Abstract
Camelid immunoglobulin variable (IGV) regions were found homologous to their human counterparts; however, the germline V repertoires of camelid heavy and light chains are still incomplete and their therapeutic potential is only beginning to be appreciated. We therefore leveraged the publicly available HTG and WGS databases of Lama pacos and Camelus ferus to retrieve the germline repertoire of V genes using human IGV genes as reference. In addition, we amplified IGKV and IGLV genes to uncover the V germline repertoire of Lama glama and sequenced BAC clones covering part of the Lama pacos IGK and IGL loci. Our in silico analysis showed that camelid counterparts of all human IGKV and IGLV families and most IGHV families could be identified, based on canonical structure and sequence homology. Interestingly, this sequence homology seemed largely restricted to the Ig V genes and was far less apparent in other genes: 6 therapeutically relevant target genes differed significantly from their human orthologs. This contributed to efficient immunization of llamas with the human proteins CD70, MET, interleukin (IL)-1β and IL-6, resulting in large panels of functional antibodies. The in silico predicted human-homologous canonical folds of camelid-derived antibodies were confirmed by X-ray crystallography solving the structure of 2 selected camelid anti-CD70 and anti-MET antibodies. These antibodies showed identical fold combinations as found in the corresponding human germline V families, yielding binding site structures closely similar to those occurring in human antibodies. In conclusion, our results indicate that active immunization of camelids can be a powerful therapeutic antibody platform.
Keywords: camelid, germline variable genes, IgG, human sequence and structural homology, antibodies, sequence mining, canonical folds, CDR, FR, biologics
Abbreviations
- BLAST
basic local alignment search tools
- CDR
complementarity-determining region
- FR
framework region
- HTG
High-Throughput Genomic database
- IGHV
immunoglobulin heavy chain variable region gene
- IGLV
immunoglobulin light chain lambda variable region gene
- IGKV
immunoglobulin light chain kappa variable region gene
- PDB
Protein Data Bank
- V family/genes
variable region family/genes
- VH
light chain variable region
- Vλ
light chain lambda variable region
- Vκ
light chain kappa variable region
- WGS
Whole Genome Shotgun database
Introduction
Over the past 20 years, therapeutic antibodies have become an important option in the treatment of malignant, infectious and auto-immune diseases. By merit of high specificity, potency, stability, solubility, clinical tolerability and relatively inexpensive manufacturing, monoclonal antibodies (mAb) made a leap in the field of targeted therapeutics.1 A growing body of genetic, molecular and structural data has explained the success of antibody specificity and potency.1
The potential for immunogenicity, i.e., the ability of a biological agent to induce a humoral or cell-mediated immune response, has long been the “Achilles' heel” of therapeutic antibodies.2,3 When administered, non-human antibodies were commonly recognized as foreign by the immune systems of treated patients, leading to neutralization and rapid clearance of the injected therapeutic antibodies or derivatives thereof, thereby limiting their efficacy. The introduction of chimeric antibodies, in which murine constant domains were replaced by human ones,4 was an early attempt to reduce immunogenicity. The humanness of variable regions was increased further in complementarity-determining region (CDR)-grafted antibodies, where rodent CDRs were joined to human FRs, generally leading to markedly improved immunogenicity profiles.5 Disappointingly, the question of how humanized and even fully human antibodies are still more or less immunogenic in patients remains to be explained.6-8
The observation that the CDRs of most humanized antibodies exhibit different structural fold combinations compared with naturally occurring human antibodies, offers a plausible explanation to the induction of undesirable immune responses against therapeutic antibodies. These “canonical structures” (canonical folds) were proven to be critical to retain affinity when humanizing antibodies,9 ultimately leading to the approach of “super humanization,” which provides the opportunity to match murine to human canonical structures.8 One important driver of the breakthrough of antibody-based therapeutics was the sequencing/characterization of human, rodent and other species germline immunoglobulin (Ig) heavy and light chain (V, (D), J and C) genes.10-12 This revealed the extensive diversity of the Ig repertoire and explained its ability to recognize and bind virtually any antigen with a different level of affinity.13 Numerous antibody discovery platforms generating human therapeutic antibodies are now available, including transgenic mouse systems as well as non-immune and synthetic human antibody libraries that can be screened by various display technologies. 4,14-16
In 1993, a surprising discovery revealed that camelids produce both conventional 4-chain structure IgGs and a non-conventional 2-chain structure IgG devoid of light chains and heavy constant region CH1.17 The heavy-chain only antibody-derived variable domain (VHH) has attracted much interest because of its stability, size (15 kDa), and cavity binding, as well as the ease with which it is engineered into manifold constructs with improved potency or broadened cross-reactivity profiles.18,19 Due to the overwhelming attention that VHHs received over the last 2 decades, camelid conventional antibodies were overlooked.
Based on published llama and dromedary in vivo matured (conventional) VH regions, we noticed the high degree of human sequence homology within the FRs, which inspired us to examine the germline VH sequences and extend these investigations to the lambda and kappa light chain variable regions for which a comprehensive analyses is missing.
We therefore sought to perform the first comprehensive analysis of the camelid germline Ig V gene repertoire and assess the homology to their human counterparts. Based on in silico analysis leveraging genomic databases, orthologs of all human IGLV and IGKV genes could be identified in camelids. Surprisingly, these turned out to have CDR1 and CDR2 canonical structure combinations identical to those of the corresponding human germline orthologs. In addition and in agreement with previous analysis,20,21 we identified an IGHV5 and IGHV7 gene family, adding to the existing knowledge of IGHV genes in camelids. Using X-ray crystallography we confirmed the presence of predicted canonical structures similar to human canonical folds on 2 selected camelid antibodies separately directed against the therapeutic targets CD70 and MET. By overlaying the structures of the camelid V regions over V regions with known canonical structures, we could confirm the presence of HCDR and LCDR canonical fold combinations identical to those found in related human V regions. Interestingly, the camelid to human homology seemed to be confined to V genes, since camelid-derived target proteins such as CD70 and MET were deviating in sequence from their human counterparts. This phylogenetic difference might explain why the camelid immune response against more foreign human antigens translates into a greater diversity of potential therapeutic antibodies.22 The combined data strongly suggests camelids as a potent source of therapeutic antibodies.
Results
Identification of camelid germline IGHV, IGLV and IGKV gene repertoires
Using in silico analysis of camel and alpaca Whole Genome Shotgun databases (WGS) and High-Throughput Genome Sequencing database (HTG), as well as (RT-)PCR-derived llama V gene sequences, we were able to retrieve camelid germline IGHV, IGLV and IGKV genes. Within the HTG database, we identified V gene specific working draft sequences of 2 BAC clones (AC232782 or CH246-176N1 ; AC232951 or CH246-77B1) covering a large part of the Lama pacos IgK and IgL loci. We sequenced these 2 BACs by Next Generation Sequencing technologies (NGS) to fully uncover camelid V genes. The newly identified and extracted V gene repertoires were annotated and classified in families and sub-families by comparison with their human V gene family counterparts. All discovered camelid V genes were collected, annotated (F. functional, and P. pseudogene) and assigned with accession numbers (Table S1). We used a set of basic local alignment search tools and the software Antibody-extractor© (antibody-extractor.net). The results depicted in the sequence alignments show that camelid and human V gene repertoires share sequence homology based on the percentage of identity of their respective frameworks FR1, FR2 and FR3 and the length of the CDR1 and CDR2, as well as their predicted canonical structures (Figs. 1-3, Table 1, Fig. S1, and Table S1). The canonical structure prediction is based on the previously defined and well-known Chothia canonical classes that, in addition to the length of CDRs, takes the presence of key residues within CDRs and FRs into account.9 Moreover, the alignments show that between different camelid species, IGHV, IGLV and IGKV genes are homologous (sometimes even identical) and share the same V family classes and sub-classes. This cross-validates the retrieved V genes from different (type and source) databases and provides a high level of confidence in the identity of the newly documented germline camelid IGHV, IGLV and IGKV repertoires.
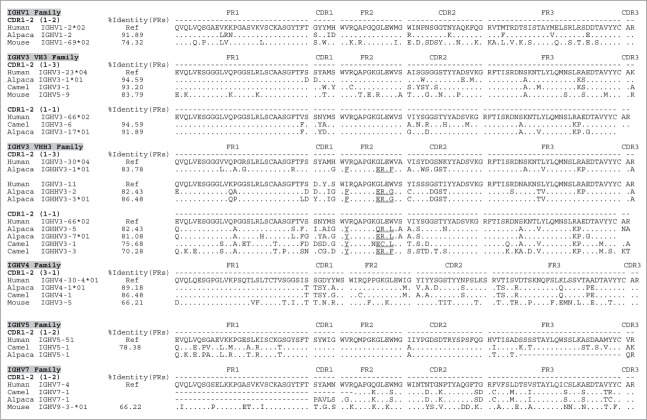
Figure 1.
Human identity of camelid IGVH germline families and their corresponding members. Similarity of each of the camelid germline families to their human counterparts is represented by a set of alignments based on the canonical folds of CDR1-2 and the % identity of their corresponding frameworks (FRs). The percentage of identity (%Identity) corresponds to identical amino acids of the V gene framework families (FR1+FR2+FR3) shared by each of the Camelus ferus (camel), Lama pacos (alpaca) and Lama glama (llama) with human IGV reference counterparts (Ref). The % identity is illustrated only for potentially functional genes. Canonical folds of CDR1-2 are indicated between parentheses. Hallmark residues in the FR2 of VHH3 that distinguish them from VH3 are underlined. Also cDNA derived IGHV5 and 7 were depicted with shading on key residues to distinguish them from one another. Complete alignments of all the VH families are illustrated in Figure S1. Stop codons are indicated by a star (*), missing or incomplete amino acid sequences are indicated by a space, and out of frame parts of V gene sequences are indicated by a dash (−).
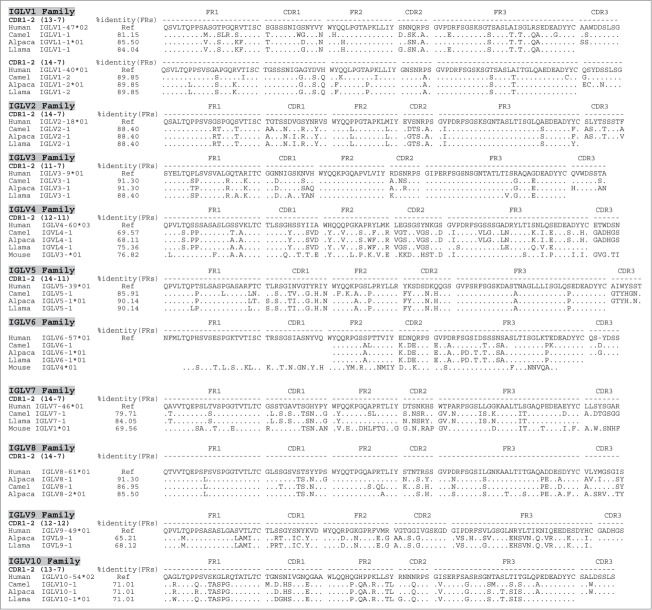
Figure 2.
Human identity of camelid IGVL germline families and their corresponding members. Similarity of each of the camelid germline families to their human counterparts is represented by a set of alignments based on the canonical folds of CDR1-2 and the % identity of their corresponding frameworks (FRs). The percentage of identity (%Identity) corresponds to identical amino acids of the V gene framework families (FR1+FR2+FR3) shared by each of the Camelus ferus (camel), Lama pacos (alpaca) and Lama glama (llama) with human IGV reference counterparts (Ref). The % identity is illustrated only for potentially functional genes. Canonical folds of CDR1-2 are indicated between parentheses. Stop codons are indicated by a star (*), missing or incomplete amino acid sequences are indicated by a space, and out of frame parts of V gene sequences are indicated by a dash (−).
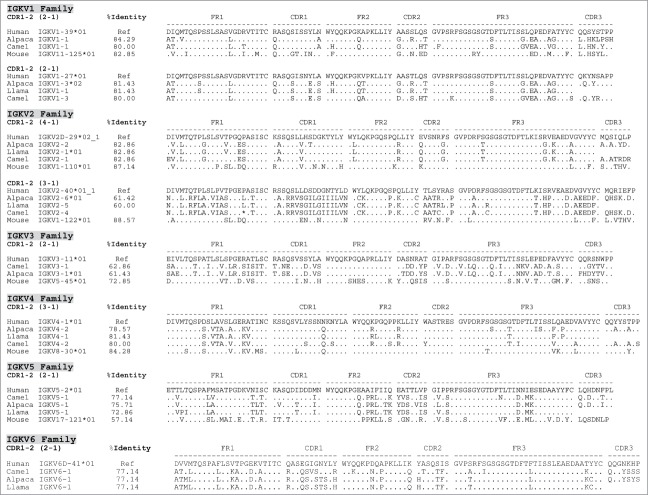
Figure 3.
Human identity of camelid IGVK germline families and their corresponding members. Similarity of each of the camelid germline families to their human counterparts is represented by a set of alignments based on the canonical folds of CDR1-2 and the % identity of their corresponding frameworks (FRs). The percentage of identity (%Identity) corresponds to identical amino acids of the V gene framework families (FR1+FR2+FR3) shared by each of the Camelus ferus (camel), Lama pacos (alpaca) and Lama glama (llama) with human IGV reference counterparts (Ref). The % identity is illustrated only for potentially functional genes. Canonical folds of CDR1-2 are indicated between parentheses. Stop codons are indicated by a star (*), missing or incomplete amino acid sequences are indicated by a space, and out of frame parts of V gene sequences are indicated by a dash (−).
Table 1.
Similarity of camelid IGV genes to human IGV counterparts
| Canonical Fold |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | Camelid IGHV | # seq | Source | Human IGHV | % Identity (FRs) | Human | Camelid | ||
| Lama pacos (Alpaca) | V1 | IGHV1-1*01 to IGHV1-3 | F | 5 | WGS Ref | IGHV1-2*02 | 81–92 | 1–2 | 1–2 |
| IGHV1-1*02; IGHV1-4 | P | 2 | WGS Ref | IGHV1-2*02 | ND | 1–2 | ND | ||
| V3 | IGHV3 | F | 45 | WGS Ref | IGHV3-23*04 | 86–94 | 1–3 | 1–3 | |
| IGHV3-7; IGHV3-8; IGHV3-10 | P | 3 | WGS Ref | IGHV3-23*04 | ND | 1–3 | ND | ||
| IGHV3-17 | F | 2 | WGS Ref | IGHV3-66*02 | 88–92 | 1–1 | 1–1 | ||
| IGHHV3-1; IGHHV3-s14 | F | 4 | WGS Ref | IGHV3-30*04 | 82–84 | 1–3 | 1–3 | ||
| IGHHV3-2 to IGHHV3-4; s2-7 | F | 6 | WGS Ref | IGHV3-11 | 82–88 | 1–3 | 1–3 | ||
| IGHHV3-5 to IGHHV3-7; s15 | F | 5 | WGS Ref | IGHV3-66*02 | 80–84 | 1–1 | 1–1 | ||
| V4 | IGHV4-1 to IGHV4-11 | F | 8 | WGS Ref | IGHV4-30-4*01 | 80–89 | 3–1 | 3–1 | |
| IGHV4-12 to IGHV4-15 | F | 5 | WGS Ref | IGHV4-30-4*01 | 77–82 | 3–1 | 3-x | ||
| V5 | IGHV5-1 | P | 1 | WGS | IGHV5-51 | ND | 1–2 | ND | |
| V7 | IGHV7-1 | P | 1 | WGS | IGHV7-4 | ND | 1–2 | ND | |
| Camelus ferus | V1 | IGHV1-1 to IGHV1-4 | P | 4 | WGS | IGHV1-2*02 | ND | 1–2 | ND |
| V3 | IGHV3-1 to IGHV3-3 | F | 3 | WGS | IGHV3-23*04 | 86–93 | 1–3 | 1–3 | |
| IGHV3-4 to IGHV3-5 | P | 2 | WGS | IGHV3-23*04 | ND | 1–3 | ND | ||
| IGHV3-6 | F | 1 | WGS | IGHV3-66*02 | 95 | 1–1 | 1–1 | ||
| IGHHV3-1 to IGHHV1-3 | F | 3 | WGS | IGHV3-66*02 | 67–74 | 1–1 | 1–1 | ||
| IGHHV3-4 to IGHHV1-5 | P | 2 | WGS | IGHV3-66*02 | ND | 1–3 | ND | ||
| V4 | IGHV4-1 | F | 1 | WGS | IGHV4-30-4*01 | 86 | 3–1 | 3–1 | |
| IGHV4-1 to IGHV3 | P | 2 | WGS | IGHV4-30-4*01 | ND | 3–1 | ND | ||
| V5 | IGHV5-1 | F | 1 | WGS | IGHV5-51 | 78 | 1–2 | 1–2 | |
| V7 | IGHV7-1 | P | 1 | WGS | IGHV7-4 | ND | 1–2 | ND | |
|
Canonical Fold |
|||||||||
| B | Camelid IGLV | #seq | Source | Human IGLV | % Identity (FRs) | Human | Camelid | ||
| Lama pacos (Alpaca) | V1 | IGLV1-1*01-*02 | F | 2 | WGS HTG | IGLV1-47*02 | 85.50 | 13–7 | 13–7 |
| IGLV1-2 to 1-6 | F | 8 | WGS HTG | IGLV1-40*01 | 86–90 | 14–7 | 14–7 | ||
| IGLV1-7 to 1-11 | P | 5 | WGS | IGLV1-40*01 | ND | 14–7 | ND | ||
| V2 | IGLV2-1 to 2-5 | F | 6 | WGS | IGLV2-18*01 | 83–88 | 14–7 | 14–7 | |
| IGLV2-6 to 2-11 | P | 6 | WGS | IGLV2-18*01 | ND | 14–7 | ND | ||
| V3 | IGLV3-1 to 3-3 | F | 3 | WGS | IGLV3-9*01 | 87–91 | 11–7 | 11–7 | |
| IGLV3-4 to 3-15 | F | 12 | WGS | IGLV3-25*02/ *03 | 81–87 | 11–7 | 11–7 | ||
| IGLV3-16 to 3-25 | P | 10 | WGS | ND | ND | 11–7 | ND | ||
| V4 | IGLV4-1 | F | 1 | WGS | IGLV4-60*03 | 68 | 12–11 | 12–11 | |
| IGLV4-2 to 4-7 | P | 6 | WGS | IGLV4-60*03 | ND | 12–11 | ND | ||
| V5 | IGLV5-1 to 5-9 | F | 9 | WGS HTG | IGLV5-39*01; 45*01, 37*01, 52*01 | 69–91 | 14–11 | 14–11 | |
| IGLV5-10 | F | 1 | HTG | IGLV5-37*01 | 84 | 14–11 | 13–11 | ||
| IGLV5-11 to 5-17 | P | 8 | WGS HTG | IGLV5-37*01 | ND | 14–11 | ND | ||
| V6 | IGLV6-1 | P | 2 | WGS HTG | IGLV6-57*01 | ND | 13–7 | ND | |
| V7 | IGLV7-1 | NA | NA | NA | NA | NA | |||
| V8 | IGLV8-1 to 8-10 | F | 10 | WGS HTG | IGLV8-61*01 | 81–91 | 14–7 | 14–7 | |
| IGLV8-11 to 8-14 | P | 7 | WGS HTG | IGLV8-61*01 | ND | 14–7 | ND | ||
| V9 | IGLV9-1*01-*02 | F | 2 | WGS HTG | IGLV9-49*01 | 65 | 12–12 | 12–12 | |
| IGLV9-2 to 5 | P | 4 | WGS | IGLV9-49*01 | ND | 12–12 | ND | ||
| V10 | IGLV10-1 | F | 1 | WGS HTG | IGLV10-54*02 | 71 | 13–7 | 13–7 | |
| Camelus ferus | V1 | IGLV1-1 | F | 1 | WGS | IGLV1-47*02 | 81 | 13–7 | 13–7 |
| IGLV1-2 to 1-7 | F | 6 | WGS | IGLV1-40*01 | 66–90 | 14–7 | 14–7 | ||
| IGLV1-8 to 1-15 | P | 8 | WGS | IGLV1-40*01 | ND | 14–7 | ND | ||
| V2 | IGLV2-1 to 2-5 | F | 7 | WGS | IGLV2-18*01 | 81–88 | 14–7 | 14–7 | |
| IGLV2-6 to 2-9 | P | 4 | WGS | IGLV2-18*01 | ND | 14–7 | ND | ||
| V3 | IGLV3-1 | F | 1 | WGS | IGLV3-9*01 | 91 | 11–7 | 11–7 | |
| IGLV3-2 to 3-6 | F | 5 | WGS | IGLV3-25*02/*03 | 81–88 | 11–7 | 11–7 | ||
| IGLV3-7 to 3-9 | P | 3 | WGS | ND | ND | 11–7 | ND | ||
| V4 | IGLV4-1 to 4-2 | F | 2 | WGS | IGLV4-60*03 | 70–74 | 12–11 | 12–11 | |
| IGLV4-3 to 4-5 | P | 3 | WGS | IGLV4-60*03 | ND | 12–11 | ND | ||
| V5 | IGLV5-1 to 5-10 | F | 9 | WGS | IGLV5-39*01; 45*01, 37*01, 52*01 | 79–86 | 14–11 | 14–11 | |
| IGLV5-11 to 5-19 | P | 9 | WGS | IGLV5-37*01 | ND | 14–11 | ND | ||
| V6 | IGLV6-1 | P | 1 | WGS | IGLV6-57*01 | ND | 13–7 | ND | |
| V7 | IGLV7-1 | F | 1 | WGS | IGLV7-46*01 | 80 | 14–7 | 14–7 | |
|
Table 1. Similarity of camelid IGV genes to human IGV counterparts | |||||||||
|
Canonical Fold |
|||||||||
| B | Camelid IGLV | #seq | Source | Human IGLV | % Identity (FRs) | Human | Camelid | ||
| V8 | IGLV8-1 to 8-7 | F | 7 | WGS | IGLV8-61*01 | 81–87 | 14–7 | 14–7 | |
| IGLV8-8 to 8-11 | P | 4 | WGS | IGLV8-61*01 | ND | 14–7 | ND | ||
| V9 | IGLV9-1 to 9-3 | P | 3 | WGS | IGLV9-49*01 | ND | 12–12 | ND | |
| V10 | IGLV10-1 to 10-2 | F | 2 | WGS | IGLV10-54*02 | 69–71 | 13–7 | 13–7 | |
| Lama glama (llama) | V1 | IGLV1-1 | F | 1 | PCR | IGLV1-47*02 | 84 | 13–7 | 13–7 |
| IGLV1-2 to 1-6 | F | 5 | PCR | IGLV1-40*01 | 84–91 | 14–7 | 14–7 | ||
| IGLV1-7 to 1-12 | P | 6 | PCR | IGLV1-40*01 | ND | ND | ND | ||
| V2 | IGLV2-1 to 2-5 | F | 7 | PCR | IGLV2-18*01 | 84–88 | 14–7 | 14–7 | |
| IGLV2-6 to 2-7 | P | 2 | PCR | IGLV2-18*01 | ND | 14–7 | ND | ||
| V3 | IGLV3-1 to 3-3 | F | 4 | PCR | IGLV3-9*01 | 87–88 | 11–7 | 11–7 | |
| IGLV3-4 to 3-11 | F | 8 | PCR | IGLV3-25*02/*03 | 81–87 | 11–7 | 11–7 | ||
| V4 | IGLV4-1 to 4-2 | F | 2 | PCR | IGLV4-60*03 | 70–75 | 12–11 | 12–11 | |
| IGLV4-3 to 4-7 | P | 5 | PCR | IGLV4-60*03 | ND | 12–11 | ND | ||
| V5 | IGLV5-1 to 5-10 | F | 10 | PCR | IGLV5-39*01; 45*01, 37*01, 52*01 | 75–90 | 14–11 | 14–11 | |
| IGLV5-11 | F | 1 | PCR | IGLV5-37*01 | 84 | 14–11 | 13–11 | ||
| IGLV5-12 to 5-20 | P | 8 | PCR | ND | ND | 14–11 | ND | ||
| V6 | IGLV6-1 | P | 2 | PCR | IGLV6-57*01 | ND | 13–7 | ND | |
| V7 | IGLV7-1 | F | 1 | PCR | IGLV7-43*01 | 84 | 14–7 | 14–7 | |
| V8 | IGLV8-1 to 8-4 | F | 4 | PCR | IGLV8-61*01 | 87–88 | 14–7 | 14–7 | |
| IGLV8-5 to 8-8 | P | 4 | PCR | IGLV8-61*01 | ND | 14–7 | ND | ||
| V9 | IGLV9-1 to 9-3 | F | 3 | PCR | IGLV9-49*01 | 65–68 | 12–12 | 12–12 | |
| V10 | IGLV10-1*01-*02 | F | 2 | WGS | IGLV10-54*02 | 69–71 | 13–7 | 13–7 | |
|
Canonical Fold |
|||||||||
| C | Camelid IGKV | #seq | Source | Human IGKV | % Identity (FRs) | Human | Camelid | ||
| Lama pacos | V1 | IGKV1-1 to 1-2 | F | 2 | HTG ; WGS | IGKV1-39*01 | 84–86 | 2–1 | 2–1 |
| IGKV1-3 to 1-4 | F | 3 | HTG ; WGS | IGKV1-27*01 | 80–86 | 2–1 | 2–1 | ||
| IGKV1-5*01-*02 | P | 2 | HTG ; WGS | IGKV1-27*01 | ND | 2–1 | ND | ||
| V2 | IGKV2-1 to 2-5 | F | 5 | HTG ; WGS | IGKV2D-29*02 | 83 | 4–1 | 4–1 | |
| IGKV2-6*01-*02 | F | 2 | HTG ; WGS | IGKV2-40*01 | 60–61 | 3–1 | 3–1 | ||
| V3 | IGKV3-1 to 3-2 | F | 3 | HTG ; WGS | IGKV3-11*01 | 60–61 | 2–1 | 2–1 | |
| IGKV3-3 to 3-4 | P | 2 | WGS | IGKV3-11*01 | ND | 2–1 | ND | ||
| V4 | IGKV4-1 to 4-2 | F | 3 | HTG ; WGS | IGKV4-1*01 | 78–80 | 3–1 | 3–1 | |
| V5 | IGKV5-1 | F | 1 | HTG ; WGS | IGKV5-2*01 | 76 | 2–1 | 2–1 | |
| V6 | IGKV6-1 | F | 1 | WGS | IGKV6D-41*01 | 77 | 2–1 | 2–1 | |
| Camelus ferus | V1 | IGKV1-1 to 1-2 | F | 2 | WGS | IGKV1-39*01 | 80 | 2–1 | 2–1 |
| IGKV1-3 | F | 1 | WGS | IGKV1-27*01 | 80 | 2–1 | 2–1 | ||
| IGKV1-3 to 1-6 | P | 3 | WGS | IGKV1-27*01 | ND | 2–1 | ND | ||
| V2 | IGKV2-1 | F | 1 | WGS | IGKV2D-29*01 | 83 | 4–1 | 4–1 | |
| IGKV2-2 to 2-4 | P | 3 | WGS | IGKV2D-29*01 | ND | 4–1 | ND | ||
| V3 | IGKV3-1 to 3-2 | F | 1 | WGS | IGKV3-11*01 | 60–62 | 2–1 | 2–1 | |
| V4 | IGKV4-1 to 4-2 | F | 2 | WGS | IGKV4-1*01 | 78–80 | 3–1 | 3–1 | |
| V5 | IGKV5-1 | F | 1 | WGS | IGKV5-2*01 | 77 | 2–1 | 2–1 | |
| V6 | IGKV6-1 | F | 1 | WGS | IGKV6D-41*01 | 77 | 2–1 | 2–1 | |
| Lama glama | V1 | IGKV1-1 | F | 1 | PCR | IGKV1-27*01 | 81 | 2–1 | 2–1 |
| V2 | IGKV2-1 to 2-4 | F | 10 | PCR | IGKV2D-29*01 | 82 | 4–1 | 4–1 | |
| IGKV2-5 | F | 1 | PCR | IGKV2-40*01 | 60 | 3–1 | 3–1 | ||
| V4 | IGKV4-1 | F | 1 | PCR | IGKV4-1*01 | 81 | 3–1 | 3–1 | |
| V5 | IGKV5-1 | F | 1 | PCR | IGKV5-2*01 | 73 | 2–1 | 2–1 | |
| V6 | IGKV6-1*01-02 | F | 2 | PCR | IGKV6D-41*01 | 77 | 2–1 | 2–1 | |
Camelid IGV germline families were compared to their human counterparts based on the canonical folds of CDR1-2 and the identity of their corresponding frameworks (FRs). The first 5 columns (camelid IGV, #seq and Source) indicate Lama pacos and Camelus ferus IGHV (A), IGLV (B) and IGKV (C) families along with their corresponding members. The numbers of sequences (#seq) are based on all sequences including allelic versions (*) of the families found similar to the human IGV gene counterparts (sixth column). The last 3 columns summarize the range of identity of the camelid frameworks and canonical folds of CDR1 and CDR2 compared to human counterparts. ND indicates genes from which the % identity and the canonical folds could not to be determined. The letters P and F indicate camelid pseudo genes and functional IGV family members, respectively. One-to-one IGHV, IGLV and IGKV comparison, including sequence ID and Genebank accession numbers, is illustrated in Table S1.
Camelid IGHV repertoires
Building on the work described in Achour et al.,20 database mining was used to determine if further information could be added to the existing dataset of known camelid IGHV, i.e., VH1, VH3, VHH3 and VH4 gene families. V genes of camelid conventional and heavy-chain only antibodies (VH and VHH) were analyzed. In addition to recovering genes belonging to IGHV families 1, 3, 4, V genes belonging to IGHV 5 and 7 families were also retrieved from the camel and alpaca WGS databases. Although the alpaca IGHV5 sequence was incomplete (Fig. 1), we designed FR1 and FR4 specific primers, and we were able (by means of PCR from cDNA) to confirm the expression of bona fide IGHV5 and IGHV7 families in Lama glama among transcripts that harbored premature stops and frame shifts (data not shown). As a negative control, murine V genes were aligned alongside their human and camelid analogs, based on canonical structure. All five V gene families were found highly homologous to their human counterparts (Fig. 1, Table 1A and Fig. S1). IGHV1 was identified based on matched canonical structures 1–2 (HCDR1 and HCDR2), and these are related to the group of human germline hIGHV1-1, 1–2, 1–3. For the IGHV3 family, 2 subfamilies were identified that could be subdivided into 4 groups (a-d) based on their CDR2 residue make-up. Camelid IGHV3 subfamilies 1 and 2 could be assigned to hIGHV3-23 and IGHV3-66 subfamilies with canonical structures 1–3 and 1–1, respectively. VHH3 is specific for camelids and is expressed as the binding fragment of heavy chain only antibodies that are not found in humans. Nevertheless, 2 subfamilies were retrieved from which the first was related to hIGHV3-30/hIGHV3-11 and the second to hIGHV3-66 subfamilies, based on the canonical structures of 1–3 and 1–1, respectively.
Within the IGHV4 family, the 2 subfamilies could be identified: one related to hIGHV4-30-4 with canonical structures of 3–1, and the other related to hIGHV4-30-4 with an identical HCDR1 length but with a different HCDR2 that might lead to different canonical folds than found in its human germline IGHV4 counterpart. It is worth noting that the latter subfamily was only found in the alpaca V gene repertoire retrieved from WGS and a previously published data set,20 but does not seem to be present in Camelus ferus and Lama glama. Finally, members belonging to the IGHV5 and IGHV7 families were found to be related to human IGHV5-51 and IGHV7-4 genes sharing the same canonical structures 1–2. The functional IGHV5 germline gene found in camel is identified as a pseudogene in alpaca with a frameshift in FR3. IGHV7 germline genes in both camel and alpaca are annotated as pseudogenes due to frameshifts in FR2. These pseudogenes (P) might result from sequencing or assembly artifacts inherent to the WGS database (Table S1).
Remarkably, camelid IGHV families 1 and 3 show an FR identity to their human counterparts of 92 to 95%, respectively, and IGHV family 4 was found to be similar by at least 82% (Fig. 1, Table 1A and Table S1). Camelid IGHV families 5 and 7 display a lower FR sequence identity, which is still 77% (Fig. 1, Table 1A). VHH3 and VH3 genes were further analyzed and shown to contain the well-described amino acid residue substitutions in FR2,23-25 being hydrophilic (F42/Y42, E49/Q49, R50/C50 and F52/G52/L52) and hydrophobic (V42/I42, G49, L50 and W52/S52), respectively.
Camelid IGLV repertoires
Located on chromosome 22q11.2, the human immunoglobulin lambda (IGL) locus consists of 3 distinct clusters, each containing members of 10 different IGLV families. All orthologs of human IGLV families were retrieved in the camelid V repertoire, including several subfamilies; sequences are listed in Fig. 2, Table 1B, Fig. S1. The Camelid IGLV1 family consists of 2 subfamilies that are related to hIGLV1-47 and hIGLV1-40, containing canonical structures 13–7 and 14–7, respectively. Most Camelid IGLV2 subfamilies found were related to hIGLV2-18 with a canonical structure of 14–7. Two camelid IGLV3 subfamilies were recovered: one related more to hIGLV3-9 while the second could be further divided in 3 that are related to human IGLV3-25 (both having canonical structures 11–7). Five camelid IGLV4 subfamilies were found that were most similar to hIGLV4-60 with canonical structures 14–11. In addition, we were able to align murine IGLV4 as a control, that in turn was found more similar to human (76%) than camel and alpaca (68–69%). Of the IGLV5 gene family, 10 Camelid subfamilies could be related to hIGLV5-39, hIGLV5-52, hIGLV5-37, hIGLV5-45, all (except subfamily 8) sharing the same canonical structures of 14–11. Camelid IGLV6 genes showed similarity to the hIGLV6-57. However, due to a premature stop codon, canonical structures could not be determined. The IGLV7 gene was only identified in llama and camel genomes and was shown to relate to hIGLV7-46 with canonical structures of 14–7. Eight camelid IGLV8 and 2 IGLV9 subfamilies were found to be related to hIGLV8-61 having canonical structures of 14–7 and to hIGLV9-49, with canonical structures of 12–12, respectively. Finally, one camelid IGLV10 family member was identified as being related to hIGLV10-54 with the same canonical structure of 13–7. As described above, the in silico analyses of the different GenBank databases and the analysis of PCR amplification-derived sequences resulted in the identification of counterparts of all human families in either or all llama, alpaca and camel species (Table 1B). The three major human IGLV families (1–3) were found to be highly homologous in camelids, with 81 to 91% sequence identity (based on FR). Interestingly, although overall identity ranged between 61 to 91%, all canonical structures matched human V family counterparts. In addition, the conservation of key residues was not only confined to the frameworks, but extended to the CDRs as well (Fig. 2). This high sequence and structural similarity of FR between camelid and human V genes is likely of great importance with regard to immunogenicity.26
Camelid IGKV repertoires
A similar approach as described for IGLV was used to identify the camelid IGKV repertoires, which was found to consist of 6 families, similar to their human counterparts (Table 1C, Fig. S1, and Table S1). Surprisingly, murine IGKV genes were closer related to their human counterparts, with the exeption of IGKV5. Three camelid IGKV1 subfamilies were found to be related to hIGKV1-27 and −39 and to share the same canonical structures 2–1. Two camelid IGKV2 subfamilies (related to hIGKV2-29 and −40) had canonical structures 4–1 and 3–1, respectively. Camelid IGKV3 and IGKV4 subfamilies were related to hIGKV3-11, having canonical structures 2–1 and to hIGKV4-1 containing canonical structures 3–1, respectively. Finally, camelid IGKV5 and IGKV6 subfamilies were found that were related to hIGKV5-1 and hIGKV6-41, based on canonical structures 2–1. The percentage of identity of camelid IGKV genes to human counterparts ranged between 60 and 85% with subfamily 2 of IGKV3 being the least similar to its human counterpart (Table 1C, Fig. S1 and Table S1). However, the identity of the residues was again not only confined to the frameworks, but also extended into the CDRs (Fig. 3).
Confirmation of predicted canonical structures in target-specific camelid antibodies determined by X-ray crystallography
To confirm the predicted high structural homology between camelid and human V regions, we sought to determine the structure of 2 camelid antibodies by X-ray crystallography. The two antibodies were isolated from antibody libraries made from actively immunized Lama glama with either human CD70 or MET over-expressing cells using phage display.27,28 Both CD70 (a membrane-bound cytokine) and MET (a tyrosine kinase receptor) play important roles in tumor growth/survival, angiogenesis and metastasis.29,30 Our acquired candidate llama-derived antibodies were subjected to structure determination by X-ray crystallography. Data collection and refinement results for the structural analysis of the Fabs derived from the anti-CD70 (PDB 4R90; VH3/Vλ7) and anti-MET (PDB 4R96; VH1/ Vκ4) antibodies are presented in Table 2.
Table 2.
X-ray crystallography, data collection and refinement statistics
| Data collection | Anti-CD70, 27B3 | Anti-MET, 48A2 |
|---|---|---|
| Beamline | Proxima 1 | Proxima 1 |
| Space group | P212121 | P212121 |
| Cell dimensions (Å/°) | 63.6, 66.9, 125.4 | 107.0, 121.5, 185.9 |
| Resolution limitsa (Å) | 65.0–1.75 (1.85–1.75) | 45.0–3.3(3.4–3.3) |
| Rmeasa (%) | 7.4 (60) | 12.0 (58.6) |
| No of observationsa | 344853 (56009) | 120361 (8817) |
| No unique reflectionsa | 55037 (8761) | 35909 (2593) |
| Mean((I)/sd(I))a | 16.4 (3.0) | 7.0 (2.4) |
| Completenessa (%) | 99.9 (99.5) | 97.5 (97.1) |
| Multiplicitya | 6.1 (6.3) | 3.3 (3.4) |
| Refinement | ||
| No of Fabs molecules | 1 | 4 |
| Resolutiona (Å) | 62.7–1.75 (1.79–1.75) | 44.0–3.3 (3.4–3.1) |
| No of reflectionsa | 52290 (3535) | 35872 (2508) |
| Atoms protein/water | 3398/567 | 13302/290 |
| No test set reflections | 2753 | 3580 |
| Rwork/Rfreea (%)a | 16.3/18.5 (21.7/22.0) | 22.1/26.0 (25.0–31.5) |
| r.m.s.d.bonds (Å)/angles (°) | 0.007/1.27 | 0.009/1.25 |
| B-average | 14.2 | 44.4 |
| Ramachandran, Preferred, | 96.2/3.8/0 | 90/6.6/3.4 |
Our structural analysis supports the presence of the same canonical structures 1–3 (red) for HCDR1 and HCDR2 in the VH domain of the camelid anti-CD70 (VH3/Vλ7) mAb, as is found in the matching human germline gene IGHV3-23 (PDB 1MFA; gray) (Fig. 4A). The light-chain domain and L1-2 loops (green) of the camelid-derived antibody superimposes perfectly with the Vλ region known for its canonical structure of 14–7. The L3 loops displays a rigid body rotation compared to its Vλ equivalent, but keeps the same conformation as the Vλ region when superimposed independently. Therefore, the predicted canonical structures that are found in the matching human germline segment Vλ7-1 (gray) were confirmed to be present in the structure of the lambda V region of the llama-derived antibody (Fig. 4B).
Figure 4.
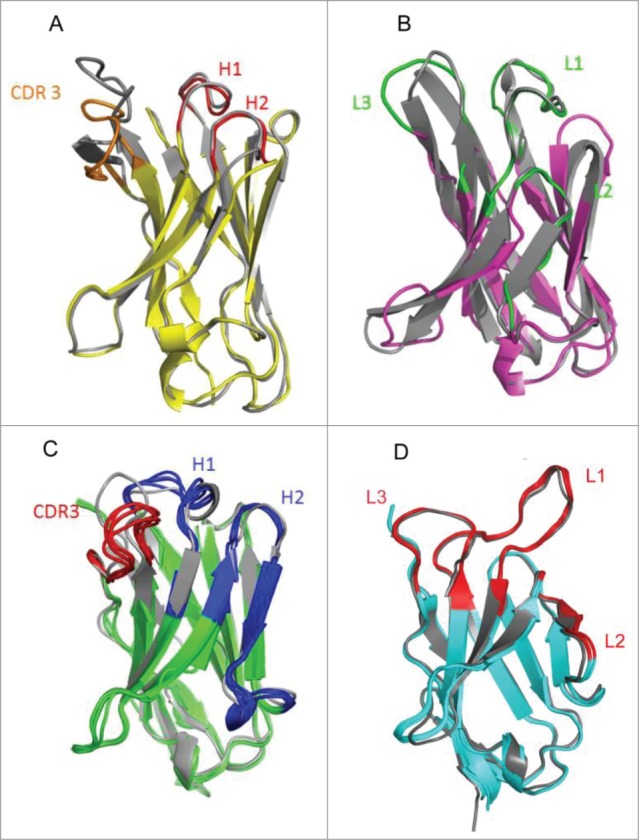
Crystal structures of heavy chain and light chain Lama glama V domains (colors) and their human counterparts (gray) depicted as an overlay. Structures are depicted as ribbons, with arrow shapes for the β-strands. (A) The llama anti-CD70 VH (yellow core, red H1 and H2, orange CDR3) is superimposed to its human counterpart (1MFA, all gray). (B) The llama anti-CD70 VL (pink core, green H1-H3) is superimposed to its human counterpart (all gray). (C) Superimposed structures of VH of llama anti-MET mAb (green core, blue H1-H2, red CDR3) onto human heavy chain references 1FVD (Vκ1) and 1HIL (Vκ4) (all gray). (D) Superimposed structures of VK of llama anti-MET mAb (green core, blue H1-H3) onto human heavy chain references 1HIL (Vκ4) (all gray).
In addition, the anti-MET (VH1/ Vκ4) mAb had a similar resemblance in its loop structures to its human heavy and light chain counterparts, for which the PDB structures 1FVD and 1HIL were used as templates (shown in gray in Fig. 4C and 4D, respectively). The canonical structures 1–2 for HCDR1 and HCDR2 present in the template 1FVD Fab fit nicely on those of the anti-MET Fab (belonging to the IGHV1 family), implying that it indeed utilizes the same canonical fold combination as found in the matching human germline VH1 segments. Also, the canonical structures 3-1-1 of LCDR1, LCDR2 and LCDR3 (blue), respectively, as present in the 1HIL template, superimposed perfectly on those of the Vκ segment of the anti-MET antibody (gray). This confirms that this llama-derived IGKV4 uses the same canonical folds as the human IGKV4 segment (Fig. 4C and D). Altogether, the structural data unequivocally confirmed the presence of predicted human canonical fold combinations in camelid-derived variable regions.
Sequence homology between human and camelid therapeutically relevant target genes
Immunizations of Lama glama using various human target proteins showed that the distant phylogenetic relationship between camelids and humans, while sharing strongly conserved IGV region genes, is beneficial for generating potent immune responses.
Immunogenicity of human (target) proteins, when administered to other species, is mostly dependent on protein complexity, size and foreignness.22 The latter increases when more residues (epitopes) differ from “self,” leading to the generation of antibodies directed against different regions of the antigen, but also affecting the magnitude of the response (i.e., high titer response). For example, a diverse set of camelid antibodies directed against a range of epitopes on human interleukin (IL)-6, IL-1β, CD70 and MET was selected from phage libraries of antibodies synthesized from immunized animals (data not shown).27,28 To reveal how foreign human therapeutic target proteins are in sequence compared to their camelid counterparts, an alignment was performed focusing on the above-mentioned human antigens (including IL-1α and tumor necrosis factor) using the sequences of camelid orthologs retrieved from WGS genomic databases (Table 3). As a reference, sequences of the macaque orthologs were also included, because they are known to have a close phylogenetic relationship to humans. Macaques have IGV genes homologous to those found in humans and are routinely used for toxicology studies, as well as for the generation of therapeutic antibodies.31 The amino acid identity of camelid to human therapeutic target genes (in the ones tested) is between 60 and 86% (based on Lama glama- or Camelus ferus-derived target sequences, when compared to their human counterparts), while macaque shows a higher identity to human of up to 90% (Table 3). These analyses support our hypothesis that high serum Ig titers observed in immunized llamas and the isolation of large panels of functional antibodies directed against diverse epitopes are (among other causes) caused by the high degree of foreignness of the target protein. However, even if immunization is performed with highly homologous human orthologs, the generation of a robust immune response may still be possible if the target antigen shows restricted expression in the llama.
Table 3.
Percentage of identity between human and camelid therapeutic target proteins, reveals low conservation in contrasting identity found between human and macaque targets (Macaca mulatta)
| Antigen | Macaca mulatta vs human identity (%) | Lama glama vs human identity (%) | Camelus ferus vs human Identity (%) | |
|---|---|---|---|---|
| CD70 | 93 | 70* | 72 | |
| IL-1α | 90 | 74 | 74 | |
| IL-1β | 90 | 63 | 65 | |
| IL-6 | 97 | 60 | 62 | |
| MET | 93 | 89* | ND | |
| TNF | 93 | 78 | 78 | |
| Antigen GenBank | human | Macaca mulatta | Lama glama | Camelus ferus |
| CD70 | BC000725.2 | XM_001088935.2 | * | NW_006210496.1 |
| Il-1α | X03833.1 | NM_001042757.1 | AB107645.1 | XM_006183588.1 |
| IL-1β | NM_000576.2 | NM_001042756.1 | AB107644.1 | XM_006183589.1 |
| IL-6 | NM_000600.3 | NC_007860.1 | AB107647.1 | XM_006179204.1 |
| MET | NC_000007.14 | NC_007860.1 | * | NW_006210492.1 |
| TNF | X02910.1 | NM_001047149.1 | AB107646.1 | XM_006178751.1 |
Human, CD70, IL1α, IL-1β, IL-6, MET, and TNF were compared to camelid and macaque counterparts using Blast (NCBI). * indicates sequence not found in GenBank, but was acquired in-house.
Discussion
To assess the potential usefulness of camelid-derived conventional antibodies as protein-based therapeutics, we characterized the camelid germline gene repertoire coding for conventional heavy and light chain V regions and determined their degree of homology to their human counterparts. First, we retrieved and annotated camelid V genes from publically available genomic databases (WGS and HTG). Here, it is worth indicating that the camel and alpaca WGS databases are yet to be fully completed and assembled. For instance, we found that the genomic sequence encoding the extracellular domain of MET deposited in the WGS database contained 2 frameshifts. Amplification of this gene segment from llama peripheral blood lymphocyte-derived RNA resulted in a sequence without frameshifts (Genbank accesion number, KF042853), indicating the database not to be 100% reliable.28
Importantly, besides the human germline V sequences, each newly identified camelid V gene was applied as reference sequence or “query” when using BLAST and Antibody-extractor©. This allowed the complete retrieval of camelid V gene repertoires, preventing bias for human homologous V genes only. Using multiple alignment algorithms and manual identification, we classified each of the camelid IGHV (VH), IGKV (Vκ) and IGLV (Vλ) repertoires in (sub-) /families based on the conservation of their canonical structure combination and sequence homology of their frameworks compared with their human counterparts. Canonical structures were identified according to the widely accepted Chothia classification. We have considered refining the analyses of the canonical structures as proposed by North et al.32 However, to provide supportive evidence that variable VH, Vκ and Vλ domains of camelid conventional antibodies are quite homologous to their human counterparts in both sequence and structure, such a refinement does not provide novel insights.
The human germline IGHV gene repertoire encompasses 51 known functional genes that belong to 7 IGHV families with one to multiple members (11 IGHV1, 3 IGHV2, 22 IGHV3, 11 IGHV4, 2 IGHV5, one IGHV6 and one IGHV7 member). Our in silico analyses uncovered all families in camelid but 2: IGHV2 and IGHV6. Strikingly, in humans, these “missing” families are known to be less frequently used than other IGHV families in VH/Vλ pairing.33,34
Surprisingly, macaque IGHV4 CDR1 and 2 contain one additional amino acid compared to humans (7–16 versus 8–17), which results in a different canonical structure (GenBank DQ437842).31 Thus, despite the fact that macaques are more closely related to humans than camelids are, the canonical structures of the IGHV4 genes of the latter may be considered more human than those of the first (the latter not having these additional residues).
Equally astounding, llama IGHV5 and IGHV7 were very difficult to find among camelids, leading us to amplify these genes from Lama glama cDNA to find just one functional transcript of each. These were accompanied by roughly 40 homologous transcripts that could be defined as pseudogenes based on the presence of premature stop codons and frameshifts. Although the exact mechanism behind this remains obscure, the high incidence of insertions and deletions found in independent amplifications points in the direction of a gene conversion-like event, as is seen in rabbit, cattle and chicken.35-37
Humans display a germline IGLV repertoire of 33 distinguishable functional genes, belonging to 10 IGLV families (5 IGLV1, 5 IGLV2, 10 IGLV3, 3 IGLV4, 4 IGLV5, one IGLV6, 2 IGLV7, one IGLV8/ IGLV9/ IGLV10). All ten camelid IGLV counterparts were retrieved with their corresponding canonical structures. The framework regions of camelid IGLV genes share 65 to 91% identity with their human counterparts. Remarkably, the highest homology to human (81–91%) was present among the 3 IGLV families (IGLV1-3) predominantly used in species that express mainly lambda light chains, like humans.11 In this respect, transgenic mouse systems could produce human mAbs when, in addition to the human heavy and kappa light chain genes, the human lambda Ig repertoire were added and kappa expression was silenced.38 When taking into account that light chain usage during the B-cell response is far more lambda-dependent in humans than, for instance, in the murine immune system (that uses close to 95% kappa light chains),39 the advantage of having a similar to human light chain repertoire, (as in camelids) becomes clear.
In contrast to the Ig gene repertoire, camelid genes orthologous to human genes encoding proteins targeted in disease display a relatively low homology. This is in contrast to the situation in macaques: despite being considered as animals from which therapeutic antibodies can be retrieved, macaques show a high human genomic sequence identity that includes genes of therapeutic interest. This makes their Ig repertoire after active immunization with human proteins prone to lower epitope coverage, skewed toward those epitopes that differ strongly.
Our study was meant to gather information about the camelid V genes that were largely unknown and to understand their sequence and structural homology to the human orthologs. In paralell, Griffin et al. 21 reported about complete conventional camelid V genes and their intra-/interspecies sequence identity. Importantly, the sequence identity found between camelid and human VH, Vλ and Vκ genes resembled our findings nicely, despite a difference in assigning interspecies counterparts (sequence identity vs canonical structure identity). For instance, IGHV3 and 4 were reported with 94% and 86% identity, respectively, where we found 86% to 94% and 79% to 89%, respectively. IGLV and IGKV were found to have 85% and 77% identity, where we found comparable numbers among our IGLV families. Of our isolated IGKV families, Vκ2-3 had a 60% human identity and Vκ5-6 had a 72–77% human identity. Most of the IGKV families (66%) had a identity of 80% and higher.
Immunization of camelids with human target proteins benefits from a more distant phylogenetic relationship between humans and camelids, resulting in a greater diversity of potential therapeutic antibodies due to the presence of more foreign epitopes.22 The importance of this “foreignness” is best described in a setting where camelid immunization against alien viral envelope proteins resulted in a highly diverse panel of binders, enabling targeting of multiple epitopes on the same viral envelope protein.40,41
The camelid V family repertoire described here serves as a template to humanize camelid V genes (for therapeutic purposes) in such a way that affinity, potency and stability are retained. Finally, to fully utilize camelid IGHV, IGLV, IGKV repertoires, an optimal design of V family (member) specific primers is now made possible. This will be beneficial for antibody discovery via non-immune and immune phage display library construction. Our study provides the necessary tools and knowledge base to allow further advances in the field of comparative immunology and applied antibody research. These data are supported by the clinical development of 2 antibodies (anti-CD70 and anti-MET) discussed here. These antibodies, ARGX-110 and ARGX-111, are now being evaluated in Phase 1b clinical studies (NCT01813539 and NCT02055066, respectively), thereby further validating the use of camelids as a powerful technology platform for the generation of antibody therapeutics.
Materials and Methods
Description of databases and data mining tools
Camelid germline V genes were retrieved from the publically available Whole Genome Shotgun database, the WGS phase I ABRR01000000 and the WGS phase II ABRR02000000 of Vicugna pacos (also referred to as Lama pacos or alpaca), the WGS phase I AGVR01000000 of Camelus ferus (also referred to as Bactrian camel) and the High-Throughput Genome Sequencing database, HTG GenBank accession number Vicugna pacos AC232951 and AC232782. The HTG alpaca database consists of sequence collections derived from the CHORI-246 alpaca BAC Library. Since the 2 HTG derived BACs are working draft sequences, we proceeded to their sequencing using NGS Ion Torrent technology (Life Technologies, New York, USA) to retrieve more complete V gene sequences. Human germline V gene sequences are derived from the publicly available human reference genome (Build 37, hg 19), Vbase2 database (www.vbase2.org/vbdownload.php) and NCBI Igblast database (www.ncbi.nlm.nih.gov/igblast/showGermline.cgi). Three NCBI basic local alignment search tools were used: BLAST with Vicugna pacos genome (www.ncbi.nlm.nih.gov/genomes/geblast.cgi?taxid=30538); BLAST with Camelus ferus genome (www.ncbi.nlm.nih.gov/genomes/geblast.cgi?taxid= 419612) and BLAST (blastn, blastx and tblastn) with the HTG selected database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). CLC Main Workbench software local blast and assembly to reference tools were used to annotate, extract and create camel and alpaca IGLV (Vλ) IGKV (Vκ), IGHV (VH/VHH) database sequences. Additionally, we used Antibody-extractor© software (antibody-extractor.net), a set of algorithms and tools dedicated to antibody DNA and protein sequence analysis using human V region knowledge base.
PCR amplifications, cloning and sequencing of llama germline V genes
Genomic DNA was extracted from a llama (Lama glama) testis. Fragments of frozen testis were ground in liquid nitrogen and incubated in 20 ml of lysis buffer, 100 mM Tris HCl pH 8.5 (Sigma-Aldrich, H1758), 5 mM EDTA (Sigma-Aldrich, E6758), 0.2% SDS (Sigma-Aldrich, L3771), 200 mM NaCl (Sigma-Aldrich, S3014), 100 µg Proteinase K/ml (Promega, V3021) at 37°C until fully dissolved. Genomic DNA was extracted using the phenol/chlorophorm/isoamyl alcohol (Sigma-Aldrich, P3803) method as previously described.42 PCR amplification was performed in the presence of forward and reverse (Vλ or Vκ) specific primers using 0.5 U Phusion high fidelity DNA polymerase in a 50 μl final reaction volume according to the manufacturer's instructions (Finnzymes, F-530L). Parameters for PCR were 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 60–65°C for 30 s, 72°C for 30 s to 1 min, and a final extension at 72°C for 10 min. PCR products were purified using NucleoSpin Gel and PCR Clean-up (Machery-Nagel, 740609.10/.50/.250), cloned into a pJET1.2 vector using a CloneJET PCR cloning kit (Thermo Fisher Scientific, K1231), and transformed by electroporation into E. coli strain TG1 DUOs (Lucigen, 60502-1). Clones were then sequenced, analyzed and gathered in distinct Vκ and Vλ gene specific datasets. Vκ and Vλ specific primers were designed based on the germline Vκ or Vλ gene sequences extracted and annotated from the publicly available databases, Vicugna pacos WGS phase I ABRR01000000, HTG AC232951 and HTG AC232782. The forward primers were based on either leader part1 (exon1 of V genes) or the framework 1 (FR1), and the reverse primers were based on the framework 3 (FR3) of germline Vλ or Vκ genes. A list of the primers used is shown in Table S2.
Amplification of VH5 and VH7 V regions, from llama cDNA
Peripheral blood lymphocytes (PBL) were isolated by centrifugation using a Ficoll (Pharmacia Biotech, A2252) discontinuous gradient. Total RNA was then isolated by an acid guanidium thiocyanate extraction, as previously described.43 The RNA integrity was verified via gel electrophoresis; concentration and purity were determined by OD measurement (wavelength scan between 230 nm 320 nm) (A260nm / A280nm ratio). Total RNA was used as template to generate cDNA using random hexamer primers and the SuperScriptIII® RT-PCR system (Invitrogen, 12574-030). Oligonucleotides VH5-leader-primer1 ATGCGGTC TGTCACAGCCATC, VH7-leader-primer1 ATGGACGGGACCTGGACAATCC and CH1 were used for PCR amplification of Lama glama V gene segments.
Identification, annotation and extraction of camel and alpaca germline V genes
Three steps were used to retrieve camel and alpaca germline V genes from the publicly available databases described above. The first step of analysis consisted of using the human germline V gene nucleotide and translated sequences as a “query” in the BLAST tools to identify contigs, scaffolds and BACs that harbor alpaca and camel germline V gene of heavy chains (VH and VHH) and light chains (Vλ and Vκ). In addition, previously published alpaca germline VH and VHH genes described in Achour et al. were used as a “query” in the first step.20 The second step consisted of downloading reads, contigs and scaffolds in the CLC Main Workbench software, after which camel and alpaca sequences were extracted from this data set as follows: a local blast, assembly to reference tool and annotation tools were applied using human V genes as a “query” while contigs and scaffolds were considered the reference sequences. The third step consisted of using the newly identified and extracted camel and alpaca germline V gene sequences, as well as the PCR amplified llama V gene sequences, as a “query” in the BLAST tools. The second and third steps were carried out several times (loop process) until no new identified V gene sequences were recovered. The last step consisted of creating distinct camel and alpaca Vκ, Vλ and VH/VHH gene specific datasets. The gene sequence data sets illustrating FR1 to FR3, leader exon 1, leader exon 2 and recombination sequence sites (RSS) were extracted to fully annotate the newly identified camel and alpaca V gene regions.
Identification and classification of the camelid V family repertoire
Multiple alignments using CLC Main Workbench software were performed for each of the camel, alpaca and llama Vκ, Vλ or VH/VHH specific datasets along with human V gene counterparts. The identification and classification of camelid V genes families and sub-families were based on the percentage of identity in FR1, FR2 and FR3, along with the size and the degree of homology of predicted CDR canonical folding (CDR1-CDR2) shared with human V genes counterparts. Camel, alpaca and llama germline V gene canonical folding were determined using Canonicals - Chothia Canonical Assignment software (www.bioinf.org.uk/abs/chothia.html). As WGS and HTG contain gaps, V genes (FR1-FR3) are designed as functional (F) when they are devoid of premature stop codons and are in frame. Pseudo-genes among V genes were designated as such when they harbored stop codons, were out of frame, or were incomplete.
Papain digestion and purification of Fabs
Eight mg of anti-CD70 and anti-MET mAbs at 4 mg/ml in Dulbecco's phosphate-buffered saline (d-PBS) pH 7.2 (Life Technologies, 70013032) were buffer-exchanged to a digestion buffer containing 20 mM cysteine-HCl (Sigma-Aldrich, Fallavier, France) on a Zeba TM Desalt Spin Column (Pierce Fab Preparation Kit) (Pierce Thermo Scientific, 44985). Samples were incubated with Immobilized Papain (Pierce Thermo Scientific, 20341) and digested for 6 hours at 37°C. The Fcs were separated from the Fabs using a CaptureSelect human Fc affinity matrix (BAC BV, 190082210) equilibrated in d-PBS. Fabs were recovered in the flow-through and Fcs were eluted using 0.1 M glycine pH 2.0 (Sigma-Aldrich, G2879). Protein concentration was determined by UV spectrometry from the absorbance at 280 nm. ˜3 mg of purified Fab were recovered and concentrated to 8–9 mg/ml on Amicon-Ultra (Merck, UFC201024) (cut-off 10 kDa).
Crystallization, X-ray diffraction and structure determination of Fabs
Initial crystallization screening of Fab derived from the anti-CD70 mAb was performed with commercial kits Structure screen 1 & 2, Wizard screen and Stura Footprint screen (Molecular Dimensions Limited, MD1-30, MD15-JCSGP-B, and MD1-20, respectively). Drops were set-up with a 1:1 (v:v) ratio of protein (8.9 mg/ml) to mother liquor in a total volume of 200 nl on Greiner 96-well plates using a Cartesian MicroSys SQ robot. A diffraction-quality crystal of Fab was obtained by sitting-drop vapor diffusion at 277 K after one week in 2 M ammonium sulfate (Sigma-Aldrich, A4418) and 0.15 M Na citrate pH 5.5 (Sigma-Aldrich, PHR1416). Diffraction data were collected at beamline Proxima-1 (Synchrotron Soleil, Saint-Aubin, France), integrated with XDS and scaled with Xscale.44 The structure was solved using molecular replacement with Molrep.45 The starting models were the CH, CL, VH and Vλ from the IL-17a/Fab complex (PDB 2VXS).46 Refinement was made with Refmac,47 alternating with manual construction/reconstruction with Coot.48
Crystallization screening of Fab prepared from the anti-MET antibody was performed with commercial kits Proplex screen, Wizard screen and Stura Footprint screen (Molecular Dimensions Limited, Suffolk, United Kingdom). Drops were set-up with a 3,2,1:1 (v:v) ratio of protein (8.2 mg/ml) to mother liquor in a total volume of 200 nl. A diffraction-quality crystal of Fab was obtained by sitting-drop vapor diffusion at 277 K after 3 weeks in 1.4 M Na Malonate pH 6.0 (Sigma-Aldrich, M4795). Diffraction data were collected and the structure was solved as described above. Detailed data collection and refinement statistics are displayed in Table 2.
Disclosure of Potential Conflicts of Interest
CB, AH, NDJ and HdH are employees of arGEN-X and AK was an employee of arGEN-X for part of the work. KEM is consultant and inventor of the Antibody-extractor©. IA, RR and TV declare no conflict of interest.
Acknowledgments
The authors would like to thank John Wijdenes for providing Lama glama testis, from which genomic DNA was extracted. Also our special thanks to Janet M. Van Bobo for linguistic corrections.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Finlay WJ, Almagro JC. Natural and man-made V-gene repertoires for antibody discovery. Front Immunol 2012; 3:342; PMID:23162556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima T, Yagi H, Takemoto E, Shibata‐Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells 2011; 16:1071-80; PMID:22023369; http://dx.doi.org/ 10.1111/j.1365-2443.2011.01552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol 2009; 157:220-33; PMID:19459844; http://dx.doi.org/ 10.1111/j.1476-5381.2009.00190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986; 4:522-5; PMID:3713831; http://dx.doi.org/ 10.1038/321522a0 [DOI] [PubMed] [Google Scholar]
- 5.Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature 1988; 332:323-7; PMID:3127726; http://dx.doi.org/ 10.1038/332323a0 [DOI] [PubMed] [Google Scholar]
- 6.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 1999; 17:2639-48; PMID:10561337 [DOI] [PubMed] [Google Scholar]
- 7.Ritter G, Cohen LS, Williams C, Richards EC, Old LJ, Welt S. Serological analysis of human anti-human antibody responses in colon cancer patients treated with repeated doses of humanized monoclonal antibody A33. Cancer Res 2001; 61:6851-9; PMID:11559561 [PubMed] [Google Scholar]
- 8.Tan P, Mitchell DA, Buss TN, Holmes MA, Anasetti C, Foote J. “Superhumanized” antibodies: reduction of immunogenic potential by complementarity-determining region grafting with human germline sequences: application to an anti-CD28. J Immunol 2002; 169:1119-25; PMID:12097421; http://dx.doi.org/ 10.4049/jimmunol.169.2.1119 [DOI] [PubMed] [Google Scholar]
- 9.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol 1987; 196:901-17; PMID:3681981; http://dx.doi.org/ 10.1016/0022-2836(87)90412-8 [DOI] [PubMed] [Google Scholar]
- 10.Chothia C, Lesk AM, Gherardi E, Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. Structural repertoire of the human VH segments. J Mol Biol 1992; 227:799-817; PMID:1404389; http://dx.doi.org/ 10.1016/0022-2836(92)90224-8 [DOI] [PubMed] [Google Scholar]
- 11.Williams SC, Frippiat JP, Tomlinson IM, Ignatovich O, Lefranc MP, Winter G. Sequence and evolution of the human germline V lambda repertoire. J Mol Biol 1996; 264:220-32; PMID:8951372; http://dx.doi.org/ 10.1006/jmbi.1996.0636 [DOI] [PubMed] [Google Scholar]
- 12.Tomlinson IM, Cox J, Gherardi E, Lesk A, Chothia C. The structural repertoire of the human V kappa domain. EMBO J 1995; 14:4628-38; PMID:7556106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol 1995; 7:248-54; PMID:7546385; http://dx.doi.org/ 10.1016/0952-7915(95)80010-7 [DOI] [PubMed] [Google Scholar]
- 14.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23:1105-16; PMID:16151404; http://dx.doi.org/ 10.1038/nbt1126 [DOI] [PubMed] [Google Scholar]
- 15.Taylor LD, Carmack CE, Schramm SR, Mashayekh R, Higgins KM, Kuo C-C, Woodhouse C, Kay RM, Lonberg N. A transgenic mouse that expresses a diversity of human sequence heavy and light chain immunoglobulins. Nucleic Acids Res 1992; 20:6287-95; PMID:1475190; http://dx.doi.org/ 10.1093/nar/20.23.6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brüggemann M, Neuberger MS. Strategies for expressing human antibody repertoires in transgenic mice. Immunol Today 1996; 17:391-7; PMID:8783501 [DOI] [PubMed] [Google Scholar]
- 17.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hammers C, Songa EB, Bendahman N, Hammers R. Naturally occurring antibodies devoid of light chains. Nature 1993; 363:446-8; PMID:8502296; http://dx.doi.org/ 10.1038/363446a0 [DOI] [PubMed] [Google Scholar]
- 18.Vincke C, Gutiérrez C, Wernery U, Devoogdt N, Hassanzadeh-Ghassabeh G, Muyldermans S. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol Biol 2012; 907:145-76; PMID:2290735021520037 [DOI] [PubMed] [Google Scholar]
- 19.Roovers RC, Vosjan MJ, Laeremans T, el Khoulati R, de Bruin RC, Ferguson KM, Verkleij AJ, van Dongen GA, van Bergen en Henegouwen PM. A biparatopic anti-EGFR nanobody efficiently inhibits solid tumour growth. Int J Cancer 2011; 129:2013-24; PMID:21520037; http://dx.doi.org/ 10.1002/ijc.26145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Achour I, Cavelier P, Tichit M, Bouchier C, Lafaye P, Rougeon F. Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J Immunol 2008; 181:2001-9; PMID:18641337; http://dx.doi.org/ 10.4049/jimmunol.181.3.2001 [DOI] [PubMed] [Google Scholar]
- 21.Griffin LM, Snowden JR, Lawson AD, Wernery U, Kinne J, Baker TS. Analysis of heavy and light chain sequences of conventional camelid antibodies from Camelus dromedarius and Camelus bactrianus species. J Immunol Methods 2014; 405:35-46; PMID:24444705 [DOI] [PubMed] [Google Scholar]
- 22.Tainer JA, Getzoff ED, Paterson Y, Olson AJ, Lerner RA. The atomic mobility component of protein antigenicity. Annu Rev Immunol 1985; 3:501-39; PMID:2415142; http://dx.doi.org/ 10.1146/annurev.iy.03.040185.002441 [DOI] [PubMed] [Google Scholar]
- 23.Maass DR, Sepulveda J, Pernthaner A, Shoemaker CB. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J Immunol Methods 2007; 324:13-25; PMID:17568607; http://dx.doi.org/ 10.1016/j.jim.2007.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol 1997; 34:1121-31; PMID:9566760; http://dx.doi.org/ 10.1016/S0161-5890(97)00146-6 [DOI] [PubMed] [Google Scholar]
- 25.Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng 1994; 7:1129-35; PMID:7831284; http://dx.doi.org/ 10.1093/protein/7.9.1129 [DOI] [PubMed] [Google Scholar]
- 26.Hwang WY, Foote J. Immunogenicity of engineered antibodies. Methods 2005; 36:3-10; PMID:15848070; http://dx.doi.org/ 10.1016/j.ymeth.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 27.Silence K, Dreier T, Moshir M, Ulrichts P, Gabriels SM, Saunders M, Wajant H, Brouckaert P, Huyghe L, Van Hauwermeiren T, et al.. ARGX-110, a highly potent antibody targeting CD70, eliminates tumors via both enhanced ADCC and immune checkpoint blockade. Mabs 2014; 6:523-32; PMID:24492296; http://dx.doi.org/ 10.4161/mabs.27398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basilico C, Hultberg A, Blanchetot C, de Jonge N, Festjens E, Hanssens V, Osepa SI, De Boeck G, Mira A, Cazzanti M, et al.. Four individually druggable MET hotspots mediate HGF-driven tumor progression. J Clin Invest 2014; 124:3172-86; PMID:24865428; http://dx.doi.org/ 10.1172/JCI72316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gherardi E, Birchmeier W, Birchmeier C, Woude GV. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012; 12:89-103; PMID:22270953 [DOI] [PubMed] [Google Scholar]
- 30.Grewal IS. CD70 as a therapeutic target in human malignancies. Expert Opin Ther Targets 2008; 12:341-51; PMID:18269343; http://dx.doi.org/ 10.1517/14728222.12.3.341 [DOI] [PubMed] [Google Scholar]
- 31.Thullier P, Chahboun S, Pelat T. A comparison of human and macaque (Macaca mulatta) immunoglobulin germline V regions and its implications for antibody engineering. Mabs 2010; 2:528-38; PMID:20562531; http://dx.doi.org/ 10.4161/mabs.2.5.12545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.North B, Lehmann A, Dunbrack RL Jr.. A new clustering of antibody CDR loop conformations. J Mol Biol 2011; 406:228-56; PMID:21035459; http://dx.doi.org/ 10.1016/j.jmb.2010.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jayaram N, Bhowmick P, Martin AC. Germline VH/VL pairing in antibodies. Prot Eng Des Sel 2012; 25:523-30; PMID:22802295; http://dx.doi.org/ 10.1093/protein/gzs043 [DOI] [PubMed] [Google Scholar]
- 34.Jiménez-Gómez G, Gómez-Perales JL, Ramos-Amaya A, González-García I, Campos-Caro A, Brieva JA. Modulated selection of IGHV gene somatic hypermutation during systemic maturation of human plasma cells. J Leukocyte Biol 2010; 87:523-30; PMID:Can't; http://dx.doi.org/ 10.1189/jlb.0709514 [DOI] [PubMed] [Google Scholar]
- 35.Knight KL, Becker RS. Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: implications for the generation of antibody diversity. Cell 1990; 60:963-70; PMID:2317867; http://dx.doi.org/ 10.1016/0092-8674(90)90344-E [DOI] [PubMed] [Google Scholar]
- 36.Parng CL, Hansal S, Goldsby RA, Osborne BA. Gene conversion contributes to Ig light chain diversity in cattle. J Immunol 1996; 157:5478-86; PMID:8955197 [PubMed] [Google Scholar]
- 37.Reynaud CA, Anquez V, Grimal H, Weill JC. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 1987; 48:379-88; PMID:3100050; http://dx.doi.org/ 10.1016/0092-8674(87)90189-9 [DOI] [PubMed] [Google Scholar]
- 38.Nicholson IC, Zou X, Popov AV, Cook GP, Corps EM, Humphries S, Ayling C, Goyenechea B, Xian J, Taussig MJ. Antibody repertoires of four-and five-feature translocus mice carrying human immunoglobulin heavy chain and κ and λ light chain yeast artificial chromosomes. J Immunol 1999; 163:6898-906; PMID:10586092 [PubMed] [Google Scholar]
- 39.Zocher I, Roschenthaler F, Kirschbaum T, Schable KF, Horlein R, Fleischmann B, Kofler R, Geley S, Hameister H, Zachau HG. Clustered and interspersed gene families in the mouse immunoglobulin kappa locus. Eur J Immunol 1995; 25:3326-31; PMID:8566019; http://dx.doi.org/ 10.1002/eji.1830251219 [DOI] [PubMed] [Google Scholar]
- 40.McCoy LE, Quigley AF, Strokappe NM, Bulmer-Thomas B, Seaman MS, Mortier D, Rutten L, Chander N, Edwards CJ, Ketteler R, et al.. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J Exp Med 2012; 209:1091-103; PMID:22641382; http://dx.doi.org/ 10.1084/jem.20112655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hultberg A, Temperton NJ, Rosseels V, Koenders M, Gonzalez-Pajuelo M, Schepens B, Ibanez LI, Vanlandschoot P, Schillemans J, Saunders M, et al.. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PloS One 2011; 6:e17665; PMID:21483777; http://dx.doi.org/ 10.1371/journal.pone.0017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maniatis T, Hardison RC, Lacy E, Lauer J, O'Connell C, Quon D, Sim GK, Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell 1978; 15:687-701; PMID:719759; http://dx.doi.org/ 10.1016/0092-8674(78)90036-3 [DOI] [PubMed] [Google Scholar]
- 43.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156-9; PMID:2440339; http://dx.doi.org/ 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallogr Sect D Biol Crystallogr 2010; 66:125-32; PMID:20124692; http://dx.doi.org/ 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr Sect D Biol Crystallogr 2010; 66:22-5; PMID:20057045; http://dx.doi.org/ 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 46.Gerhardt S, Abbott WM, Hargreaves D, Pauptit RA, Davies RA, Needham MR, Langham C, Barker W, Aziz A, Snow MJ, et al.. Structure of IL-17A in complex with a potent, fully human neutralizing antibody. J Mol Biol 2009; 394:905-21; PMID:19835883; http://dx.doi.org/ 10.1016/j.jmb.2009.10.008 [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 2003; 374:300-21; PMID:14696379; http://dx.doi.org/ 10.1016/S0076-6879(03)74014-2 [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr 2010; 66:486-501; PMID:20383002; http://dx.doi.org/ 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.