156 | 1 | 1 |
| 6. SNP313 | g/a | promoter | 0.075 | 0.078 | 1 | 0.445 |
| 7. SNP363 | a/g | promoter | 0.05 | 0 | 0.150 | 1 |
| 8. SNP433 | c/t | promoter | 0 | 0.011 | 1 | 1 |
| 9. SNP440 | c/a | promoter | 0.262 | 0.211 | 0.476 | 0.573 |
| 10. SNP478 | c/t | promoter | 0.275 | 0.300 | 0.755 | 1 |
| 11. SNP613 | g/t | promoter | 0.295 | 0.350 | 1 | 0.972 |
Identification of CpG islands in great tit
Approximately 60% of gene promoters are associated with DNA sequences rich in CpG dinucleotides, termed CpG islands. These genomic features are usually unmethylated and permissive to transcription. Tissue-specific difference in expression can be explained by different methylation profiles with highly methylated regions being associated with robust gene silencing. To determine the methylation profiles of the DRD4 and SERT promoters, we first performed a bioinformatics screen for CpG islands. This revealed that the DRD4 locus contains 3 CpG islands. The largest encompasses the first exon and 5′UTR (1094 bp; GC 66%), whereas 2 smaller CpG island are intergenic overlapping exon 3 (350 bp; GC 52%) and within intron 3 (262 bp; GC 86%), respectively. Interestingly the same analysis on SERT sequence failed to identify any CpG islands, the interval being CpG sparse, containing only 2 CpG dinucleotides.
Methylation profiling of the DRD4 promoter and exon 3 CpG islands
To determine the methylation profile of the 2 largest CpG islands in the DRD4 locus in brain and blood-derived DNA, we performed bisulphite PCR and sequencing. To ensure that we would identify any genotype-dependent methylation effects, we performed the analysis on birds with different genotype combinations. This revealed that the DRD4 promoter interval was robustly unmethylated in both blood and brain, whereas the CpG island overlapping exon 3 was fully methylated. The overall methylation profiles in samples with different genotypes were similar; however, 2 of the SNPs abolish CpG dinucleotides (SNP1878 and SNP1884) that would indirectly affect methylation at these sites. We observed no association between the synonymous SNP830 genotype and the methylation surrounding exon 3, suggesting that variation in methylation due to genotype does not explain the function for this SNP in relation to personality.
To allow for accurate quantification of methylation in an extended cohort of birds from different environments, we optimized the bisulphite PCRs for pyrosequencing. This allowed for 7 of the 41 CpGs included in the DRD4 promoter PCR amplicons to be measured, while 5 of the 14 CpGs within the exon 3 CpG island PCR product would be quantified. The average DNA methylation level across all CpGs was 8% (SE = 0.16%) for DRD4 promoter and 93% (SE = 0.08%) for the DRD4 exon 3. Furthermore this analysis revealed, despite similar methylation profiles at these 2 genomic locations, that urban-dwelling great tits exhibited ∼1% higher methylation levels than forest-dwelling birds (DRD4 promoter: urban = 8.61±0.24, forest = 7.38 ± 0.18, F = 16.48, df = 1,89, P < 0.005; DRD4 exon3: urban = 93.82 ± 0.09, forest = 93.19 ± 0.13, F = 14, df = 1,93, P < 0.005).
Allele-specific methylation at the SERT promoter in great tit
Bisulphite PCR encompassing the 2 CpG dinucleotides within the SERT gene promoter revealed that both sites were fully methylated in brain and blood-derived DNA samples consistent with reports for CpG-poor promoters.38 One of the CpG dinucleotides, located 288 bp from the transcriptional start site (TSS), is abolished by a genomic variant (SNP290) in which the G nucleotide is replaced by an A variant, therefore behaving as a site of ASM. Bisulphite pyrosequencing enabled us to quantify the methylation of the 2 CpG dinucleotides in our extended cohort. Consistent with the allele-specific bisulphite PCR results, average methylation at the first CpG site within the SERT promoter was 83% (SE = 0.26%), with methylation level being ∼2% higher in urban-dwelling than in forest great tits (F = 8.9, df = 1,94, P < 0.005). However, the methylation levels of the second CpG site clearly stratified into three groups dictated by SNP290 genotype: birds homozygous for the A allele were ∼8.1% (SE = 0.55%) methylated, GA heterozygous individuals were ∼31.3% methylated (SE = 1.03%) and homozygous G birds were ∼49.0% methylated (SE = 0.85%) (Fig. 2c). Methylation percentage at SERT second CpG site was not only significantly affected by genotype but also by habitat, so that city birds showed ∼3.9% hypermethylation than forest birds (urban: 46.89 ± 1.40%; forest: 43.01 ±1.49 %) (habitat: F = 5.69, df = 1,78, P < 0.05; SNP290 genotype: F = 116.74, df = 1,78, P < 0.005; habitat × genotype: F = 0.26, df = 1,78, P = 0.60) (we only considered here GA and GG genotypes because the AA genotype had a too small frequency in our populations to provide enough sample size). A similar genotype-associated methylation profile was also observed for SNP440 and SNP478, located 150–188 bp from SNP290, presumably because the variants are in linkage disequilibrium and the methylation profile is dictated by SNP290.
Trait association
DNA methylation in the DRD4 promoter or in DRD4 exon 3 was not significantly associated with personality scores, measured as time to approach to a novel object and exploration score in a novel environment (Table 3a and b). However, we found that percentage of methylation in SERT second CpG site was related to exploration score. Nevertheless, the relationship was complex, with exploration score showing a significant interaction between habitat and methylation percentage (Table 3d). Although no relationship was found between exploration score and methylation for forest birds (Fig. 3b), the relationship was marginally significant for urban birds (Table 4, Fig. 3a). No relationship was found for SERT first CpG site (Table 3c).
Table 3.
Result of GLM on the variation in exploration score (season corrected) in relation to methylation. Results for the methylation percentage of (A) DRD4 promoter, (B) methylation percentage of DRD4 exon 3, (C) methylation percentage of SERT first CpG site and (D) methylation percentage of second SERT CpG site and habitat (forest vs. urban).
| F | Df | P | ||
|---|---|---|---|---|
| A) | ||||
| Habitat | 0.04 | 1,58 | 0.85 | |
| methylation of DRD4 promoter | 0.3 | 1,58 | 0.58 | |
| Habitat x methylation of DRD4 promoter | 0.11 | 1,58 | 0.74 | |
| B) | ||||
| Habitat | 2.77 | 1,62 | 0.10 | |
| methylation of DRD4 exon3 | 3.08 | 1,62 | 0.08 | |
| Habitat x methylation of DRD4 exon3 | 2.82 | 1,62 | 0.10 | |
| C) | ||||
| Habitat | 0.36 | 1,63 | 0.55 | |
| methylation of SERT first CpG site | 0.12 | 1,63 | 0.73 | |
| Habitat x methylation of SERT first CpG site | 0.42 | 1,63 | 0.52 | |
| D) | ||||
| Habitat | 2.84 | 1,59 | 0.10 | |
| methylation of SERT second CpG | 0.49 | 1,59 | 0.49 | |
| Habitat x methylation of SERT second CpG site | 3.9 | 1,59 | <0.05 |
Figure 3.
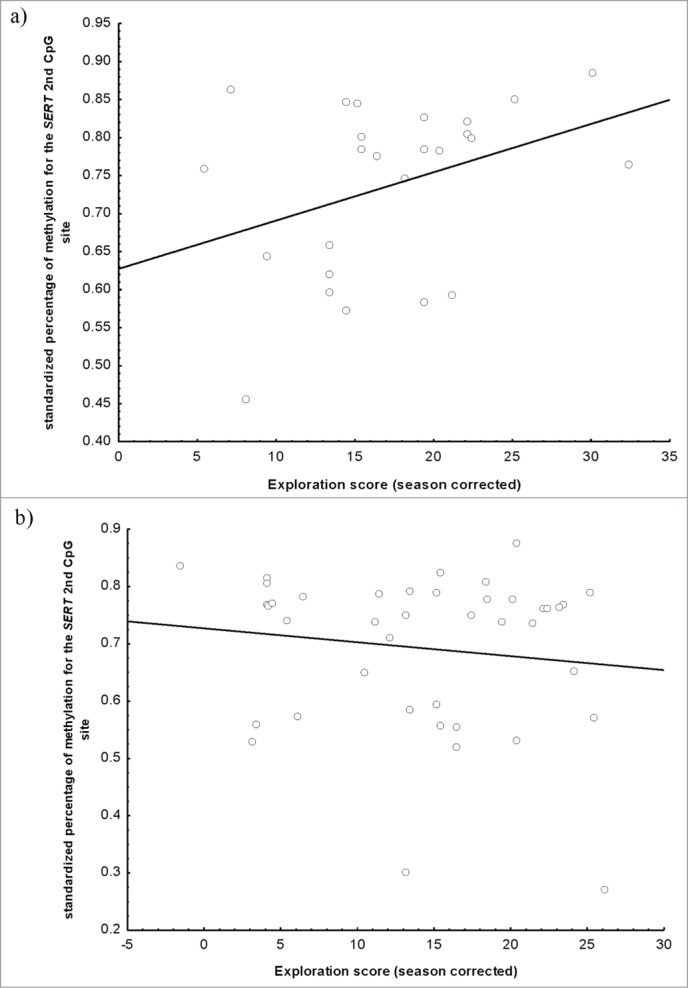
Exploration score of wild great tits, measured as number of movements during 2 min in a standard room with five artificial trees, in relation to methylation level of SERT second CpG site. We provide independent figures for urban (a) and forest (b) great tits (urban: r = 0.37, P = 0.06; forest: r = −0.13, P = 0.40).
Table 4.
Result of GLM on the variation in exploration score (season corrected) in relation to methylation percentage of SERT second CpG site. The results are provided for the 2 sampled populations: forest and urban.
| Population | F | Df | P |
|---|---|---|---|
| Forest | 1.03 | 1,38 | 0.32 |
| Urban | 3.57 | 1,23 | 0.07 |
No SNPs in the DRD4 gene or in the SERT promoter were significantly associated with the behavioral traits tested (all P > 0.42), with one remarkable exception: SERT SNP234. This SNP showed highly significant association with the novel object test, with birds with TT genotype approaching faster to the novel object (Cox analysis Z = 2.03; P < 0.05; Fig. 4). The explained variance in response to novel object by SNP234 was 7%. Accordingly, and as shown before, the TT genotype was also significantly more frequent in the urban population while TA heterozygotes were more abundant in forest birds. The Tajima's D neutrality tests were not significant for SERT SNP234 in either population.
Figure 4.
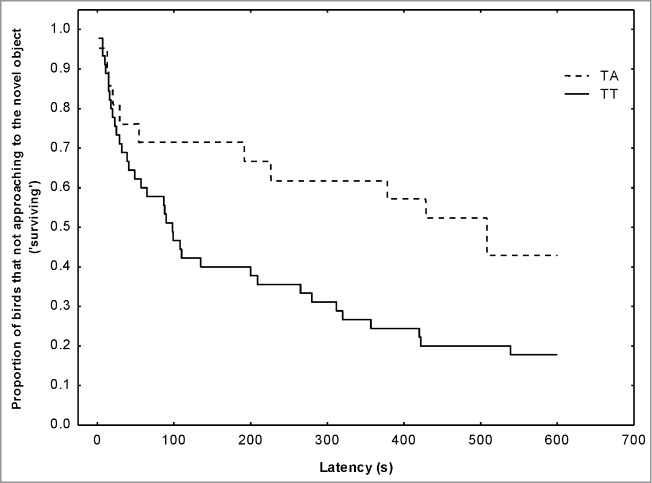
Survivorship plot function for latency to approach a novel object (a penlight battery on the feeder) by wild great tits according to SERT SNP234 genotypes. Black line represents TT birds and dash line represents TA birds. The figure shows the proportion of great tits that do not approach the object (‘surviving’) up to the respective time interval.
Discussion
Recent studies in human and laboratory animals have found a relationship between methylation changes and intraspecific variation in personality traits. For instance, rats treated with endocrine-disrupting chemicals showed changes in mate choice behavior due to methylation variation, even after 3 generations.39 Here we found, for the first time in a free-living bird species, that variations in methylation levels, via a region of allele-specific methylation (ASM) within the SERT promoter, may be related to personality traits and may modify exploratory behavior in the urban-dwelling great tits. In addition, our results showed that methylation variation could be relevant in urbanized areas.
Methylation, habitat, and novelty seeking behavior
DRD4 and SERT have both previously been suggested as 2 prime candidate genes for personality traits and, therefore, have been the focus of our research. Regarding the DRD4 gene, the only epigenetic difference we observed was associated with habitat and not tissue-specificity or underlying genotype. The ∼1% difference in methylation between urban and forest-dwelling birds did not explain personality variations between habitats.
In SERT promoter, we found a similar ∼2% hypermethylation in urban compared to forest birds at CpG dinucleotide located nearest transcriptional start site (TSS). However, intraspecific variation in methylation was far more extreme at the second CpG site, with levels dictated by the genotype of a single SNP irrespective of the habitat. The higher level of methylation within DRD4 and SERT loci in urban birds might be due to some environmental conditions, for instance pollution. In support of this, air pollution has been previously shown to be related to whole genome hypermethylation in the sperm DNA of mice.25 It remains to be determined whether additional loci show this habitat-associated gain in methylation, and if this occurs genome-wide in our great tit population.
Interestingly, we found that the relationship between methylation levels of the SERT second CpG site and exploration score interacted significantly with habitat, with more explorative urban birds tending to have higher levels of methylation at this site; however, no relationship was found for forest birds. Although there is no available study on wild birds, studies in human have shown how early environmental conditions affect SERT methylation differences between individuals and, hence, account for some inter-individual phenotypic variation.40,41 This could explain habitat differences we have found in this study. Furthermore, the importance of early stress in shaping higher methylation in serotonin transporter gene and, consequently, health composite scores has been reported recently in monkeys.42 Therefore, we suggest that early environmental conditions in the city may adjust methylation at this particular CpG site in the SERT gene, and this may affect adult behavior.
Differential methylation in the promoter of the SERT gene may lead to alteration in different gene expression levels (mRNA and protein). Nevertheless, the mechanism by which allele-specific methylation can affect SERT gene expression and, consequently, personality is still unclear. Future studies should provide further insights into the underlying mechanisms by which genetic and epigenetic modification in the SERT gene interact to shape personality traits.
SERT SNPs and novelty seeking behavior
Previous studies had tried to link novelty seeking behavior and genetic variability. However, although earlier works found an association between DRD4 SNP830 polymorphism and novelty seeking in great tits,10 this association was only found in one population.11,12,43 More recently, the SERT gene has been suggested as one of the candidate genes for local adaptation to novel conditions and urbanization.13 Since we found that one of the main differences between urban and forest great tits probably relied on personality,27,44 we thought that it could be valuable to look for a direct relationship between SERT polymorphisms, in addition to DRD4, and personality. Out of 21 SNPs (10 SNPs in DRD4 and 11 SNPs in SERT), we found a highly significant association between SERT SNP234 and novelty seeking behavior: TT birds approached faster to a novel object than TA birds. Accordingly, TT birds were more frequent within the urban habitat. To explore whether this polymorphism is indeed adaptive, we performed Tajima's D neutrality tests, which were not significant in either population. In this case, adaptation would mean an increase in a pre-existing polymorphism, and natural selection based on standing variation is notoriously difficult to detect by neutrality testing.45 The actual mechanism by which SNP324 affects personality remains to be known. Non-synonymous substitutions and distinctive codon bias have been reported in the same region of DRD4 in Passeriform species, which may have contributed to the degree of behavioral diversity and potential for adapting to the novel environments such as big cities.37
Final considerations
As we know, natural selection can be triggered by epigenetic as well as genetic variation. As a result, DNA methylation is a potential source of inter-individual phenotypic variation.46 Recent studies comparing plants in different environments came to the conclusion that the genome contains single site methylation polymorphisms in addition to SNPs.47 These polymorphisms at the epigenetic level allow individuals in the populations to react differently to fluctuating environments.48 Considering that, epigenetic modifications can occur much faster than genetic divergence,49 methylation might have a role in shaping personality traits in newly formed environments. We believe that this can be a fructiferous future avenue of new research.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
We would like to thank Mohammad Ali Sabbaghi and Robert Carreras Torres for their helpful comments and we are thankful to Emilio Pagani-Núñez for his help in capturing the birds. We are also grateful to the Botanical Institute of Barcelona for allowing us to use their molecular lab and to Alfonso Susana and Sara López-Vinyallonga for their support and help.
Funding
The present study was funded by research project CGL2012–38262 to JCS from the Ministry of Science and Innovation, Spanish Research Council. Birds were handled and kept in captivity with the permission of the Environment Department of the Generalitat de Catalunya (licenses number 2012-SF/518, 2013-SF/677 and 2014-SF/090).
References
- 1.Felthous A, Sass H. International Handbook on Psychopathic Disorders and the Law; Chichester: John Wiley & Sons: 2007; Vol. 1, Diagnosis and Treatment. [Google Scholar]
- 2.Reif A, Lesch KP. Toward a molecular architecture of personality. Behav Brain Res 2003; 139(1):1-20; PMID:12642172; http://dx.doi.org/ 10.1016/S0166-4328(02)00267-X [DOI] [PubMed] [Google Scholar]
- 3.Bailey JN, Breidenthal SE, Jorgensen MJ, McCracken JT, Fairbanks LA. The association of DRD4 and novelty seeking is found in a nonhuman primate model. Psychiat Genet 2007; 17(1):23-7; PMID:17167341; http://dx.doi.org/ 10.1097/YPG.0b013e32801140f2 [DOI] [PubMed] [Google Scholar]
- 4.Van Gestel S, Van Broeckhoven C. Genetics of personality: are we making progress? Mol Psychiat 2003; 8(10):840-52; PMID:14515135; http://dx.doi.org/ 10.1038/sj.mp.4001367 [DOI] [PubMed] [Google Scholar]
- 5.Schinka JA, Letsch EA, Crawford FC. DRD4 and novelty seeking: results of meta-analyses. Am J Med Genet 2002; 114(6):643-8; PMID:12210280; http://dx.doi.org/ 10.1002/ajmg.10649 [DOI] [PubMed] [Google Scholar]
- 6.Savitz JB, Ramesar RS. Genetic variants implicated in personality: a review of the more promising candidates. Am J Med Genet Part B: Neuropsychiatr Genet 2004; 131(1):20-32; PMID:15389772; http://dx.doi.org/ 10.1002/ajmg.b.20155 [DOI] [PubMed] [Google Scholar]
- 7.Ebstein RP. The molecular genetic architecture of human personality: beyond self-report questionnaires. Mol Psychiat 2006; 11(5):427-45; PMID:16534505; http://dx.doi.org/ 10.1038/sj.mp.4001814 [DOI] [PubMed] [Google Scholar]
- 8.Momozawa Y, Takeuchi Y, Kusunose R, Kikusui T, Mori Y. Association between equine temperament and polymorphisms in dopamine D4 receptor gene. Mamm Genome 2005; 16(7):538-44; PMID:16151699; http://dx.doi.org/ 10.1007/s00335-005-0021-3 [DOI] [PubMed] [Google Scholar]
- 9.Kluger AN, Siegfried Z, Ebstein RP. A meta-analysis of the association between DRD4 polymorphism and novelty seeking. Mol Psychiat 2002; 7(7):712-7; PMID:12192615; http://dx.doi.org/ 10.1038/sj.mp.4001082 [DOI] [PubMed] [Google Scholar]
- 10.Fidler AE, van Oers K, Drent PJ, Kuhn S, Mueller JC, Kempenaers B. Drd4 gene polymorphisms are associated with personality variation in a passerine bird. Proc R Soc Lond B 2007; 274(1619):1685-91; PMID:17472912; http://dx.doi.org/ 10.1098/rspb.2007.0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korsten P, Mueller JC, Hermannstadter C, Bouwman KM, Dingemanse NJ, Drent PJ, Liedvogel M, Matthysen E, Van Oers K, Van Overveld T, et al.. Association between DRD4 gene polymorphism and personality variation in great tits: a test across four wild populations. Mol Ecol 2010; 19(4):832-43; PMID:20070517; http://dx.doi.org/ 10.1111/j.1365-294X.2009.04518.x [DOI] [PubMed] [Google Scholar]
- 12.Mueller JC, Korsten P, Hermannstaedter C, Feulner T, Dingemanse NJ, Matthysen E, van Oers K, van Overveld T, Patrick SC, Quinn JL, et al.. Haplotype structure, adaptive history and associations with exploratory behaviour of the DRD4 gene region in four great tit (Parus major) populations. Mol Ecol 2013; 22(10):2797-809; PMID:23506506; http://dx.doi.org/ 10.1111/mec.12282 [DOI] [PubMed] [Google Scholar]
- 13.Mueller JC, Partecke J, Hatchwell BJ, Gaston KJ, Evans KL. Candidate gene polymorphisms for behavioural adaptations during urbanization in blackbirds. Mol Ecol 2013; 22(13):3629-37; PMID:23495914; http://dx.doi.org/ 10.1111/mec.12288 [DOI] [PubMed] [Google Scholar]
- 14.Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet 2006; 2(4), e49; PMID:16604157; http://dx.doi.org/ 10.1371/journal.pgen.0020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusco G, Minelli A. Phenotypic plasticity in development and evolution: facts and concepts. Philos Trans Royal Soc B: Biol Sci 2010; 365(1540):547-56; PMID:20083631; http://dx.doi.org/ 10.1098/rstb.2009.0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008; 319(5871):1827-30; PMID:18339900; http://dx.doi.org/ 10.1126/science.1153069 [DOI] [PubMed] [Google Scholar]
- 17.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 2008; 40(7):904-8; PMID:18568024; http://dx.doi.org/ 10.1038/ng.174 [DOI] [PubMed] [Google Scholar]
- 18.Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, Yen CA, Lin S, Lin Y, Qiu Y. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 2015; 518(7539):350-4; PMID:25693566; http://dx.doi.org/ 10.1038/nature14217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet 2010; 86(2):196-212; PMID:20159110; http://dx.doi.org/ 10.1016/j.ajhg.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devlin AM, Brain U, Austin J, Oberlander TF. Prenatal exposure to maternal depressed mood and the MTHFR C677T variant affect SLC6A4 methylation in infants at birth. Plos One 2010; 5(8):e12201; PMID:20808944; http://dx.doi.org/ 10.1371/journal.pone.0012201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 2008; 3(2):97-106; PMID:18536531; http://dx.doi.org/ 10.4161/epi.3.2.6034 [DOI] [PubMed] [Google Scholar]
- 22.Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA. Methylation at SLC6A4 is linked to family history of child abuse: an examination of the Iowa Adoptee sample. Am J Med Genet Part B: Neuropsychiatr Genet 2010; 153(2):710-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Mil NH, Steegers-Theunissen RgP, Bouwland-Both MI, Verbiest MM, Rijlaarsdam J, Hofman A, Steegers EA, Heijmans BT, Jaddoe VW, Verhulst FC. DNA methylation profiles at birth and child ADHD symptoms. J Psychiatr Res 2014; 49:51-9; PMID:24290898; http://dx.doi.org/ 10.1016/j.jpsychires.2013.10.017 [DOI] [PubMed] [Google Scholar]
- 24.Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics 2010; 5(6):516-26; PMID:20505345; http://dx.doi.org/ 10.4161/epi.5.6.12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yauk C, Polyzos A, Rowan-Carroll A, Somers CM, Godschalk RW, Van Schooten FJ, Berndt ML, Pogribny IP, Koturbash I, Williams A. Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci 2008; 105(2):605-10; PMID:18195365; http://dx.doi.org/ 10.1073/pnas.0705896105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Björklund M, Ruiz I, Senar JC. Genetic differentiation in the urban habitat: the great tits (Parus major) of the parks of Barcelona city. Biol J Linn Soc 2010; 99(1):9-19; PMID:24843434; http://dx.doi.org/ 10.1111/j.1095-8312.2009.01335.x24843434 [DOI] [Google Scholar]
- 27.Senar JC, Conroy MJ, Quesada J, Mateos-Gonzalez F. Selection based on the size of the black tie of the great tit may be reversed in urban habitats. Ecol Evol 2014; 4:2625-32; PMID:25077014; http://dx.doi.org/ 10.1002/ece3.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drent PJ, Van Oers K, Van Noordwijk AJ. Realized heritability of personalities in the great tit (Parus major). Proc R Soc Lond B 2003; 270:45-51; PMID:12590770; http://dx.doi.org/ 10.1098/rspb.2002.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verbeek MEM, Drent PJ, Wiepkema PR. Consistent individual differences in early exploratory behaviour of male great tits. Anim Behav 1994; 48:1113-21; PMID:15520517; http://dx.doi.org/ 10.1006/anbe.1994.134415520517 [DOI] [Google Scholar]
- 30.Dingemanse NJ, Both C, Drent PJ, Van Oers K, Van Noordwijk AJ. Repeatability and heritability of exploratory behaviour in great tits from the wild. Anim Behav 2002; 64:929-38; PMID:15002773; http://dx.doi.org/ 10.1006/anbe.2002.200615002773 [DOI] [Google Scholar]
- 31.Quinn JL, Patrick SC, Bouwhuis S, Wilkin TA, Sheldon BC. Heterogeneous selection on a heritable temperament trait in a variable environment. J Anim Ecol 2009; 78(6):1203-15; PMID:19558612; http://dx.doi.org/ 10.1111/j.1365-2656.2009.01585.x [DOI] [PubMed] [Google Scholar]
- 32.Budaev SV. The statistical analysis of behavioural latency measures. ISCP Newslett 1997; 14:1-4 [Google Scholar]
- 33.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68(4):978-89; PMID:11254454; http://dx.doi.org/ 10.1086/319501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009; 25(11):1451-2; PMID:19346325; http://dx.doi.org/ 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 35.Excoffier L, Laval G, Schneider S. Arlequin. (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 2005; 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller JDMJ, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21(2):263-5; PMID:15297300; http://dx.doi.org/ 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 37.Abe H, Ito S, Inoue-Murayama M. Polymorphisms in the extracellular region of dopamine receptor D4 within and among avian orders. J Mol Evol 2011; 72(3):253-64; PMID:21286696; http://dx.doi.org/ 10.1007/s00239-011-9432-9 [DOI] [PubMed] [Google Scholar]
- 38.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39(4):457-66; PMID:17334365 [DOI] [PubMed] [Google Scholar]
- 39.Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci 2007; 104(14):5942-6; PMID:17389367; http://dx.doi.org/ 10.1073/pnas.0610410104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouellet-Morin I, Wong CCY, Danese A, Pariante CM, Papadopoulos AS, Mill J, Arseneault L. Increased serotonin transporter gene (SERT) DNA methylation is associated with bullying victimization and blunted cortisol response to stress in childhood: a longitudinal study of discordant monozygotic twins. Psychol Med 2013; 43(09):1813-23; PMID:23217646; http://dx.doi.org/ 10.1017/S0033291712002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong CCY, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics 2010; 5(6):516-26; PMID:20505345; http://dx.doi.org/ 10.4161/epi.5.6.12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinnally EL. Epigenetic plasticity following early stress predicts long-term health outcomes in rhesus macaques. Am J Phys Anthropol 2014; 155(2):192-9; PMID:25100197; http://dx.doi.org/ 10.1002/ajpa.22565 [DOI] [PubMed] [Google Scholar]
- 43.Tschirren B, Bensch S. Genetics of personalities: no simple answers for complex traits. Mol Ecol 2010; 19(4):624-6; PMID:20456219; http://dx.doi.org/ 10.1111/j.1365-294X.2009.04519.x [DOI] [PubMed] [Google Scholar]
- 44.Torné-Noguera A, Pagani-Núñez E, Senar JC. Great Tit (Parus major) breath rate in response to handling stress: urban and forest birds differ. J Orn 2014; 155:315-8; http://dx.doi.org/ 10.1007/s10336-013-1025-5 [DOI] [Google Scholar]
- 45.Prezeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution 2005; 59(11):2312-23; PMID:16396172; http://dx.doi.org/ 10.1111/j.0014-3820.2005.tb00941.x [DOI] [PubMed] [Google Scholar]
- 46.Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett 2008; 11(2):106-15 [DOI] [PubMed] [Google Scholar]
- 47.Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ. Patterns of population epigenomic diversity. Nature 2013; 495(7440):193-8; PMID:23467092; http://dx.doi.org/ 10.1038/nature11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan EJ, Gluckman PD, Dearden PK. Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype? J Exp Zool Part B: Mol Dev Evol 2014; 322(4):208-20; PMID:24719220; http://dx.doi.org/ 10.1002/jez.b.22571 [DOI] [PubMed] [Google Scholar]
- 49.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell 2007; 128(4):655-68; PMID:17320504; http://dx.doi.org/ 10.1016/j.cell.2007.01.023 [DOI] [PubMed] [Google Scholar]