Abstract
Plant root systems are critical for survival, acting as the primary interface for nutrient and water acquisition, as well as anchoring the plant to the ground. As plants grow, their root systems become more elaborate, which is largely mediated by the formation of root branches, or lateral roots. Lateral roots initiate deep within the root in the pericycle cell layer, and their development is controlled by a wide range of internal signaling factors and environmental cues, as well as mechanical feedback from the surrounding cells. The endodermal cell layer, which overlies the pericycle, has emerged as an important tissue regulating LR initiation and formation. We recently identified the AtMYB93 transcription factor as a negative regulator of lateral root development in Arabidopsis. Interestingly, AtMYB93 expression is highly restricted to the few endodermal cells overlying developing lateral root primordia, suggesting that this transcriptional regulator might play a key role in mediating the effect of the endodermis on lateral root development. Here we discuss our recent findings in the wider context of root system development – with a particular focus on the role of the endodermis - and propose several potential models to explain AtMYB93 function during lateral root organogenesis.
Keywords: lateral root, endodermis, MYB transcription factor, Auxin, Armadillo-repeat proteins
Lateral root development
The ability of a plant to regulate its root architecture in response to environmental change is key to its survival. In flowering plants, root architecture is shaped to a large extent by the number and length of its root branches, or lateral roots (LRs). LRs initiate via division of certain cells of the pericycle, the cell layer between the vasculature and the endodermis (Fig. 1A). Pericycle cell divisions are stimulated by localized, periodic auxin maxima1-4 and require cell cycle activation.5 The newly-divided pericycle cells form a lateral root primordium (LRP), which then continues to divide and grow, emerging from between the outer cell layers of the primary root, which must separate to allow its passage. The mechanism by which LRPs initiate from the pericycle has been well studied.5-7 The plant hormone auxin is key in regulating both initiation and subsequent emergence,5,8 but nearly all plant hormones so far investigated can affect LR development in an integrated signaling network.7, 9-15
Figure 1.
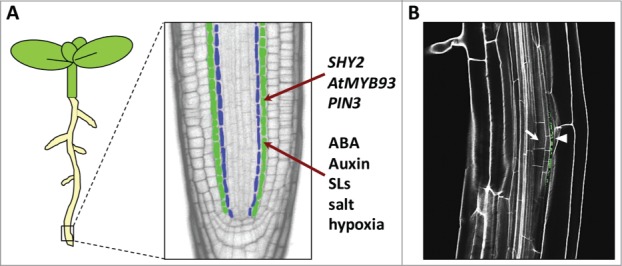
The endodermis and its role in lateral root regulation. (A) An Arabidopsis seedling (left) showing magnification of a confocal section of the root tip (right) with the endodermal cell layer highlighted in green and the pericycle, from which lateral roots arise, in blue. The genes, phytohormones and environmental/stress signals currently known to impinge on the endodermis during lateral root development are shown. ABA, abscisic acid; SLs, strigolactones. (B) Confocal section of an Arabidopsis root showing a developing lateral root primordium (arrow) with expression of pAtMYB93::GUS (green) immediately adjacent to the primordium in the overlying endodermal cell (arrowhead). Image courtesy of George Bassel.
Manipulation of root system architecture is a key focus for the development of crops with improved performance. Identification and characterization of novel genes regulating LR development is therefore extremely important. One crucial aim will be to identify factors that are highly specific to the LR developmental process, which will represent targets for plant biotechnologists and crop breeders to modulate root responses without affecting other aspects of plant growth and development.
The endodermis – an emerging regulator of LR development
In recent years it has become apparent that the endodermis – the cell layer overlying the pericycle – plays a key role in regulating both LR initiation and subsequent emergence. Very early on during LR initiation, pericycle cells acquire founder cell (FC) identity in response to the accumulation of auxin. Expression of the auxin-efflux carrier PIN3 in the endodermis plays a key role in this process, directing the flow of auxin to help generate the requisite auxin maximum that promotes the transition from FC to LR initiation status: signaling from the endodermis therefore acts as an important checkpoint for the development of LRs.16 The endodermis also plays an important role during subsequent LRP emergence. It was found that endodermal cells undergo a number of changes as the LRP develops – including a reduction in volume, shape alterations, and regulated breakdown of the Casparian strip.17 These changes are tightly coordinated, and are absolutely required for normal LRP progression. Targeted manipulation of the properties of outer tissues surrounding developing LRPs (including the endodermis) has also been shown to impact on LRP morphogenesis, further lending support to the role of the endodermis in coordinating LR emergence.18
In addition to acting as a mediator of intrinsic LR development, recent evidence suggests that the endodermis also coordinates LR formation in response to environmental signals.9 The endodermis was shown to be the primary site of ABA-mediated inhibition of LR development in response to salt stress, indicating that this tissue layer may play a key role in controlling root system architecture in response to a wide range of exogenous stress signals9 (Fig. 1A). Collectively, these studies highlight the importance of endodermis as an important regulator of LR development.
Despite the clear role the endodermis plays in LR organogenesis, endodermis-specific factors that might contribute to these LR responses have remained elusive. One candidate is SHY2/IAA3, an endodermally-expressed auxin signaling inhibitor, which is part of an auxin signaling module that promotes LR emergence and may also feed back to inhibit LR initiation.17,19,20 Interestingly, SHY2 also functions in the endodermis to mediate the negative-regulatory effect of strigolactones on LR development,21 suggesting a role for endodermal SHY2 in integrating auxin- and other hormone signals. However, SHY2 is not root-specific, as it has functions throughout the plant and multiple roles in auxin signaling.22
AtMYB93 is a novel endodermis-specific regulator of LR development
We have recently identified the AtMYB93 gene as the first auxin-induced negative regulator of both LR initiation and subsequent development.23 AtMYB93 expression is confined exclusively to roots and is induced specifically during LR initiation and the early stages of LRP formation. Atmyb93 mutants show increased LR initiation and faster developmental progression, while AtMYB93-overexpressing lines have fewer LRs that transit more slowly through development. Moreover, Atmyb93 mutant LRs are somewhat insensitive to exogenously applied auxin, while the primary root responds similarly to wild type plants, indicating that AtMYB93 activity is specific to root branching.23 We speculate that AtMYB93 is part of an auxin-induced negative feedback module, which may allow the plant to make a LR in response to local auxin fluctuations only when they reach a certain threshold, ensuring that LRs only develop when absolutely required.
AtMYB93 promoter activity is exquisitely localized, being confined to just a few cells in the endodermis that overlie the initiating LRP 23 (Fig. 1B). This provides us with new mechanistic detail about how the endodermis may feedback on to the LR initiation process, and AtMYB93 therefore represents a promising target for future studies into the molecular mechanisms underlying the control of LR development by the endodermis. Is AtMYB93 RNA/protein active in the endodermis, or does either move into the pericycle? Does AtMYB93 enable endodermal remodeling and cell separation through controlling gene expression? How does AtMYB93 interact with other known signals and proteins that mediate LR organogenesis? These questions remain to be answered, but future work analyzing AtMYB93 protein activity and gene targets will help to define more accurately its mode of action.
We also found that AtMYB93 expression is upregulated by ABA.23 Interestingly, it was previously shown that AtMYB93 is a direct target of endodermis-specific transcription factor SCARECROW (SCR), which is itself implicated in ABA responses, with scr mutants being ABA-hypersensitive.24 As mentioned previously, the endodermis regulates later LR development in response to salt stress.9 Moreover, AtMYB93 translation is downregulated by hypoxia.25 Thus, AtMYB93 represents a very interesting new target for root stress biologists, as it may provide a further potential link between environmental change and root system plasticity (Fig. 1A).
Potential molecular mechanisms for AtMYB93 function
AtMYB93 was originally identified as an interaction partner of ARABIDILLO (Arabidopsis Armadillo-related) proteins. ARABIDILLOs are “intrinsic” LR regulators, which promote LR development independently from any known hormones or environmental cues.26,27 Despite their rooting function, ARABIDILLO homologues are also found in early evolving plants that lack branched root systems.28 ARABIDILLOs are F-box proteins, meaning they are implicated in proteasomal degradation of proteins.29 Thus, we initially hypothesized that ARABIDILLOs might promote LR development through targeting a negative regulator of LR development – e.g., AtMYB93 – for degradation26,27 (Fig. 2A). However, our studies found that AtMYB93 stability was not enhanced in the arabidillo1/2 double mutant, nor in response to the pharmacological proteasome inhibitor MG132.23 Moreover, our genetic analysis showed that an arabidillo1/2/Atmyb93 triple mutant had a similar reduction in LRs to the arabidillo1/2 double mutant – opposite to the increased LR numbers observed in the Atmyb93 single mutant. Collectively these data indicate that AtMYB93 is not a ubiquitination target of ARABIDIILOs, suggesting that our initial hypothesis is incorrect.
Figure 2.
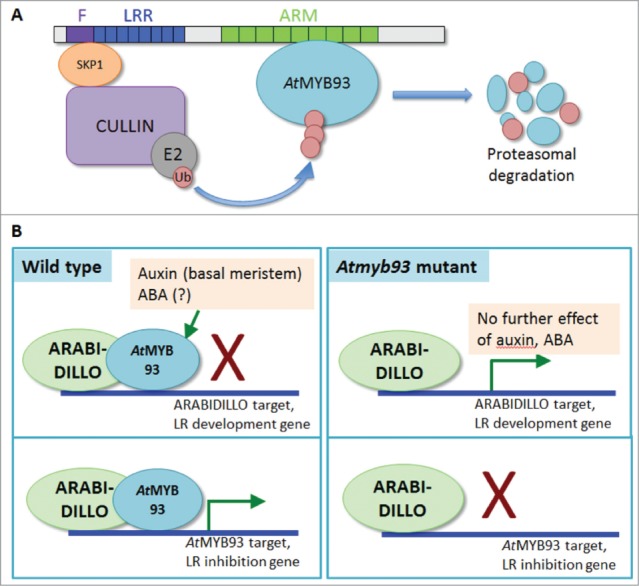
Potential models for AtMYB93 activity and its interaction with ARABIDILLOs. (A) Model for ARABIDILLO-AtMYB93 interaction based on the hypothesis that ARABIDILLOs act as F-box components of an SCF E3 ligase complex during LR development. In this scenario, ARABIDILLOs would ubiquitinate AtMYB93, an inhibitor of LR development. This would promote AtMYB93 degradation by the proteasome and promote LR formation. An arabidillo mutant would accumulate AtMYB93 protein, and hence have fewer LRs, as has been reported (Coates et al., 2006). F, F-box; LRR, Leucine-Rich Repeats; ARM, Armadillo repeats; Ub, ubiquitin. (B) Model for the ARABIDILLO-AtMYB93 interaction based on their action as a bipartite transcription factor, supported by data in Gibbs et al., (2014). Top left: AtMYB93 could act as an inhibitor of LR development genes, blocking a positive action of ARABIDILLO. Auxin and ABA increase AtMYB93 transcript levels. Top right: in the Atmyb93 mutant, absence of AtMYB93 would allow ARABIDILLO to activate LR development genes, and auxin and ABA would have no further effect. Bottom left: in this scenario, AtMYB93 activates genes that inhibit LR development, acting as part of a bipartite transcription factor withARABIDILLO. Bottom right: In the Atmyb93 mutant, ARABIDILLO cannot function by itself, so no LR inhibition genes are activated.
ARABIDILLOs share structural similarity, in the form of an Arm-repeat domain, to animal Armadillo/β-catenin proteins,30 which interact with HMG-domain transcription factors of the LEF/TCF family to activate gene expression in a complex context-dependent manner.31-33 Therefore, an alternative model for the potential functional interaction of ARABIDILLO and AtMYB93 is that the 2 proteins act as a bipartite transcription factor, activating or repressing genes required for LR formation (Fig. 2B). For example, in this scenario, AtMYB93 could act as a repressor, masking a positive role for ARABIDILLOs in inducing LR gene expression. Alternatively, ARABIDILLO and AtMYB93 may act together as a bipartite transcription factor, activating a gene required for LR inhibition. In either case, in the Atmyb93 mutant, loss of AtMYB93 would lead to promotion of LR development, either directly by ARABIDILLOs being derepressed and activating LR target gene expression, or by ARABIDILLO failing to activate a LR inhibition gene in the absence of its interaction partner (Fig. 2B). Future identification of ARABIDILLO/AtMYB93 transcriptional targets will help to resolve these models.
Outlook
Recent work from several labs has demonstrated a key role for the endodermis in regulating LR development in response to auxin, other hormones and environmental signals. We have identified an endodermis-specific transcription factor, AtMYB93, which controls a novel auxin-induced negative feedback loop that inhibits LR development. It will be of great interest to identify genetic targets of AtMYB93, and to integrate its function into the known network of proteins and signaling factors regulating LR formation. As AtMYB93 also interfaces with stress responses, it represents an exciting new target for understanding how changing environmental conditions regulate root architecture. Furthermore, studies of AtMYB93 homologues in crops may provide new ways of regulating or manipulating root architecture in a highly specific and targeted manner.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 2010; 20:1697-706; PMID:20888232; http://dx.doi.org/ 10.1016/j.cub.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 2. De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 2007; 134:681-90; PMID:17215297; http://dx.doi.org/ 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- 3. Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010; 329:1306-11; PMID:20829477; http://dx.doi.org/ 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Norman JM, Xuan W, Beeckman T, Benfey PN. To branch or not to branch: the role of pre-patterning in lateral root formation. Development 2013; 140:4301-10; PMID:24130327; http://dx.doi.org/ 10.1242/dev.090548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. Arabidopsis lateral root development: an emerging story. Trends Plant Sci 2009; 14:399-408; PMID:19559642; http://dx.doi.org/ 10.1016/j.tplants.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 6. De Smet I, Vanneste S, Inze D, Beeckman T. Lateral root initiation or the birth of a new meristem. Plant Mol Biol 2006; 60:871-87; PMID:16724258 [DOI] [PubMed] [Google Scholar]
- 7. Nibau C, Gibbs DJ, Coates JC. Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 2008; 179:595-614; PMID:1842506; http://dx.doi.org/ 10.1111/j.1469-8137.2008.02472.x [DOI] [PubMed] [Google Scholar]
- 8. Lavenus J, Goh T, Roberts I, Guyomarc'h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 2013; 18:450-8; PMID:23701908; http://dx.doi.org/ 10.1016/j.tplants.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 9. Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 2013; 25:324-41; PMID:23341337; http://dx.doi.org/ 10.1105/tpc.112.107227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fukaki H, Tasaka M. Hormone interactions during lateral root formation. Plant Mol Biol 2009; 69:437-49; PMID:18982413; http://dx.doi.org/ 10.1007/s11103-008-9417-2 [DOI] [PubMed] [Google Scholar]
- 11. Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Séjalon-Delmas N, Combier JP, Bécard G, Belausov E, et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011; 233:209-16; PMID:21080198; http://dx.doi.org/ 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- 12. Osmont KS, Sibout R, Hardtke CS. Hidden branches: developments in root system architecture. Annu Rev Plant Biol 2007; 58:93-113; PMID:17177637; http://dx.doi.org/ 10.1146/annurev.arplant.58.032806.104006 [DOI] [PubMed] [Google Scholar]
- 13. Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 2011; 155:721-34; PMID:21119044; http://dx.doi.org/ 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009; 21:1495-511; PMID:19435934; http://dx.doi.org/ 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xue R, Zhang B. Increased endogenous methyl jasmonate altered leaf and root development in transgenic soybean plants. J Genet Genomics 2007; 34:339-46; PMID:17498632; http://dx.doi.org/ 10.1016/S1673-8527(07)60036-8 [DOI] [PubMed] [Google Scholar]
- 16. Marhavy P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benková E. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J 2013; 32:149-58; PMID:23178590; http://dx.doi.org/ 10.1038/emboj.2012.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 2014; 343:178-83; PMID:24408432; http://dx.doi.org/ 10.1126/science.1245871 [DOI] [PubMed] [Google Scholar]
- 18. Lucas M, Kenobi K, von Wangenheim D, Voβ U, Swarup K, De Smet I, Van Damme D, Lawrence T, Péret B, Moscardi E, et al. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 2013; 110:5229-34; PMID:23479644; http://dx.doi.org/ 10.1073/pnas.1210807110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci 2012; 367:1461-8; PMID:22527388; http://dx.doi.org/ 10.1098/rstb.2011.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swarup K, Benkova E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 2008; 10:946-54; PMID:18622388; http://dx.doi.org/ 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- 21. Koren D, Resnick N, Mayzlish Gati E, Belausov E, Weininger S, Kapulnik Y, Koltai H. Strigolactone signaling in the endodermis is sufficient to restore root responses and involves SHORT HYPOCOTYL 2 (SHY2) activity. New Phytol 2013; 198:866-74; PMID:23425316; http://dx.doi.org/ 10.1111/nph.12189 [DOI] [PubMed] [Google Scholar]
- 22. Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 1999; 126:711-21; PMID:9895319 [DOI] [PubMed] [Google Scholar]
- 23. Gibbs DJ, Voss U, Harding SA, Fannon J, Moody LA, Yamada E, Swarup K, Nibau C, Bassel GW, Choudhary A, et al. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol 2014; 203:1194-207; PMID:24902892; http://dx.doi.org/ 10.1111/nph.12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 2011; 21:770-82; PMID:22014526; http://dx.doi.org/ 10.1016/j.devcel.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mustroph A, Zanetti ME, Jang CJ, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 2009; 106:18843-8; PMID:19843695; http://dx.doi.org/ 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coates JC, Laplaze L, Haseloff J. Armadillo-related proteins promote lateral root development in Arabidopsis. Proc Natl Acad Sci USA 2006; 103:1621-6; PMID:16434475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nibau C, Gibbs DJ, Bunting KA, Moody LA, Smiles EJ, Tubby JA, Bradshaw SJ, Coates JC. ARABIDILLO proteins have a novel and conserved domain structure important for the regulation of their stability. Plant Mol Biol 2011; 75:77-92; PMID:21052782; http://dx.doi.org/ 10.1007/s11103-010-9709-1 [DOI] [PubMed] [Google Scholar]
- 28. Moody LA, Saidi Y, Smiles EJ, Bradshaw SJ, Meddings M, Winn PJ, Coates JC. ARABIDILLO gene homologues in basal land plants: species-specific gene duplication and likely functional redundancy. Planta 2012; 236:1927-41; PMID:22945313; http://dx.doi.org/ 10.1007/s00425-012-1742-7 [DOI] [PubMed] [Google Scholar]
- 29. Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013; 14:369-81; PMID:23657496; http://dx.doi.org/ 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol 2003; 13:463-71; PMID:12946625 [DOI] [PubMed] [Google Scholar]
- 31. Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 2012; 4:PMID:23024173; http://dx.doi.org/ 10.1101/cshperspect.a007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cadigan KM. TCFs and Wnt/beta-catenin signaling: more than one way to throw the switch. Curr Top Dev Biol 2012; 98:1-34; PMID:22305157; http://dx.doi.org/ 10.1016/B978-0-12-386499-4.00001-X [DOI] [PubMed] [Google Scholar]
- 33. Mao CD, Byers SW. Cell-context dependent TCF/LEF expression and function: alternative tales of repression, de-repression and activation potentials. Crit Rev Eukaryot Gene Expr 2011; 21:207-36; PMID:22111711; http://dx.doi.org/ 10.1615/CritRevEukarGeneExpr.v21.i3.10 [DOI] [PMC free article] [PubMed] [Google Scholar]


