Abstract
The growth of plant organ to its characteristic size is a fundamental developmental process, but the mechanism is still poorly understood. Plant hormones play a great role in organ size control by modulating cell division and/or cell expansion. ETHYLENE INSENSITVE 2 (EIN2) was first identified by a genetic screen for ethylene insensitivity and is regarded as a central component of ethylene signaling, but its role in cell growth has not been reported. Here we demonstrate that changed expression of EIN2 led to abnormity of cell expansion by morphological and cytological analyses of EIN2 loss-of-function mutants and the overexpressing transgenic plant. Our findings suggest that EIN2 controls final organ size by restricting cell expansion.
Keywords: Arabidopsis thaliana, cell expansion, EIN2, organ size
Introduction
The plant organs develop from meristems, the reservoirs of pluripotent cells of the plant, and then further grow to their specific size by coordinating cell proliferation and cell expansion.1 The relative constancy of organ size within a given species, but its tremendous variance among species, suggests that the final size of an organ is monitored by intrinsically developmental programs.2 Given their sessile and post-embryonic organogenesis lifestyle, the growth of plants are greatly influenced by the environmental signals, such as plant hormones, light, temperature and nutrients.3,4
Genetic studies have identified a number of factors influencing either cell proliferation or cell expansion. In plant, alteration of cell number within an organ usually results from a change in the duration of cell proliferation.5 This process is modulated by positive factors, including AINTEGUMENTA (ANT), GROWTH-REGULATING FACTORS (AtGRFs), GRF-INTERACTING FACTORS (AtGIFs), ARGOS, ORGAN SIZE RELATED 1 (OSR1), JAGGED and KLUH.6-13 Overexpression of these genes results in enlarged organs with increased cells due to extended duration of cell proliferation. In contrast, several factors restricting organ size by limiting the period of cell proliferation have been characterized, such as CINCINNATA (CIN), BIG BROTHER (BB), AUXIN RESPONSE FACTOR 2 (ARF2), DA1, DA2 and MEDIATOR COMPLEX SUBUNIT 25 (MED25).14-19 The loss-of-function mutants of these genes exhibit enlarged organs. However, a few of factors influencing plant organ size by regulating cell expansion have been identified. For example, AtEXP10, ANGUSTIFOLIA (AN), ROTUNDIFOLIA 3 (ROT3), ARGOS-LIKE (ARL) and KUODA1 (KUA1) enhance organ growth by promoting cell expansion.20-25 Whereas, BIGPETALp (BPEp) and its interacting protein AUXIN RESPONSE FACTOR8 (ARF8), control petal size by limiting cell expansion.26,27
The gas hormone ethylene plays numerous roles in the development and environmental responses of the plant, including seed germination, seedling growth and senescence, fruit ripening, stress and pathogen responses.11 ETHYLENE INSENSITVE 2 (EIN2), acting downstream of CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), is the central regulator of ethylene signaling pathway.28,29 The EIN2 protein consists of a hydrophobic N-terminus with the 12-transmembrane domains and a hydrophilic C-terminal domain that contains a conserved nuclear localization sequence.29 EIN2 is localized at the ER membrane and physically interacts with the kinase domain of the ethylene receptors in the absence of ethylene, and is cleaved in ethylene-induced manner with the C-terminal fragment translocated into nucleus to stabilize the short-lived protein ETHYLENE INSENSITVE 3 (EIN3).30,31
Previous works about EIN2 primarily focused on its pivotal roles in ethylene signal transduction and functions in abiotic and biotic stress. Its effects on organ size regulation and cell growth control were always neglected. In order to explore these roles of EIN2, we analyzed the loss-of-function mutants of EIN2 and its overexpressing transgenic plant, finding that mutants of ein2-1 and ein2-5 exhibit enlarged organs due to expanded cells, whereas the EIN2 overexpressing transgenic plants which show constitutive ethylene response produce small organs with smaller cells. The marker genes related to cell expansion process display relevant change within the mutants and transgenic plants. These results suggest that EIN2 plays a role in restricting cell expansion and keeping plant final organ size in check.
Materials and Methods
Plant materials and growth conditions
Wild-type Arabidopsis ecotype Columbia Col-0 (WT), ein2-1 and ein2-5 mutant were used in this study. Sterilized seeds were plated on 1/2 MS medium containing 1% sucrose and 0.6% agar, and then vernalized at 4°C in darkness for 2 d For seed germination, the plate was then transferred to a culture room at 22 ± 1°C with illumination of 80–90 μmol m-2 s-1 with a 16-h light /8-h dark photoperiod. The 7-day old seedlings after germination were planted in soil for further growth.32
Morphological and cytological analyses
The fully expanded leaves were used to determine the size of leaf and palisade cells. They were excised and photographed, and then cleared with chloral hydrate as previously described.32 The palisade cells at the central position of leaf were visualized under a microscope and photographed. Areas of leaves and cells were measured with IMAGE J software (http://rsbweb.nih.gov/ij/), and the total number of palisade cells per leaf was estimated by the total leaf area multiplied by the average cell number per area.
Plant transformation
The 3885-bp EIN2 coding sequence was amplified by reverse transcription polymerase chain reaction (RT-PCR) and cloned into pVIP96 for generation of the 35S-EIN2 construct. The 35S-EIN2 transgenic plants were generated by Agrobacterium tumefaciens-mediated transformation.34 Twenty independently transgenic lines harboring a single T-DNA insertion were used for morphological analyses and three of T3 generation plants were used for gene expression analysis.
Gene expression analysis
Total RNA was isolated with a guanidine thiocyanate extraction buffer, and the reverse-transcribed PCR (RT-PCR) was performed to monitor the expression of cell expansion related marker genes as described previously.33 Real-time quantitative RT-PCR (qRT-PCR) was carried out using a Rotor-Gene 3000 thermocycler (Corbett Research, Sydney, Australia) with the SYBR® Premix ExTM Taq II kit (Takara, Dalian, China). The normalization and relative values of expression level for each gene were calculated from three biological replicates, as previously described.35
Flow cytometric assay
The fully expanded (25 d after initiation) fifth leaves of WT, ein2-1, ein2-5 and 35S-EIN2 were chopped with a razor, suspended in cold nuclear isolation buffer and flow cytometric analysis was carried out as described with a FACS Caliber flow cytometer (BD Biosciences, http://www.bdbiosciences.com/).34
Results
The enlarged organs of ein2-1 and ein2-5 and small organs of 35S-EIN2 transgenic plant
In order to investigate the roles of EIN2 in plant organ size control, the well-known EIN2 loss-of-function mutant ein2-1 and ein2-5 were measured for the morphological analyses. The areas of fully expanded cotyledons of ein2-1 and ein2-5 increased by 47% and 55% respectively, when compared with those of wild-type (WT) plant (Fig. 1A). Detailed characterization of the fifth rosette leaf showed that the average leaf blade areas of ein2-1 and ein2-5 increased by 56% and 48% when compared with those in the WT, respectively. The other aerial organs, including stems, flowers and siliques, were also enlarged to some extent, resulting in bigger plants compared with WT plants (Fig. 1; Table 1). These observations demonstrate that the mutation in EIN2 results in the excessive growth of aerial organs.
Figure 1.
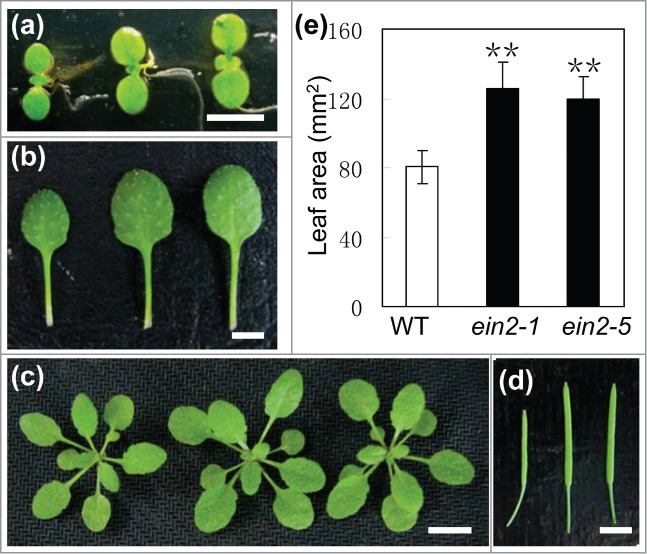
Mutation in EIN2 led to enlarged organs. (A) Cotyledons, (B) fully expanded fifth leaves, (C) whole plants and (D) siliques of Wild-type (WT) (left), ein2-1(middle) and ein2-5 (right). Bar, 5 mm in (A, B, D) and 1 cm in (C). (E) The areas of fully expanded fifth leaf blades in WT, ein2-1 and ein2-5 plants. Mean ± SD; Student's t-test: **, P < 0.01.
Table 1.
Phenotype of ein2-1 and ein2-5 Plants
| Variable | WT | ein2-1 | ein2-5 |
|---|---|---|---|
| Cotyledon area (mm2) | 2.16 ± 0.31 | 3.17 ± 0.34** | 3.35 ± 0.42** |
| Petal area (mm2) | 1.35 ± 0.16 | 1.87 ± 0.19* | 1.74 ± 0.15* |
| Flowering time (days) | 25.1 ± 1.15 | 32.3 ± 1.32 | 31.5 ± 1.31 |
| Silique length (mm) | 10.6 ± 0.92 | 13.3 ± 1.57* | 12.8 ± 1.44* |
| Plant height (cm) | 27.6 ± 1.92 | 36.1 ± 3.04* | 34.9 ± 3.32* |
Mean ± SD; Student's t-test: **, P < 0.01; *, P < 0.05.
To further determine the organ size regulating role of EIN2, we generated 35S-EIN2 transgenic plants. All of 20 independently transgenic lines overexpressing EIN2 displayed obvious smaller organs. The areas of fully expanded cotyledons and the fifth rosette leaves of 35S-EIN2 transgenic plants decreased by 39% and 62% respectively, when compared with those of WT plants (Fig. 2). These results, together with the above morphological analyses of ein2 mutants, state clearly that EIN2 impedes organ growth during plant development.
Figure 2.
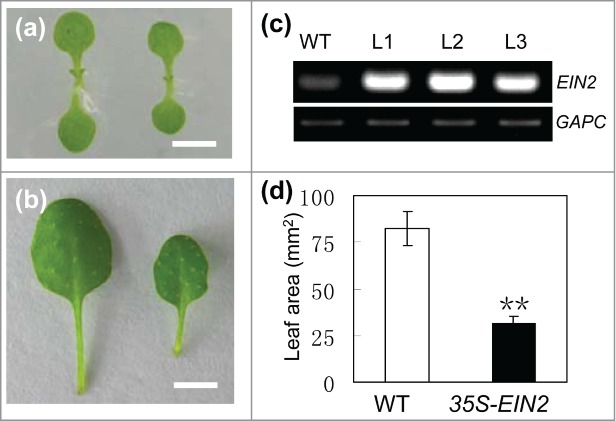
Morphology of 35S-EIN2 transgenic plants. (A) Cotyledons and (B) fully expanded fifth leaves of Wild-type (WT) (left), and 35S-EIN2 (right) plants. Bar = 5 mm. (C) Expression analyses of EIN2 in three independent lines (L1 to L3) of 35S-EIN2 transgenic plants. The transcript abundance of GAPC was used as an internal control in the RT-PCR. (D) The areas of fully expanded fifth leaf blades in WT and 35S-EIN2 plants. Mean ± SD; Student's t-test: **, P < 0.01.
EIN2 controls organ size by limiting cell expansion
To assess the contributions of cell division and cell expansion to the phenotypes of EIN2 loss-of-function mutants and overexpressing transgenic plants, their palisade cells of the fully expanded fifth leaves were visualized under a microscope. As shown in Fig. 3, the average size of palisade cells in ein2-1 and ein2-5 increased by 52% and 45% respectively when compared with that of WT plants, while the average size of palisade cells in 35S-EIN2 transgenic plants decreased to 41% of that of WT plants. Furthermore, the estimated palisade cell number per leaf of all these lines remains comparable, demonstrating that EIN2 control organ size by limiting cell expansion and not by manipulating cell proliferation.
Figure 3.
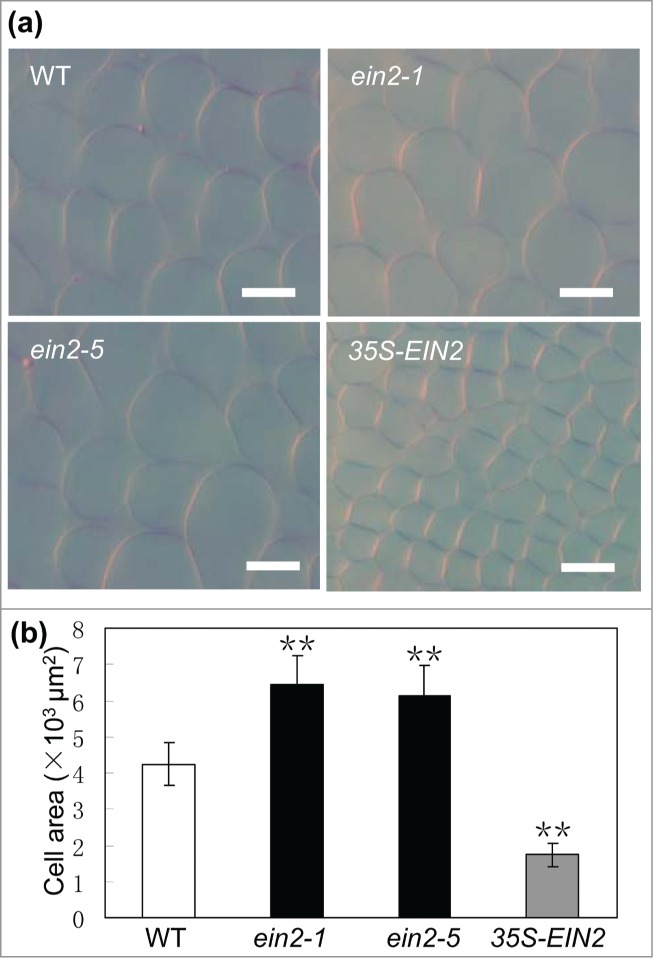
Cytological characterization of ein2-1, ein2-5 and 35S-EIN2 transgenic plants. (A) Palisade cells of the fully expanded fifth leaf in WT, ein2-1, ein2-5 and 35S-EIN2 transgenic plants. Bars = 100 μm. (B) Estimated palisade cell area in WT, ein2-1, ein2-5 and 35S-EIN2 transgenic plants. Mean ± SD; Student's t-test: **, P < 0.01.
Cell expansion restricted by EIN2 is related to expansins but not to endoreduplication
The final size of a plant cell is often associated with nuclear polyploidy, which results from nuclear endoreduplication when a cell undergoes differentiation and cell expansion.37 Analysis of the DNA content of fully expanded fifth leaves by flow cytometry with nuclei indicated that no enhanced endoreduplication in the fifth rosette leaves of ein2-1 and ein2-5 or reduced endoreduplication in 35S-EIN2 transgenic plant were observed when compared to WT plants (Fig. 4). These findings demonstrated that the cell size increase or decrease observed in leaves of the ein2-1 and ein2-5 or 35S-EIN2 transgenic plants was not resulting from the changed endoreduplication.
Figure 4.
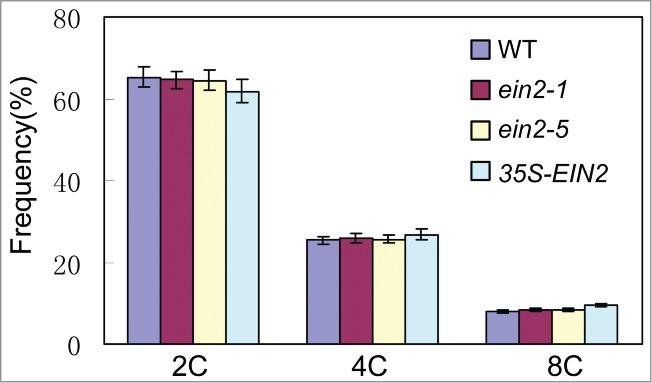
Ploidy levels of WT, ein2-1, ein2-5 and 35S-EIN2 transgenic plants. Percentage of cells with different classes of nuclear ploidy. Mean ± SD.
Expansins play essential roles in cell enlargement as key regulators of cell wall extension.37,38 In the Arabidopsis genome, there are 26 α-expansin genes and at least five β-expansin genes, which might act redundantly to regulate plant growth and development.37,38 In order to monitor the cell expansion processes within the EIN2 loss-of function mutants and 35S-EIN2 transgenic plants, we examined the expression levels of the expansin genes. AtEXP3, AtEXP5 and AtEXP10 showed a higher expression level in the ein2-1 and ein2-5, while the expression of AtEXP5 decreased in 35S-EIN2 transgenic plants compared to WT plants (Fig. 5), and other expansin genes didn't show any significant expressive changes (data not shown), suggesting that repressing the expression of particular expansin genes accounts for cell expansion restricting by EIN2.
Figure 5.
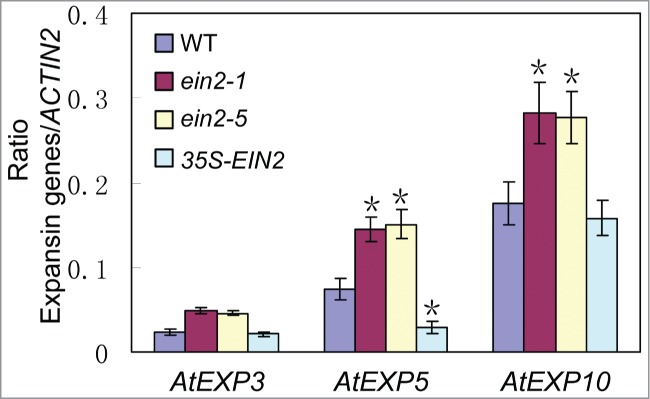
The expressions of expansin genes in WT, ein2-1, ein2-5 and 35S-EIN2 transgenic plants. Expression levels of AtEXP3, AtEXP5, AtEXP10 are higher in ein2-1 and ein2-5 leaves than in wild-type, and decreased expression levels of AtEXP5 in 35S-EIN2 transgenic plants. Mean ± SD. Student's t-test: *, P < 0.05.
Discussion
To date, many genes participating in plant organ size control have been identified, and they are referred to as intrinsic yield genes (IYGs) due to their potential to boost the biomass and yield of agronomic plants.34,39 However, relatively few of these genes coalesce into pathways, indicating that the mechanisms of plant organ size control are more complicated than those in animals.5 In this study, we characterized EIN2 as a player in plant organ size control, with loss and gain of function producing opposite effects on organ size. Failure to maintain correct cell proliferation and/or expansion often, but not always, results in an alteration of plant organ size, because of compensation.40,41 The enlarged cells in EIN2 loss-of-function mutants and the smaller cells in 35S-EIN2 transgenic plants suggests that EIN2 restricts organ growth by limiting cell expansion, and the unchanged cell numbers in the organs of different lines illustrates that compensation does not take place. Our report discovered a novel role of EIN2 in regulating organ growth by affecting cell expansion in a general manner.
Within an organ, the final size of cells is often, but not always, associated with the ploidy level owning to the nuclear endoreduplication, which occurs when cells undergo differentiation and expansion.2 A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4(6):533–539. Although the mechanism is still obscure, the ploidy level has been regarded as an important factor in determining cell size.36,42 Alteration of cell size in the plants with abnormal expression of EIN2 provides another example that cell size is not correlated to the ploidy level. Expansion of plant cells necessarily requires a loosening of the cell wall and concomitant water uptake to a vacuole of high osmolarity to maintain turgor, and the involvement of expansins make this process more convenient.43 The increased expression levels of AtEXP3, AtEXP5 and AtEXP10 in the ein2-1 and ein2-5 mutants and decreased expression levels of AtEXP5 in 35S-EIN2 transgenic plants indicate that EIN2 might act on some unknown factors to reduce expansins expression level to restrict cell expansion.
Plant hormones, such as auxin, cytokinin, ethylene and abscisic acid, intensely influence organ growth and have been demonstrated to play a significant role in cell proliferation and/or cell expansion regulation.41 However, how these hormonal signals are integrated into the program of plant organ size control remains largely unclear. the loss-of-function mutants of EIN2 are not only insensitive to ethylene, but also have been found in screens for the mutants resistant to auxin transport inhibitors, cytokinins, or abscisic acid,28 giving a hint that EIN2 might integrate these hormones signaling to fine-tune plant cell growth considering its role in cell expansion regulation presented by our results. Future work on investigation of molecular mechanism of EIN2 from the point view of cell growth regulation will bring about more interesting discoveries.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for ein2-1 and ein2-5 lines.
Funding
This work was supported by the National Natural Science Foundation of China (31260066), the Jiangxi Provincial Natural Science Fund (20122BAB214028), and the Education Department of Jiangxi province science and technology project (GJJ13534).
References
- 1. Ha CM, Jun JH, Fletcher JC. Shoot apical meristem form and function. Curr Top Dev Biol 2010; 91:103-140; PMID:20705180; http://dx.doi.org/ 10.1016/S0070-2153(10)91004-1 [DOI] [PubMed] [Google Scholar]
- 2. Mizukami Y. A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 2001; 4(6):533-539; PMID:11641070 [DOI] [PubMed] [Google Scholar]
- 3. Tsukaya H. Organ shape and size: a lesson from studies of leaf morphogenesis. Curr Opin Plant Biol 2003; 6(1):57-62; PMID:12495752 [DOI] [PubMed] [Google Scholar]
- 4. Tsukaya H. Leaf shape: genetic controls and environmental factors. Int J Dev Biol 2005; 49(5-6):547-555 [DOI] [PubMed] [Google Scholar]
- 5. Powell AE, Lenhard M. Control of organ size in plants. Curr Biol 2012; 22(9):R360-367; PMID:22575478 [DOI] [PubMed] [Google Scholar]
- 6. Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 2000; 97(2):942-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. The role of JAGGED in shaping lateral organs. Development 2004; 131(5):1101-1110; PMID:14973282 [DOI] [PubMed] [Google Scholar]
- 8. Ohno CK, Reddy GV, Heisler MG, Meyerowitz EM. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 2004; 131(5):1111-1122 [DOI] [PubMed] [Google Scholar]
- 9. Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 2005; 43(1):68-78; PMID:15960617 [DOI] [PubMed] [Google Scholar]
- 10. Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 2007; 13(6):843-856; PMID:18061566 [DOI] [PubMed] [Google Scholar]
- 11. Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol 2009; 151(2):655-668; PMID:19648231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 2003; 15(9):1951-1961; PMID:12953103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng G, Qin Z, Yan J, Zhang X, Hu Y. Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol 2011; 191(3):635-646; PMID:21457262 [DOI] [PubMed] [Google Scholar]
- 14. Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science 2003; 299(5611):1404-1407; PMID:12610308 [DOI] [PubMed] [Google Scholar]
- 15. Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 2006; 16(3):272-279; PMID:16461280 [DOI] [PubMed] [Google Scholar]
- 16. Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The Auxin response factor 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006; 133(2):251-261; PMID:16339187 [DOI] [PubMed] [Google Scholar]
- 17. Li Y, Zheng L, Corke F, Smith C, Bevan MW. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 2008; 22(10):1331-1336; PMID:18483219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng Y, Ma W, Chen L, Yang L, Li S, Zhao H, Zhao Y, Jin W, Li N, Bevan MW, et al. Control of root meristem size by DA1-RELATED PROTEIN2 in Arabidopsis. Plant Physiol 2013; 161(3):1542-1556; PMID:23296689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu R, Li Y. Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 2011; 138(20):4545-4554; PMID:21903673 [DOI] [PubMed] [Google Scholar]
- 20. Tsuge T, Tsukaya H, Uchimiya H. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 1996; 122(5):1589-1600 [DOI] [PubMed] [Google Scholar]
- 21. Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 1998; 12(15):2381-2391; PMID:9694802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J 2002; 21(6):1267-1279; PMID:11889033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 2000; 97(17):9783-9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Y, Poh HM, Chua NH. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 2006; 47(1):1-9; PMID:16824178 [DOI] [PubMed] [Google Scholar]
- 25. Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JH. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat Commu 2013; 5:3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 2006; 25(16):3912-3920; PMID:16902407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varaud E, Brioudes F, Szecsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011; 23(3):973-983; PMID:21421811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999; 284(5423):2148-2152; PMID:10381874 [DOI] [PubMed] [Google Scholar]
- 29. Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 2012; 22(11):1613-1616; PMID:23070300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bisson MM, Groth G. New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol Plant 2010; 3(5):882-889; PMID:20591837 [DOI] [PubMed] [Google Scholar]
- 31. An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010; 22(7):2384-2401; PMID:20647342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jing Y, Cui D, Bao F, Hu Z, Qin Z, Hu Y. Tryptophan deficiency affects organ growth by retarding cell expansion in Arabidopsis. Plant J 2009; 57(3):511-521; PMID:18980661 [DOI] [PubMed] [Google Scholar]
- 33. Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 2006; 1(2): 641-646.; PMID:1740629219119056 [DOI] [PubMed] [Google Scholar]
- 34. Gonzalez N, Beemster GT, Inze D. David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol 2009; 12(2):157-164; PMID:19119056 [DOI] [PubMed] [Google Scholar]
- 35. Hu Z, Qin Z, Wang M, Xu C, Feng G, Liu J, Meng Z, Hu Y. The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J 2010; 61(4):600-610 [DOI] [PubMed] [Google Scholar]
- 36. Sugimoto-Shirasu K, Roberts K. "Big it up": endoreduplication and cell-size control in plants. Curr Opin Plant Biol 2003; 6(6):544-553; PMID:14611952 [DOI] [PubMed] [Google Scholar]
- 37. Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 2001; 4(6):527-532; PMID:11641069 [DOI] [PubMed] [Google Scholar]
- 38. Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. The growing world of expansins. Plant Cell Physiol 2002; 43(12):1436-1444; PMID:12514240 [DOI] [PubMed] [Google Scholar]
- 39. Krizek BA. Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol 2009; 12(1):17-22; PMID:18951836 [DOI] [PubMed] [Google Scholar]
- 40. Ferjani A, Horiguchi G, Yano S, Tsukaya H. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 2007; 144(2):988-999; PMID:17468216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsukaya H. Mechanism of leaf-shape determination. Ann Rev Plant Biol 2006; 57:477-496; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105320 [DOI] [PubMed] [Google Scholar]
- 42. Tsukaya H. Controlling size in multicellular organs: focus on the leaf. PLoS Biol 2008; 6(7):e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cosgrove DJ. Growth of the plant cell wall. Nat Rev 2005; 6(11):850-861; PMID:16261190 [DOI] [PubMed] [Google Scholar]


