Abstract
The recently developed transparent soil consists of particles of Nafion, a polymer with a low refractive index (RI), which is prepared by milling and chemical treatment for use as a soil analog. After the addition of a RI-matched solution, confocal imaging can be carried out in vivo and without destructive sampling. In a previous study, we showed that the new substrate provides a good approximation of plant growth conditions found in natural soils. In this paper, we present further development of the techniques for detailed quantitative analysis of images of root-microbe interactions in situ. Using this system it was possible for the first time to analyze bacterial distribution along the roots and in the bulk substrate in vivo. These findings indicate that the coupling of transparent soil with light microscopy is an important advance toward the discovery of the mechanisms of microbial colonisation of the rhizosphere.
Keywords: plant roots, Rhizobia, confocal, Nafion, refractive index matching, lettuce, image analysis, PGPR, Pseudomonas fluorescens, GFP
Abbreviations
- PGPR
plant growth promoting rhizobacteria
- RI
refractive index
- GFP
green fluorescent protein
- CFU
colony forming units
- FISH
fluorescent in situ hybridization
- T-RFLP
terminal restriction fragment length polymorphism
Plant growth promoting rhizobacteria (PGPR) enhance plant health and yield via complex interactions with the roots and soil.1-3 Rhizobacteria can offer the plant protection from pathogenic microorganisms by outcompeting them and through the promotion of plant growth via the release of plant hormones.4 They can also aid plant uptake of nutrients via the rhizosphere, for example by releasing iron-scavenging siderophores.4,5 The spatial and temporal heterogeneity of soil and the rhizosphere undoubtedly influences the communities and function of bacteria which inhabit niches where nutrients are available in soil.6 However, studying the interactions between soil bacteria and their physical habitat is currently very challenging partly due to the lack of conventional laboratory techniques and protocols. Light microscopy cannot be used to observe soil in depth because soil is opaque. X-ray imaging techniques are suitable for studying the soil structure but cannot simultaneously resolve microorganisms.7 Although many molecular methods can be used to identify the structure of soil microbial communities,8 most do not provide insight into their spatial arrangements. In contrast, recent applications of FISH (fluorescent in situ hybridization) have proved successful to analyze spatial distribution of microorganisms in soil, but the method is not suitable to study dynamic processes because samples need to be fixed prior to imaging.9
Previously, we published a study describing a new transparent soil analog for imaging plant roots using optical microscopy.10 It consists of a matrix of solid particles of the low refractive index (RI) ionomer, Nafion, water with plant nutrients and air. Transparent soil can be saturated with a RI matched liquid to reveal biological structures within. Further to this work, we have applied transparent soil to the observation of PGPR spatial interactions with roots and soil particles non-destructively, in vivo and in situ. Quantitative analysis methods were developed to study the spatial distribution of PGPR Pseudomonas fluorescens SBW25 in transparent soil, on the surface of Lactuca sativa (lettuce) roots and in the surrounding transparent soil, in relation to the pore geometry. The effect of substrate parameters on the colonisation of roots was also tested by varying the substrate particle size. The aims were to measure the effect of plants and substrate on the abundance of PGPR both on root and on the surrounding particles. After inoculation of the transparent substrate with a culture of GFP-tagged P. fluorescens, one day old L. sativa seedlings were added to the microcosms. The microcosms were sealed and incubated for 5 d allowing the plants to grow and the bacteria to colonise the roots. The transparency of the substrate allowed images to be captured on a 3D grid using confocal microscopy, thus sampling the microbial abundance at points along the roots and in the bulk soil at 2 distances from the root (supplementary information, Fig. S1). Fluorescent labeling with a range of fluorophores allowed discrimination of bacteria (GFP), root tissue (calcofluor) and the surfaces of solid Nafion particles (sulphorhodamine-B) (Fig. 1), which facilitated image analysis (Fig. 2).
Figure 1.
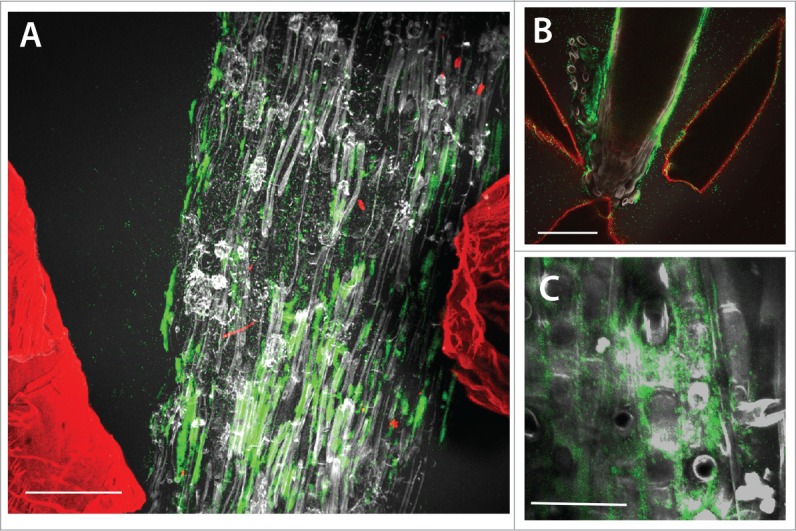
Maximum projection confocal images of GFP-labeled Pseudomonas fluorescens colonies (green) on the surface of lettuce root tissues (gray) in situ in transparent soil with Nafion particles from the substrate labeled with sulphorhodamine B fluorescent dye also visible (red). (A) The majority of the bacterial fluorescence is associated with the root tissue. Scale bar = 150 μm. (B) Bacteria are present on the root tip and in this case also the surfaces of Nafion particles in close proximity to the root have bacterial fluorescence associated with them. Scale bar = 150 μm. (C) At higher resolution, bacterial colonisation was predominantly observed in the intercellular junctions of root epithelial cells. Scale bar = 45 μm.
Figure 2.
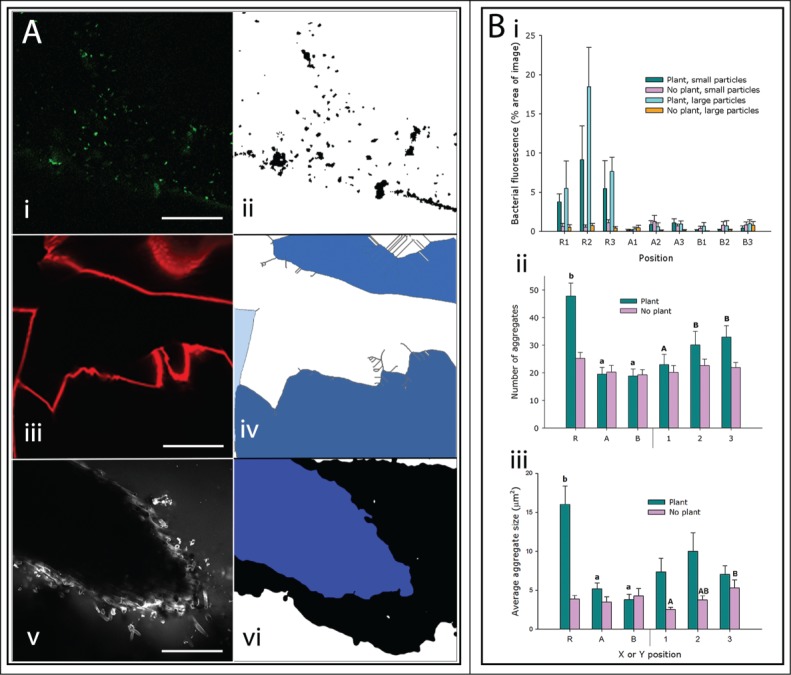
Quantification of Pseudomonas fluorescens in the rhizosphere. (A) Bacteria, Nafion particles and roots were processed sequentially to allow quantification. (i-ii) Bacterial fluorescence before and after processing with a median filter and thresholding facilitated measuring the bacterial abundance. Scale bar = 40 μm. (iii-iv) Original images of particle surfaces were processed and skeletonised. Gray lines in (iv) represent skeleton of particle surfaces in (iii). It was then possible to select the volumes inside particles (shown here in blue) to measure them to correct for available area (pore space). Scale bar = 200 μm. (v-vi) Example image of a section of lettuce root before and after the application of a median filter and subsequent thresholding were applied. This allowed the selection of the internal volume of the root for measurement (shown in blue). Scale bar = 200 μm. (B) Quantification of bacterial distribution in transparent soil with small (500–850 μm) and large particles (850–1200 μm). The positions R1 to B3 represent a 3 × 3 grid of points on and around the roots, where R is on the root and A and B are at intervals perpendicular to the root. 1 is the root tip and 2 and 3 are closer to the shoot. See Figure S1 for schematic. (i) There was higher bacterial abundance in images that include a section of plant root. At all other positions, there was a consistent area of bacterial fluorescence as a proportion of the area of backgound in images without plant roots. These values were corrected for available area. (ii) Number and (iii) average size of bacterial aggregates at the 3 horizontal (X) positions (R, A & B) and at the 3 vertical (Y) positions (1, 2 and 3) in samples with or without plants. Letters above the bars indicate the results of Fisher's protected LSD tests.
Bacteria were most abundant on the root surfaces, or rhizoplane, and on the surfaces of Nafion particles (Fig. 1). Colonisation on the root surface was concentrated in the intercellular junctions of the root epidermal cells (visual observation in 3 samples, e.g., Fig. 1C), which was similar to observations of field-grown wheat roots.11 Watt et al. quantified the fraction of the volume of soil occupied by Pseudomonas spp. found in wheat rhizospheres. Results showed that on average 15%11 of the soil volume was occupied by Pseudomonas spp. We did not characterize the colonisation of lettuce root by Pseudomonas spp. in soil, however, the overall mean rhizosphere volume occupied by P. fluorescens in the present study is of the same order of magnitude (10%) as those measured by Watt et al. Further studies comparing rhizosphere colonisation with the same plant and bacterial species in both soil and transparent soil would allow a more accurate comparison of the 2 substrates for this application. Bacterial fluorescence was detected in the pore spaces of the substrate, although at a lower level than on the surfaces (Fig. 1 and 2A). Image analysis also revealed that the abundance of bacteria in positions with no roots (Fig. 2Bi, positions A1–3 and B1–3), was constant and independent of image position, particle size and whether a plant was present or not in the chamber. This may indicate that the effect of the plants on soil microbial abundance could be limited to the substrate directly adjacent (i.e. <1.5 mm) to the root. Along the X axis (horizontal), in samples with plants, the number of discrete bacterial aggregates and the average size of the aggregates was greater on the root (position R) than at 1.5 mm (position A) and 3 mm (position B) from the root, and there was no significant difference in bacterial abundance or aggregate number between positions A and B (Fig. 2Bii). In samples with no plants, there was no difference in bacterial abundance along the X axis (horizontal positions). Along the Y axis (vertical), the number of bacterial aggregates was lower at the root tip (position 1, Fig. 2Bii) than the 2 positions further from the tip (position 2 and 3, Fig. 2Bii) but when the percentage area of the image with bacterial fluorescence was used to quantify abundance, there was no difference along the roots (data not shown). In samples with no plants present, the average size of bacterial aggregate was lowest at position 1 and highest at position 3, therefore the points closest to the surface of the substrate had the largest bacterial aggregates (Fig. 2Biii). This could be due to a higher concentration of dissolved oxygen closer to the surface, which has been observed in sludge with better bacterial flocculation at high dissolved oxygen concentrations.12
Several studies have described the distribution of PGPR on the surface of plant roots with a range of, and sometimes contrasting results. High bacterial abundance was found on the root tips11,13-15 and at root branching zones.13 Yet other studies reported an absence or scarcity of bacterial colonisation at the root tips16-20 perhaps caused by the high turnover of mucilage and border cells at the root apex.20 It is likely that the choice of the technique used to determine bacterial numbers along the root has a strong influence on bacterial count estimates. Methods based on colony forming units (CFU) are inaccurate because they rely on taking samples and this is difficult on the root tip, and only bacteria that grow well in lab cultures can be quantified. Microscopy techniques such as SEM are usually limited to detect bacteria embedded within the mucilage,16 and methods that requires fixing of samples, e.g., FISH, are susceptible to perturbation for example when washing the roots prior to imaging.11 The method described in the current study involved the addition and removal of liquids to and from the substrate. Although fluxes of water are common in soil due to rainfall or irrigation, the filling of soil samples by the matching liquid has the potential to induce anaerobic stress in the plant and bacteria over long periods. This effect was minimised by using fresh aerated solutions and by limiting the length of time during which the substrate was saturated. There are numerous non-destructive methods to image in soil, e.g., X-rays, Neutron and Magnetic Resonance Imaging.21-23 These do not rely on filling samples in liquid, but the methods are not able to resolve many micro-organisms, and imaging of biological processes such as gene expression or cell division is not possible. Molecular methods are developing rapidly, but currently these are either destructive,9 or unable to resolve spatial or temporal processes e.g., T-RFLP.24
The rhizosphere hosts large and diverse bacterial communities that establish sophisticated modes of interactions with plant roots. To date, it has been difficult to characterize such interactions because observation of roots and bacteria in depth and over time has been limiting.25 The model system described here overcomes many previous technological limitations. It combines the ability to grow biological organisms in a physically complex soil-like environment with optical microscopy25 and to detect multiple fluorescent signals in situ. The application of transparent soil microcosms is not limited to the study of roots and soil bacteria and it holds potential for studying the function of other soil organisms. Future developments could see the introduction of a diversity of microorganisms such as mycorrhizal fungi, nematodes, small invertebrates, or the incorporation of bacterial communities composed of several functional types (e.g., predators and prey). Exploiting this potential now requires exploring, testing and analyzing biological activity in transparent soil microcosms to better understand the benefits and limitations of the technology.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
HFD received funding from a joint studentship from Abertay University and The James Hutton Institute. The James Hutton Institute receives support from the Scottish Government Rural and Environment Science and Analytical Services Division (RESAS, Workpackage 3.3 and 3.4).
References
- 1. Urashima Y, Hori K. Selection of PGPR which promotes the growth of spinach. Japanese J Soil Sci Plant Nutr 2003; 74:157-62. [Google Scholar]
- 2. Abbas-Zadeh P, Saleh-Rastin N, Asadi-Rahmani H, Khavazi K, Soltani A, Shoary-Nejati AR, Miransari M. Plant growth-promoting activities of fluorescent pseudomonads, isolated from the Iranian soils. Acta Physiol Plant 2010; 32:281-8; http://dx.doi.org/ 10.1007/s11738-009-0405-1 [DOI] [Google Scholar]
- 3. Dey R, Pal KK, Bhatt DM, Chauhan SM. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol Res 2004; 159:371-94; PMID:15646384; http://dx.doi.org/ 10.1016/j.micres.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 4. Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 2010; 60:579-98; http://dx.doi.org/ 10.1007/s13213-010-0117-1 [DOI] [Google Scholar]
- 5. Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol 2013; 53:303-17; PMID:22733623; http://dx.doi.org/ 10.1002/jobm.201100552 [DOI] [PubMed] [Google Scholar]
- 6. Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol Rev 2013; 37:936-54; PMID:23550883 [DOI] [PubMed] [Google Scholar]
- 7. Fischer D, Pagenkemper S, Nellesen J, Peth S, Horn R, Schloter M. Influence of non-invasive X-ray computed tomography (XRCT) on the microbial community structure and function in soil. J Microbiol Meth 2013; 93:121-3; PMID:23499670; http://dx.doi.org/ 10.1016/j.mimet.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 8. Ranjard L, Poly F, Nazaret S. Monitoring complex bacterial communities using culture-independent molecular techniques: application to soil environment. Res Microbiol 2000; 151:167-77; PMID:10865943; http://dx.doi.org/ 10.1016/S0923-2508(00)00136-4 [DOI] [PubMed] [Google Scholar]
- 9. Eickhorst T, Tippkotter R. Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH). Soil Biol Biochem 2008; 40:1883-91; http://dx.doi.org/ 10.1016/j.soilbio.2008.03.024 [DOI] [Google Scholar]
- 10. Downie H, Holden N, Otten W, Spiers AJ, Valentine TA, Dupuy LX. Transparent soil for imaging the rhizosphere. Plos One 2012; 7:e44276; PMID:22984484; http://dx.doi.org/ 10.1371/journal.pone.0044276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watt M, Hugenholtz P, White R, Vinall K. Numbers and locations of native bacteria on field-grown wheat roots quantified by fluorescence in situ hybridization (FISH). Environ Microbiol 2006; 8:871-84; PMID:16623744; http://dx.doi.org/ 10.1111/j.1462-2920.2005.00973.x [DOI] [PubMed] [Google Scholar]
- 12. Liao BQ, Lin HJ, Langevin SP, Gao WJ, Leppard GG. Effects of temperature and dissolved oxygen on sludge properties and their role in bioflocculation and settling. Water Res 2011; 45:509-20; PMID:20875910; http://dx.doi.org/ 10.1016/j.watres.2010.09.010]- [DOI] [PubMed] [Google Scholar]
- 13. Jaeger CH, Lindow SE, Miller W, Clark E, Firestone MK. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol 1999; 65:2685-90; PMID:10347061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darwent MJ, Paterson E, McDonald AJS, Tomos AD. Biosensor reporting of root exudation from Hordeum vulgare in relation to shoot nitrate concentration. J Exp Bot 2003; 54:325-34; PMID:12493860; http://dx.doi.org/ 10.1093/jxb/erg017 [DOI] [PubMed] [Google Scholar]
- 15. Paterson E, Sim A, Standing D, Dorward M, McDonald AJS. Root exudation from Hordeum vulgare in response to localized nitrate supply. J Exp Bot 2006; 57:2413-20; PMID:16766600; http://dx.doi.org/ 10.1093/jxb/erj214 [DOI] [PubMed] [Google Scholar]
- 16. Lugtenberg BJJ, Dekkers L, Bloemberg GV. Molecular determinants of rhizosphere colonization by Pseudomonas. Ann Rev Phytopathol 2001; 39:461-90; http://dx.doi.org/ 10.1146/annurev.phyto.39.1.461 [DOI] [PubMed] [Google Scholar]
- 17. Kragelund L, Nybroe O. Competition between Pseudomonas fluorescens Ag1 and Alcaligenes eutrophus JMP134 (pJP4) during colonization of barley roots. FEMS Microbiol Ecol 1996; 20:41-51; http://dx.doi.org/ 10.1111/j.1574-6941.1996.tb00303.x [DOI] [Google Scholar]
- 18. Gamalero E, Lingua G, Tombolini R, Avidano L, Pivato B, Berta G. Colonization of tomato root seedling by Pseudomonas fluorescens 92rkG5: Spatio-temporal dynamics, localization, organization, viability, and culturability. Microb Ecol 2005; 50:289-97; PMID:16211326; http://dx.doi.org/ 10.1007/s00248-004-0149-9 [DOI] [PubMed] [Google Scholar]
- 19. Simons M, vanderBij AJ, Brand I, deWeger LA, Wijffelman CA, Lugtenberg BJJ. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol Plant-Microbe Interact 1996; 9:600-7; PMID:8810075; http://dx.doi.org/ 10.1094/MPMI-9-0600 [DOI] [PubMed] [Google Scholar]
- 20. Humphris SN, Bengough AG, Griffiths BS, Kilham K, Rodger S, Stubbs V, Valentine TA, Young IM. Root cap influences root colonisation by Pseudomonas fluorescens SBW25 on maize. FEMS Microbiol Ecol 2005; 54:123-30; PMID:16329978; http://dx.doi.org/ 10.1016/j.femsec.2005.03.005 [DOI] [PubMed] [Google Scholar]
- 21. Davey E, Wigand C, Johnson R, Sundberg K, Morris J, Roman CT. Use of computed tomography imaging for quantifying coarse roots, rhizomes, peat, and particle densities in marsh soils. Ecol Appl 2011; 21:2156-71; PMID:21939051; http://dx.doi.org/ 10.1890/10-2037.1 [DOI] [PubMed] [Google Scholar]
- 22. Pohlmeier A, Oros-Peusquens A, Javaux M, Menzel MI, Vanderborght J, Kaffanke J, Romanzettib S, Lindenmairc J, Vereeckena H, Shahb NJ. Changes in soil water content resulting from Ricinus root uptake monitored by Magnetic Resonance Imaging. Vadose Zone J 2008; 7:1010-7; http://dx.doi.org/ 10.2136/vzj2007.0110 [DOI] [Google Scholar]
- 23. Olson MS, Ford RM, Smith JA, Fernandez EJ. Quantification of bacterial chemotaxis in porous media using Magnetic Resonance Imaging. Environ Sci Technol 2004; 38:3864-70; http://dx.doi.org/ 10.1021/es035236s [DOI] [PubMed] [Google Scholar]
- 24. Gao DW, Tao Y. Current molecular biologic techniques for characterizing environmental microbial community. Front Env Sci Eng 2012; 6:82-97; http://dx.doi.org/ 10.1007/s11783-011-0306-6 [DOI] [Google Scholar]
- 25. Yang Z, Downie H, Rozbicki E, Dupuy LX, MacDonald MP. Light Sheet Tomography (LST) for in situ imaging of plant roots. Opt Express 2013; 21:16239-47; PMID:23938474; http://dx.doi.org/ 10.1364/OE.21.016239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


