Abstract
The degree of methylesterification (DM) of homogalacturonans (HGs), the main constituent of pectins in Arabidopsis thaliana, can be modified by pectin methylesterases (PMEs). Regulation of PME activity occurs through interaction with PME inhibitors (PMEIs) and subtilases (SBTs). Considering the size of the gene families encoding PMEs, PMEIs and SBTs, it is highly likely that specific pairs mediate localized changes in pectin structure with consequences on cell wall rheology and plant development. We previously reported that PME17, a group 2 PME expressed in root, could be processed by SBT3.5, a co-expressed subtilisin-like serine protease, to mediate changes in pectin properties and root growth. Here, we further report that a PMEI, PMEI4, is co-expressed with PME17 and is likely to regulate its activity. This sheds new light on the possible interplay of specific PMEs, PMEIs and SBTs in the fine-tuning of pectin structure.
Keywords: Arabidopsis thaliana, cell wall, co-expression, growth, pectin, pectin methylesterase, pectin methylesterase inhibitor, subtilase, root
Abbreviations
- ARF
Auxin response factor
- BES1/BIM1-3
BRI1 EMS suppressor 1/BES1 interaction MYC-like 1-3
- Col-0
Columbia-0
- DM
Degree of methylesterification
- Gal-A
Galacturonic acid
- HG
Homogalacturonan
- IEF
Isoelectric focusing
- KO
Knock-out
- OG
Oligogalacturonide
- PG
Polygalacturonase
- PM
Plasma membrane
- PME
Pectin methylesterase
- PMEI
Pectin methylesterase inhibitor
- PL
Pectate lyase
- RLK
Receptor-like kinase
- SBT
Subtilase
- TF
Transcription factor
- WAK
Wall-associated kinase
In the plant cell wall of dicotyledonous plants such as Arabidopsis thaliana, pectins are major non-cellulosic compounds, which play a key role in mediating changes in the mechanical properties of the cell wall.1,2 They are constituted of several polysaccharides including the simplest and most abundant homogalacturonan (HG), a linear chain of galacturonic acids (gal-A) linked in α-(1-4).3-6 HGs are synthesized in the Golgi apparatus and secreted into the cell wall in a fully methylesterified form, where they are demethylesterified by pectin methylesterases (PMEs), which constitute a multigenic family of 66 members in Arabidopsis.7-11 Within the PME family, the analysis of protein sequences has shown that PMEs can be classified into 2 groups: group 1 PMEs, which possess a mature part (PME domain, Pfam01095), and group 2 PMEs, which contain both the mature part and an N-terminal extension, the so-called PRO part (PMEI domain, Pfam04043).7,12 Conserved RR(K)LL basic motifs can be recognized by subtilases (SBTs), enabling cleavage of the PRO part of group 2 PMEs, and thus the release of active isoforms into the apoplasm.12 Consequently, in addition to the reported control of PME activity by PME inhibitors (PMEIs) through the formation of PME:PMEI stoichiometric complexes, the degree of methylesterification (DM) of HGs is likely to be controlled by the SBT-mediated processing of PMEs.13,14
Over recent years, the control of plant development by changes in the structure of pectins, mediated by PME-PMEI and PME-SBT, has been demonstrated using genetic approaches. In particular, PMEIs have been shown to be involved in developmental processes as diverse as pollen tube growth,15 seed mucilage extrusion,16 radicle emergence at the onset of germination,17 elongation of root and etiolated hypocotyl18,19 and primordia emergence at the shoot apical meristem.20-22 In parallel, a role for SBTs in the regulation of PME activity during seed mucilage extrusion14 and root development23 has been described. In the latter study, the contribution of PME17 and SBT3.5, which are both co-expressed in roots, to changing pectin structure and affecting root development was investigated. The characterization of pme17 and sbt3.5 mutants highlighted a role for PME17 and SBT3.5 in modulating pectin properties and affecting root growth. The processing of PME17 by SBT3.5 was also demonstrated,23 suggesting that SBTs co-expressed with group 2 PMEs could play a role in the processing of the proteins in planta. Among PME and PMEI genes known to be expressed in root,18,24 PMEI4 was strongly up-regulated in the pme17 KO mutant, suggesting a possible interplay between PME17 and PMEI4.23 Despite the wealth of publications concerning the regulation of plant development by PME-PMEI, much remains to be discovered, notably the identification of specific PME-PMEI pairs and the role of the PME-PMEI balance in the control of total PME activity.
Following previous results showing a role for PME17 (and SBT3.5) in the control of root growth,23 we now report a possible contribution of PMEI4 to the regulation of PME17.
Potential Regulation of PME17 Activity by PMEI4
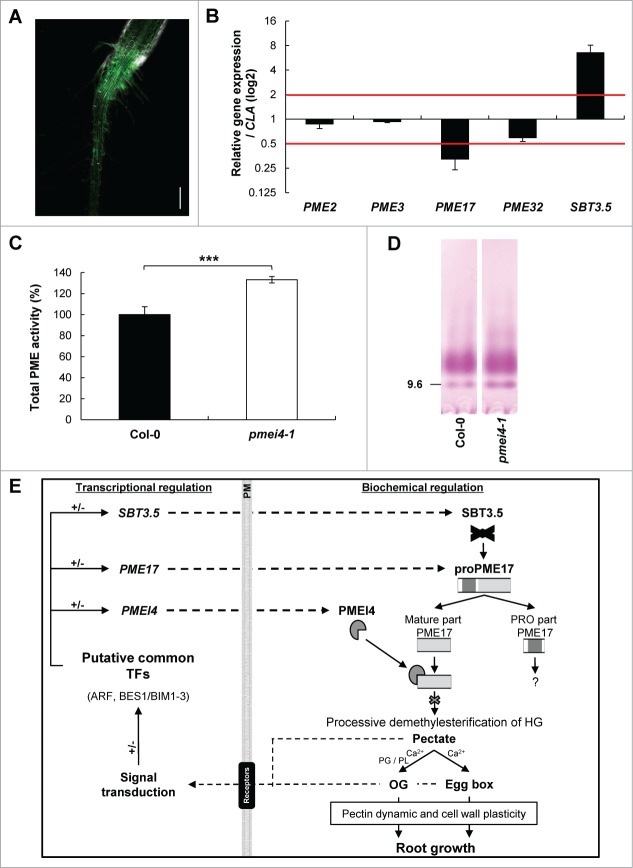
We first localized the expression of PMEI4 in an Arabidopsis transgenic plant harboring the construct where the PMEI4 promoter drives the expression of the PMEI4 coding sequence fused with GFP (pPMEI4::GFP).18 Using confocal laser microscopy, the PMEI4 promoter was active, similarly to the results for PME17 and SBT3.523 in the outer cell layer of roots from light-grown seedlings, particularly in the root-hair zone (Fig. 1A). The analysis of the PMEI4 promoter sequence with AtcisDB software (Arabidopsis cis-regulatory element database, http://arabidopsis.med.ohio-state.edu/AtcisDB/) revealed that specific transcription factor binding sites were conserved when PME17 and SBT3.5 promoters were compared.23 These transcription factors (TFs), which are known to regulate the expression of genes encoding proteins involved in the control of cell-wall modifications and plant development through auxin and brassinosteroid signaling pathways,25-29 include ARF (auxin response factor) and BES1/BIM1-3 (BRI1 EMS suppressor 1/BES1 interaction MYC-like 1-3), which could partly explain the overlapping expression patterns of all 3 genes.
Figure 1.
For figure legend, see page 3.Figure 1 (See privous page). Localization of PMEI4 expression, characterization of the pmei4-1 KO mutant and hypothetical model of root growth regulation by PMEI4, PME17 and SBT3.5. (A) Confocal laser microscopy on 4-day-old light-grown seedlings of pPMEI4::GFP transgenic plants. Localization of PMEI4, tagged with GFP, was achieved by imaging with a confocal laser-scanning microscope Zeiss LSM 780 (Zeiss Jena, Germany). References Images of GFP were collected by excitation at 488 nm with emissions collected at 493-549 nm. Captured images were analyzed by the ZEN 2011 software (Zeiss), and assigned false coloring: green for GFP fluorescence. Autofluorescence of the wild type was subtracted. Scale bar: 200 μm. (B) Relative expression of root-expressed PMEs and SBT3.5. Relative gene expression of PME2, PME3, PME17, PME32 and SBT3.5 was quantified in 10-day-old light-grown roots of the pmei4-1 KO mutant compared to the wild type (Col-0), using the reference gene CLA. Data represent the means ± SE. (C) Total PME activity in cell wall-enriched protein extracts from 10-day-old light-grown roots of Col-0 and the pmei4-1 KO mutant. Data represent the means ± SE. Significant differences were determined by the non-parametric Mann-Whitney test (***, p < 0.001). (D) Isoelectric focusing (IEF) of cell wall-enriched protein extracts from 10-day-old light-grown roots of Col-0 and the pmei4-1 KO mutant. The same PME activities (15 mU) were loaded for each condition. After IEF, PME activities were detected by incubation in a pectin (DM> 85 %) solution, followed by ruthenium red staining. (E) Hypothetical model of root growth regulation by PMEI4, PME17 and SBT3.5. At the biochemical level, proPME17 is processed by cleavage of the PRO part by SBT3.5. Processed PME17 could act on methylesterified HG to perform processive demethylesterification and produce pectate. In the presence of Ca2+ ion, processively demethylesterified HG could become the target of Ca2+-dependent pectin-degrading enzymes, such as some polygalacturonases (PGs) and pectate lyases (PLs), to form oligogalacturonides (OGs), or bind through the Ca2+ link forming the so-called “egg box." Several plasma membrane (PM) receptor-like kinases, such as WAKs (wall-associated kinases) and RLKs (receptor-like kinases), could be involved directly or indirectly in signal perception of OGs, pectate and cross-linked HGs,30,31 subsequently activating a signal transduction cascade in the cell. At the transcriptional level, signal transduction could lead to activation and/or inactivation of common TFs (ARF, BES1/BIM1-3) to regulate the expression of PMEI4, PME17 and SBT3.5. Secreted PMEI4 could inhibit PME17 activity. This transcriptional and biochemical pathway would allow the fine-tuning of HG structure with consequent effects on cell wall plasticity and root growth.
The overlapping promoter activity of PME17 and PMEI4 prompted us to investigate the potential regulation of PME17 by PMEI4. For this purpose, we used pmei4-1, a KO mutant line previously described in Pelletier et al.18 We first showed that, while PME17 expression was lower in the pmei4-1 KO line compared to the wild type (Col-0), SBT3.5 expression showed the opposite trend (Fig. 1B). In contrast, the expression of other PMEs (PME2, PME3, PME32), known to be expressed in roots at the transcript and protein levels, was not modified. The deregulation of the expression of one gene (PMEI4) is likely to have some consequences on the transcriptional regulation of other genes involved in the same biochemical pathway. If confirmed at the protein level, the increased expression of SBT3.5 could contribute to releasing a higher amount of processed, active, PME17 isoforms.
In root cell wall-enriched protein extracts, an increase in total PME activity (+33%) was measured in pmei4-1 compared to the wild type (Fig. 1C). Together with the results for dark-grown hypocotyls,18 this strongly suggests that PMEI4 can target root-expressed PMEs. Next, we separated proteins according to pH by isoelectric focusing (IEF), then the PME activities of the separated isoforms were revealed by incubation with pectin and staining with ruthenium red (Fig. 1D). At basic pH (9.6), a band of PME activity, corresponding to the presumed PME17 isoform, which was absent in pme17-1 and was characterized by proteomic analyses,23 was shown to be more intense in pmei4-1. This strongly suggests that in the wild type, PMEI4 could regulate PME17 activity, among other PME isoforms. A significant increase in root length was measured in pmei4-1 compared to the wild type (wild type = 5.75 cm ± 0.13 and pmei4-1 = 6.18 ± 0.12), further showing that PME-mediated changes in pectin structure had consequences on plant development. In particular, depending on the organs considered and on the balance between deregulated PMEs and PMEIs, an alteration in total PME activity is likely to modulate pectin properties and plant growth differentially.23,24 Considering these results and the role of SBT3.5 in mediating the processing of PME17,23 the regulation of PME is likely to be highly complex, involving both transcriptional regulation of potential specific partners (PME, PMEI, SBT) and protein-protein interactions to mediate the fine-tuning of its activity. This complex mechanism is probably a way to mediate highly localized, temporally and spatially, changes in pectin structure, with consequent effects on cell wall rheology and growth. A schematic diagram of the potential roles of the interplay between PME17, PMEI4 and SBT3.5 in the control of pectin structure affecting root development is shown (Fig. 1E). Considering the size of the gene families encoding PMEs (66 members), PMEIs (76 members) and SBTs (56 members), this opens up new exciting perspectives in the identification of interacting isoforms, both in vitro and in planta.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Herman Höfte from the Institut Jean Pierre Bourgin (INRA Versailles) for the kind gift of pPMEI4::GFP Arabidopsis transgenic plants and the pmei4-1 mutant.
Funding
This work was supported by a grant from the Agence Nationale de la Recherche (ANR-09-BLANC-0007-01, GROWPEC project) and by the Conseil Régional de Picardie through a PhD studentship awarded to F.S. This work was partly supported by the University of Rouen and the “Trans Channel Wallnet” project that has been selected by the INTERREG IVA program France (Channel) - England European cross-border cooperation programme, which is co-financed by the ERDF.
References
- 1. Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol 2008; 11:301-7; PMID:18434239 [DOI] [PubMed] [Google Scholar]
- 2. Doblin MS, Pettolino F, Bacic A. Evans Review: Plant cell walls: the skeleton of the plant world. Funct Plant Biol 2010; 37:357-81; http://dx.doi.org/ 10.1071/FP09279 [DOI] [Google Scholar]
- 3. Harholt J, Suttangkakul A, Vibe Scheller H. Biosynthesis of Pectin. Plant Physiol 2010; 153:384-95; PMID:20427466; http://dx.doi.org/ 10.1104/pp.110.156588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol 2008; 11:266-77; PMID:18486536; http://dx.doi.org/ 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 5. Willats WG, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 2001; 47:9-27; PMID:11554482; http://dx.doi.org/ 10.1023/A:1010662911148 [DOI] [PubMed] [Google Scholar]
- 6. Willats WGT, Knox JP, Mikkelsen JD. Pectin: new insights into an old polymer are starting to gel. Trends Food Sci Tech 2006; 17:97-104; http://dx.doi.org/ 10.1016/j.tifs.2005.10.008 [DOI] [Google Scholar]
- 7. Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci 2007; 12:267-77; PMID:17499007; http://dx.doi.org/ 10.1016/j.tplants.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 8. Atmodjo MA, Hao Z, Mohnen D. Evolving views of pectin biosynthesis. Annu Rev Plant Biol 2013; 64:747-79; PMID:23451775; http://dx.doi.org/ 10.1146/annurev-arplant-042811-105534 [DOI] [PubMed] [Google Scholar]
- 9. Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 2009; 344:1879-900; PMID:19616198; http://dx.doi.org/ 10.1016/j.carres.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 10. Voragen AG, Coenen G-J, Verhoef RP, Schols HA. Pectin, a versatile polysaccharide present in plant cell walls. Struct Chem 2009; 20:263-75; http://dx.doi.org/ 10.1007/s11224-009-9442-z [DOI] [Google Scholar]
- 11. Wolf S, Mouille G, Pelloux J. Homogalacturonan methyl-esterification and plant development. Mol Plant 2009; 2:851-60; PMID:19825662; http://dx.doi.org/ 10.1093/mp/ssp066 [DOI] [PubMed] [Google Scholar]
- 12. Wolf S, Rausch T, Greiner S. The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J 2009; 58:361-75; PMID:19144003; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03784.x [DOI] [PubMed] [Google Scholar]
- 13. Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D. Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 2005; 17:849-58; PMID:15722470; http://dx.doi.org/ 10.1105/tpc.104.028886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 2008; 54:466-80; PMID:18266922; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03437.x [DOI] [PubMed] [Google Scholar]
- 15. Röckel N, Wolf S, Kost B, Rausch T, Greiner S. Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J 2008; 53:133-43; PMID:17971035; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03325.x [DOI] [PubMed] [Google Scholar]
- 16. Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM. Pectin Methylesterase Inhibitor6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 2013; 25:308-23; PMID:23362209; http://dx.doi.org/ 10.1105/tpc.112.106575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muller K, Levesque-Tremblay G, Bartels S, Weitbrecht K, Wormit A, Usadel B, Haughn G, Kermode AR. Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol 2012; 161:305-16; PMID:23129203; http://dx.doi.org/ 10.1104/pp.112.205724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pelletier S, Van Orden J, Wolf S, Vissenberg K, Delacourt J, Ndong YA, Pelloux J, Bischoff V, Urbain A, Mouille G, et al. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol 2010; 188:726-39; PMID:20819179; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03409.x [DOI] [PubMed] [Google Scholar]
- 19. Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 2007; 143:1871-80; PMID:17277091; http://dx.doi.org/ 10.1104/pp.106.090803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol 2008; 18:1943-8; PMID:19097903; http://dx.doi.org/ 10.1016/j.cub.2008.10.065 [DOI] [PubMed] [Google Scholar]
- 21. Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 2011; 21:1720-6; PMID:21982593; http://dx.doi.org/ 10.1016/j.cub.2011.08.057 [DOI] [PubMed] [Google Scholar]
- 22. Peaucelle A, Louvet R, Johansen JN, Salsac F, Morin H, Fournet F, Belcram K, Gillet F, Hofte H, Laufs P, et al. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 2011; 138:4733-41; PMID:21965608; http://dx.doi.org/ 10.1242/dev.072496 [DOI] [PubMed] [Google Scholar]
- 23. Sénéchal F, Graff L, Surcouf O, Marcelo P, Rayon C, Bouton S, Mareck A, Mouille G, Stintzi A, Höfte H, et al. Arabidopsis PECTIN METHYLESTERASE17 is co-expressed with and processed by SBT3.5, a subtilisin-like serine protease. Ann Bot 2014; 114:1161-75; PMID:2466510921692803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guénin S, Mareck A, Rayon C, Lamour R, Assoumou Ndong Y, Domon J-M, Sénéchal F, Fournet F, Jamet E, Canut H, et al. Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol 2011; 192:114-26; PMID:21692803; http://dx.doi.org/ 10.1111/j.1469-8137.2011.03797.x [DOI] [PubMed] [Google Scholar]
- 25. Paque S, Mouille G, Grandont L, Alabadi D, Gaertner C, Goyallon A, Muller P, Primard-Brisset C, Sormani R, Blazquez MA, et al. AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in Arabidopsis. Plant Cell 2014; 26:280-95; PMID:24424095; http://dx.doi.org/ 10.1105/tpc.113.120048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, Clarke TH, Reed JW, van der Knaap E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J Exp Bot 2014; 65:2507-20; PMID:24723401; http://dx.doi.org/ 10.1093/jxb/eru141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perrot-Rechenmann C. Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2010; 2:a001446-6; PMID:20452959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 2012; 22:1-6; PMID:22197242; http://dx.doi.org/ 10.1016/j.cub.2012.07.036 [DOI] [PubMed] [Google Scholar]
- 29. Fàbregas N, Caño-Delgado AI. Turning on the microscope turret: a new view for the study of brassinosteroid signaling in plant development. Physiol Plant 2014; 151:172-83; PMID:24547704; http://dx.doi.org/ 10.1111/ppl.12130 [DOI] [PubMed] [Google Scholar]
- 30. Wolf S, Hématy K, Höfte H. Growth control and cell wall signaling in plants. Annu Rev Plant Biol 2012; 63:381-407; PMID:22224451; http://dx.doi.org/ 10.1146/annurev-arplant-042811-105449 [DOI] [PubMed] [Google Scholar]
- 31. Wolf S, Hofte H. Growth control: a saga of cell walls, ROS, and peptide receptors. Plant Cell 2014; PMID:24808052; http://dx.doi.org/ 10.1105/tpc.114.125518 [DOI] [PMC free article] [PubMed] [Google Scholar]