Abstract
Mercury and arsenic are known developmental toxicants. Prenatal exposures are associated with adverse childhood health outcomes that could be in part mediated by epigenetic alterations that may also contribute to altered immune profiles. In this study, we examined the association between prenatal mercury exposure on both DNA methylation and white blood cell composition of cord blood, and evaluated the interaction with prenatal arsenic exposure. A total of 138 mother-infant pairs with postpartum maternal toenail mercury, prenatal urinary arsenic concentrations, and newborn cord blood were assessed using the Illumina Infinium Methylation450 array. White blood cell composition was inferred from DNA methylation measurements. A doubling in toenail mercury concentration was associated with a 2.5% decrease (95% CI: 5.0%, 1.0%) in the estimated monocyte proportion. An increase of 3.5% (95% CI: 1.0, 7.0) in B-cell proportion was observed for females only. Among the top 100 CpGs associated with toenail mercury levels (ranked on P-value), there was a significant enrichment of loci located in North shore regions of CpG islands (P = 0.049), and the majority of these loci were hypermethylated (85%). Among the top 100 CpGs for the interaction between arsenic and mercury, there was a greater than expected proportion of loci located in CpG islands (P = 0.045) and in South shore regions (P = 0.009) and all of these loci were hypermethylated. This work supports the hypothesis that mercury may be contributing to epigenetic variability and immune cell proportion changes, and suggests that in utero exposure to mercury and arsenic, even at low levels, may interact to impact the epigenome.
Keywords: arsenic, DNA methylation, epigenetics, illumina 450K, in-utero exposure, mercury, neurodevelopment
Introduction
Mercury (Hg) enters the environment from natural and anthropogenic sources and its concentration has increased at least three-fold in the ocean since the industrial revolution.1 The largest anthropogenic source of mercury emissions in the US comes from coal-burning power plants; others sources include byproducts of chlorine production, dental fillings using amalgams, thermometers, broken fluorescent bulbs, and batteries.2 Mercury that is deposited in aquatic systems is biotransformed to methylmercury (MeHg) by anaerobic microbes. MeHg can then bioaccumulate in species of high trophic level. For the general population, consumption of fish is the primary route of human exposure to MeHg. Since MeHg can cross the placenta, maternal consumption of fish, especially large predatory fish such as shark, tile fish and king mackerel, leads to fetal exposure in utero.3,4
Epidemiological studies have found that prenatal exposure to mercury in utero is associated with poor cognitive development and behavioral disorders in children.5-8 A recent review of the literature on low level prenatal MeHg exposure found consistent evidence for early childhood neurocognitive dysfunction, limited evidence for cardiovascular effects and possible associations with fetal growth among susceptible groups.9 Furthermore, mercury exposure at low levels was found to be associated with subclinical signs of autoimmunity among women of reproductive age in the US population.10 The latency of health effects observed in prospective epidemiological studies of prenatal Hg exposures is suggestive of an epigenetic mode of action and emerging evidence suggests that placental epigenetic disruption can contribute to Hg neurodevelopmental toxicity.11 However, no studies have investigated the effects of prenatal Hg exposure on DNA methylation in cord blood.12
Additionally, there is evidence that MeHg exposure is correlated with other metals, such as arsenic (As), that are also known epigenetic toxicants. For instance, epidemiological studies in pregnant women report a moderate positive correlation between biomarkers of Hg and As exposure.13,14 Few studies, however, have examined the health effects of co-exposures to toxicants. A growing body of evidence indicates that prenatal exposure to arsenic may lead to neurotoxic effects and is most severe during brain development and fetal growth.15,16 Emerging studies have provided evidence that prenatal exposure to As may disrupt the epigenome of newborns, a potential mechanism that might explain its latent health effects.17-20
The ability of some environmental exposures to disrupt epigenetic programming during fetal development provides a mechanism to link in utero exposure to toxicants and chronic disease.21 The epigenome is thought to be most susceptible to environmental toxicants during embryogenesis, a developmental phase characterized by a rapid increase in cell division and epigenetic remodeling of the genomic landscape.22 In vitro studies in human neurons have shown that methylmercury is cytotoxic.23 However, the exact mechanism for human toxicity remains ill defined.24 Prenatal MeHg exposure has been hypothesized to disrupt calcium homeostasis, alter glutamate homeostasis, and generate oxidative stress along with increased reactive oxygen species.24 Metal induced DNA methylation changes have been hypothesized to occur through oxidative stress and production of reactive oxygen species as the unifying process for DNA methylation disruption.25 Oxidative DNA damage has been shown to interfere with DNA methyltransferase activity resulting in altered methylation patterns of cytosine residues at CpG sites.26 Both As and Hg can alter DNA methyltransferase activity suggesting this as a potential mechanism for epigenetic disruption.27,28 Moreover, inorganic arsenic undergoes methylation to facilitate excretion, using S-adenosylmethionine (SAMe) as a methyl donor, the same methyl donor used in all cellular methylation reactions. 29 Mercury has been shown to have strong inhibitory action in the methionine synthase enzyme crucial for SAMe regeneration.30 Thus, the oxidative stress action of both As and Hg, along with the possibility of mercury exposure depleting levels of SAMe, raises the question as to whether co-exposure to these two common environmental contaminants can disrupt the epigenome synergistically.
Studies that investigate the association between mercury exposure and DNA methylation are sparse and are limited to using a candidate gene approach.31,32 There is also limited data on the potential impact of prenatal Hg exposure on the distribution of leukocytes. Moreover, as even less is known about the effects of co-exposure to Hg and As on DNA methylation, we investigated both the main effect of prenatal exposure to Hg and its interaction with As exposure in utero and evaluated their ability to disrupt DNA methylation in cord blood. Furthermore, we estimated white blood cell composition using reference methylomes of isolated leukocyte subtypes to investigate the association with Hg and co-exposure to As.
Results
A total of 138 mother-infant pairs were included in the analysis. The majority of mothers were white (98%), had some level of college education (89%), reported not smoking during pregnancy (83%) and were on average 31 y old at delivery. Maternal toenail Hg ranged from 0.001 to 1.44 μg/g with a median level of 0.07 μg/g. Total maternal urinary arsenic ranged from 0.34 to 17.9 μg/L with a median urinary arsenic concentration of 3.19 μg/L. Demographic characteristic are summarized in Table 1.
Table 1.
Selected maternal and infant characteristics and Hg and As concentrations
| Characteristics | |
|---|---|
| Mother-Infant Pairs (n) | 138 |
| Maternal age at delivery (years) | 31 ± 4.6 |
| Gestational age (weeks) | 39.3 ± 1.39 |
| Pre-pregnancy BMI (kg/m2) | 24.3 ± 4.2 |
| Infant Birth weight (grams) | 3415 ± 562 |
| Infant Sex | |
| Male | 68 (49) |
| Female | 70 (51) |
| Maternal Smoking During Pregnancy | |
| No | 115 (83) |
| Yes | 7 (5) |
| Unknown | 16 (12) |
| Maternal Race | |
| White | 135 (98) |
| Other | 3 (2) |
| Education | |
| Less than 11th grade | 2 (2) |
| High school graduate or equivalent | 13 (9) |
| Junior college graduate or some college | 26 (19) |
| College graduate | 52 (38) |
| Post-graduate education | 28 (20) |
| Unknown | 17 (12) |
| Exposure | Median (min-max) |
| Maternal Toenail Hg (μg/g) | 0.07 (0.001–1.44) |
| aMaternal Urinary Arsenic (μg/L) | 3.19 (0.34–17.9) |
Maternal Urinary Arsenic=AsIII+AsV+MMA+DMA. Data are expressed as Mean ± SD or n (%), unless otherwise indicated.
An overall decrease in the proportion of imputed monocytes was observed in relationship to increasing levels of Hg exposure (β = −2.5%; 95% CI: −5.0, −1.0). After stratifying by gender, the association remained significant for females (β = −2.6%; 95% CI: −5.0, −1.0) but not males (β = −1.9%; 95% CI: −8.0, 4.0). An increase in the proportion of B-cells was also observed with increasing levels of Hg exposure in females only (β = 3.5%; 95% CI: 1.0, 7.0). Results for the relationship between log2-transformed toenail Hg and imputed white blood cells distribution in cord blood are summarized in Table 2. The interaction between Hg and As was not statistically significant for any of the estimated cell types (Table S2).
Table 2.
Estimateda change in the proportion of leukocyte composition (95% CI) in cord blood in relationship to log2 toenail Hg concentrations
| Overall Association |
Stratified by Sex |
|||||
|---|---|---|---|---|---|---|
| Males |
Females |
|||||
| Cell type | % Change (95% CI) | P-value | % Change (95% CI) | P-value | % Change (95% CI) | P-value |
| CD8T | 0.5 (−3.0, 4.0) | 0.742 | −3.5 (−11.0, 5.0) | 0.414 | 1.4 (−2.0, 5.0) | 0.395 |
| CD4T | 1.1 (−3.0, 6.0) | 0.643 | −6.6 (−18.0, 5.0) | 0.274 | 2.3 (−3.0, 7.0) | 0.367 |
| NK cells | 0.6 (−3.0, 5.0) | 0.762 | 2.7 (−8.0, 1.3) | 0.621 | 0.1 (−4.0, 5.0) | 0.991 |
| B-cells | 2.3 (−1.0, 5.0) | 0.116 | −4.9 (−13.0, 3.0) | 0.211 | 3.5 (1.0, 6.7) | 0.027 |
| Monocytes | −2.5 (−5.0, −1.0) | 0.019 | −1.9 (−8.0, 4.0) | 0.508 | −2.6 (−5.0, −1.0) | 0.026 |
| Granulocytes | −2.2 (−9.0, 5.0) | 0.543 | 1.3 (−6.0, 3.3) | 0.173 | −5.0 (−1.3, 3.0) | 0.196 |
Estimates from a multivariate linear regression model adjusted for infant sex and maternal age at delivery
In the locus-specific analysis, 11,327 (3.2%) of the 348,569 CpG loci were observed to be differentially methylated in relation to toenail Hg with a nominal P-value < 0.05 and 9 CpG loci were differentially methylated with a nominal P-value < 0.0001, after controlling for the imputed leukocyte distribution, infant sex and maternal age at enrollment. Results are shown in Figure 1A, which depicts the −log10(P-value) on the y-axis along with regression coefficients for log2-transformed Hg exposure on the x-axis for each individual CpG loci. However, no observed relationships remained significant after controlling for multiple comparisons using a Bonferroni threshold. Among the top 100 differentially methylated CpGs ranked on lowest P-value for Hg exposure, there was a greater than expected proportion of loci located in North shore regions of CpG islands (P = 0.049; Fig. 1B). Of these loci located in North shore regions of CpG islands (n = 20), the majority was observed to have an increase in methylation (85%).
Figure 1.
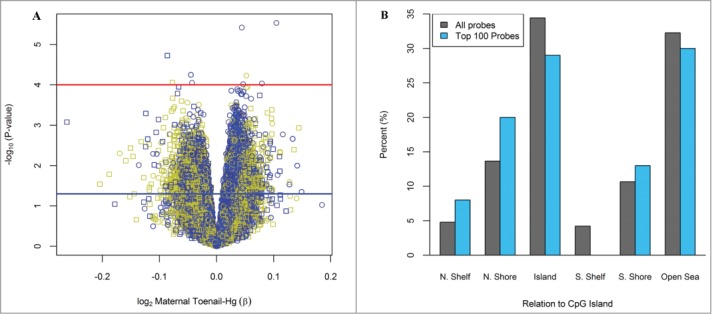
Differentially methylated loci based on maternal toenail Hg. (A) Volcano plot for the relationship between log2 toenail Hg on DNA methylation at all 348,569 CpGs analyzed. Red and blue lines indicate −log10(1 × 10−4) and −log10(0.05) P-values, respectively. Colors: Yellow=CpG island, Black=CpG Shore, Blue=Shelfs and Open sea. Symbols: Circle = Infinium Type II, Square = Infinium Type I (B) Location of the top 100-CpGs on the basis of P-values compared to all CpGs on the methylation array. Among the top 100 CpGs with lowest P-value, there was a significant enrichment in the N. Shore regions of CpG islands (P = 0.049).
For the multiplicative interaction between Hg and As in the locus-specific analysis, 15,620 (4.5%) of the 348,569 CpG loci were differentially methylated with a nominal P-value < 0.05 and 37 CpG loci were differentially methylated with a nominal P-value < 0.0001, after controlling for the imputed leukocyte distribution, infant sex, maternal age at delivery, urinary creatinine and the main effects of Hg and As. The multiplicative interaction results for all loci are shown in Figure 2A. The observed associations did not reach statistical significance using a Bonferroni correction. Among the top 100 differentially methylated CpGs ranked on lowest P-value for the multiplicative interaction between Hg and As, there was a greater than expected proportion of loci located in CpG islands (P = 0.045) and in South shore regions of CpG islands (P = 0.009), Figure 2B. All CpG loci located within CpG islands (n = 43) showed increased methylation (100%). Similarly, among the top 100 loci located in South shore regions of CpG islands (n = 19), all were observed to be hypermethylated (100%). The top 100 differentially methylated CpG loci based on toenail Hg exposure and interaction with urinary As are provided in Table S4 and Table S5, respectively.
Figure 2.
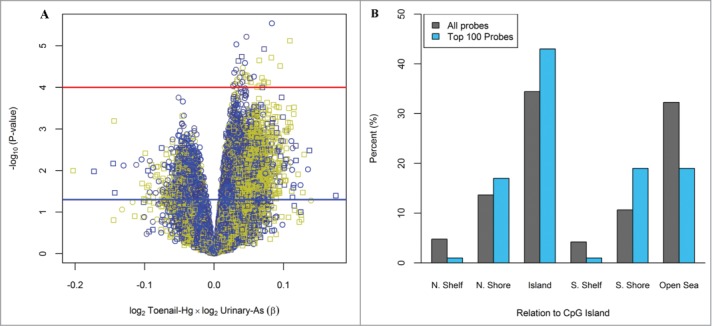
Differentially methylated loci based on the interaction between maternal toenail Hg and urinary As. (A) Volcano plot for the relationship between the multiplicative interaction of log2 toenail Hg and log2 urinary As on DNA methylation at all 348,569 CpGs analyzed. Red and blue lines indicate −log10(1 × 10−4) and −log10(0.05) P-values, respectively. Colors: Yellow = CpG island, Black = CpG Shore, Blue=Shelfs and Open sea. Symbols: Circle = Infinium Type II, Square = Infinium Type I (B) Location of the top 100-CpGs on the basis of P-values compared to all CpGs on the methylation array. Among the top 100 CpGs with lowest P-value, there was a significant enrichment of CpG islands (P = 0.045) and S. Shore regions (P = 0.009).
Discussion
Our study provides evidence that Hg exposure in utero shifted the underlying leukocyte composition in cord blood leading to a decrease in the proportion of monocytes and an increase in the proportion B-cells in female infants. Prenatal Hg exposure, as well as, co-exposure to Hg and As may also have the potential to influence the epigenome of cord blood. The effect of Hg, as well as co-exposure to Hg and As, on the epigenome of cord blood was not statistically significant after accounting for the potential for false positives using a conservative Bonferroni correction. It is important to note, however, that this population was exposed to low Hg levels. For instance, the median concentration of toenail Hg in our study was 0.07 μg/g whereas toenail Hg levels measured in 27 healthy adults within the same study area of New Hampshire had median toenail Hg concentrations of 0.16 μg/g, ranging from 0.04 to 1.15 μg/g.33 In another exposure study of 54 healthy Japanese pregnant women, the geometric mean of toenail Hg was 0.46 μg/g and ranged from 0.36 to 0.62 μg/g.34 This could be due in part by dietary counseling pregnant women in the US receive which includes fish consumption advisories by health care providers and federal agencies to reduce prenatal MeHg exposure.35 Subsequently, the low levels of exposure found in this study during pregnancy are not surprising given the high level of literacy and education reported among the study population. Future studies may benefit from examining the epigenetic effects of prenatal Hg exposure in other populations with elevated exposure levels.
Several animal models have shown that mercury exposure is an immunotoxicant.36 Particularly, studies have observed both monocyte and lymphocyte apoptosis in vitro.37 This is consistent with our results where prenatal toenail-Hg exposure was associated with a decrease in the proportion of monocytes. Interestingly, the observed association was stronger for females where an increase in the estimated proportion of B-cells was also observed. The observed increase in B-cells is consistent with in vivo studies of mice exposed to Hg were both B-cell activation and autoantibody production have been documented.38 The sex differences observed in response to Hg exposure warrant further investigation, and indicate that Hg should be considered in studies examining outcomes including autoimmune disorders which are consistent with B-cell activation and have sexual dichotomy.39 The potential health implications for the observed decrease in monocytes remain to be determined.
While we know of no prior study that investigated the effect of prenatal Hg exposure on the epigenome, one study did examine the relationship between urinary Hg and DNA methylation changes in blood using a cancer-focused array (GoldenGate Cancer Panel I) among 58 women undergoing ovarian stimulation for in vitro fertilization. These researchers observed a significant increase in DNA methylation of promoter regions of the GSTM1/5 genes among women with high urinary Hg levels.40 A second study among dental health professionals using a candidate gene approach to examine DNA methylation from buccal cells found that increasing levels of Hg exposure was associated with hypomethylation of CpG islands in the promoter region of the SEPP1 gene.32 There is also evidence from animal models that epigenetic regulation of the Brain-Derived Neurotrophic Factor (BDNF) gene mediates gene suppression linked to behavioral changes in mice exposed to MeHg.41
To evaluate these previous findings in our cohort, we performed an individual CpG lookup based of the SEPP1, GSTM5and BDNF genes using a nominal P-value < 0.05 for significance. The GSTM1 gene is polymorphic in the human population and therefore excluded a priori from our epigenome-wide analysis.42 Two loci (cg09606766; cg01636003) located in South shore regions of CpG islands of the BDNF gene were hypomethylated (P = 0.004) while one loci (cg18595174) located in an open sea region was hypermethylated (P = 0.015) with increasing toenail mercury concentrations. For the SEPP1 gene, one loci (cg04502814) located in an open sea region increase in methylation with increasing toenail mercury concentrations (P = 0.004). No other CpGs in the SEPP1 or GSTM5 genes were found to be differentially methylated. Results from the reverse CpG look up are summarized in Table S3.
The observed hypermethylation of CpG islands among the top ranked CpG loci for the interaction between As and Hg is of particular interest given that methylation of CpG islands in promoter regions has been established as a mechanism of gene silencing.43 Furthermore, the observed hypermethylation of North shore regions with increasing Hg exposure levels and South shore regions in relation to Hg and As co-exposure could be biologically relevant as shore regions of CpG islands have been shown to be highly variable, play an important role in tissue differentiation and potentially drive pathogenesis.44,45
While our study did not measure functional gene expression, some CpGs were located near genes that have been previously implicated in epigenetic mechanisms for disease. For example, among the top 100 differentially methylated CpGs in relationship to prenatal Hg levels, 2 loci (cg27458888; cg05881762) located in South shore regions of CpG islands of the Ubiquitin protein ligase E3A gene (UBE3A) were observed to be hypermethylated. This gene is subject to methylation-dependent genomic imprinting and has been previously associated with Angelman syndrome, a neurodevelopmental disorder characterized by severe intellectual disability.46 Among the top 100 loci that were differentially methylated for the interaction between Hg and As, 2 loci (cg12419685; cg17250863) located in CpG islands of the Gamma-Glutamyltransferase 7 gene (GGT7) were hypermethylated. The GGT7gene is involved in the metabolism of glutathione (GSH) and adequate levels of GSH have been shown to protect against MeHg neurotoxicity in vivo.47,48 In neurons, glutathione has been shown to be a physiological reservoir of glutamate, an important neurotransmitter involve in cognitive processes and previously implicated in MeHg-induced neurotoxicity.24,49
There are a number of limitations to our current study. First, the study population was mostly white with a high level of education, which could limit the generalizability of our findings. The exposure levels to Hg found are relatively low but might reflect common levels of exposure among pregnant women in the US. Although the exposure assessment relies on toenail Hg concentrations, this is an objective biomarker of exposure previously validated and reflective of general patterns of dietary MeHg intake.33 Moreover, Hg concentrations in toenails at birth have been shown to capture MeHg levels approximately 5 months retroactively, reflecting MeHg exposure throughout the third trimester.34 The leukocyte composition was projected from DNA methylation measurements taken from adult males and this might not accurately depict the white blood cell composition of newborns. Although DNA Methylation changes in blood might serve as a biomarker of exposure and potentially disease, the biological relevance of the observed changes in cord blood is unknown and might not necessarily result in functional changes in gene expression. As demonstrated in previous studies, only a fraction of DNA methylation changes may result in differential gene expression.17,50 However, some consistent epigenetic variation between blood and brain tissue of humans have outlined the utility of whole blood in epidemiological studies.51 While many differentially methylated loci had a relatively small P-value none met the Bonferroni correction criteria of statistical significance. Therefore, these results need to be replicated and confirmed in separate studies.
In conclusion, this study provides evidence that in utero exposure to mercury can affect leukocyte composition and may disrupt the epigenome even at low levels. Furthermore, exposure to both arsenic and mercury in utero may interact jointly to affect the epigenome by hypermethylating relevant CpG regions that have the potential to influence neurodevelopment and other childhood health outcomes. This would suggest that epigenomic alterations should also be considered in order to understand the toxic mechanisms of these exposures and their impact on children's health.
Materials & Methods
Study population
The study population consisted of the 138 initial participants enrolled in the ongoing New Hampshire Birth Cohort Study (NHBCS), which is focused on pregnant women from New Hampshire whose primary household drinking water source was a private well.52 Eligibility criteria included English speaking, English literate, and mentally competent pregnant women 18–45 y of age. Subjects who changed their residence since their last menstrual period or whose home water supply was from a source other than from a private well were excluded from the study. Information about the mother and newborn were ascertained through questionnaires and review of the prenatal and delivery medical records. This study was approved by the Committee for the Protection of Human Subjects at Dartmouth College. All study participants provided written informed consent prior to the study.
Toenail Hg assessment
At two weeks postpartum, an information packet was mailed to study participants requesting maternal toenail clipping samples within 8 weeks of birth; toenails were stored at room temperature until analysis. After careful cleaning and washing to remove external contaminants, trace elements were quantified at the Trace Element Analysis Core (Dartmouth College, Hanover, New Hampshire, USA), using inductively coupled plasma mass spectrometry (ICP-MS). Toenails were acid digested with Optima nitric acid (Fisher Scientific, St Louis, Missouri, USA) at 105°C followed by the addition of hydrogen peroxide and further heating the dilution with deionized water. All sample preparation steps were recorded gravimetrically. As a quality control, each batch of analyses included 6 standard reference material (SRM) samples with known trace element content (GBW 07601, powdered human hair) and 6 analytical blanks, along with the study samples. The majority of participants (89%) had toenail Hg levels above the limit of detection (LOD) of 0.10 ng/g. Samples that were below the detection limit were replaced with the LOD divided by the square root of 2.
Urinary arsenic assessment
Spot urine samples were collected at approximately 24–28 weeks gestation and analyzed for individual species of urinary arsenic using a high performance liquid chromatography (HPLC) inductively coupled plasma mass spectrometry (ICP-MS) system as previously described.18,52 This method determined 5 urinary arsenic species: arsenite (AsIII), arsenate (AsV), dimethylarsinic acid (DMAV) and monomethylarsonic acid (MMAV). The detection limits ranged from 0.04 to 0.21 μg/L for the individual arsenic species. Values for samples with measurements below the LOD were imputed to be the metabolites' LOD divided by the square root of 2. The proportion of samples below the LOD was 93% for AsV, 58% for AsIII, 18% for MMAV and 0% and DMAV. Subsequently, total urinary arsenic concentration (Urinary-As) was calculated by summing inorganic arsenic (AsIII and AsV) and the metabolic products MMAV and DMAV. We used total urinary-As as a measure of in utero exposure to arsenic because urinary arsenic levels have been shown to be a useful indicator of internal dose.53 Urinary creatinine levels were measured using Cayman's creatinine assay kit and protocol to control for urine dilution.
DNA methylation assessment and preprocessing
DNA was isolated from cord blood samples using DNeasy® blood and tissue kits (Qiagen, Valencia, CA) and bisulfite converted using the EZ DNA Methylation kit (Zymo, Irvine, CA). Samples were randomized across plates and subsequently subjected to epigenome-wide DNA methylation analysis using the Illumina Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA), which simultaneously profiles the methylation status for > 485,000 CpG sites at single nucleotide resolution. Microarrays were processed at the Biomedical Genomics Center at the University of Minnesota, following standard protocols. Data were extracted and processed from the raw methylation image files using functional normalization, a normalization method to correct for variations between arrays found in the minfi package of R.54 This normalization method removes unwanted technical variation using the internal control probes as surrogates of batch effects.
The methylation status for each individual CpG locus was calculated as the ratio of fluorescent signals, referred to as a β-value. This is an interval scale quantity between zero and one interpreted as the fraction of DNA molecules whose target CpG is methylated. We further removed any residual plate/BeadChip effects using ComBat.55 All CpG loci on X and Y chromosomes were excluded from the analysis, to avoid gender-specific methylation bias. Furthermore, we excluded non-specific probes, cross-reactive probes and polymorphic CpGs (at ≥ 5% of the minor allele frequency) previously identified in the 450K array to avoid spurious associations.56 Finally, all CpG loci with high background signal that had non-significant detection P-values (P > 0.01) indicating poor detection were excluded from the analysis, leaving a total of 348,569 autosomal CpG loci measured in 138 cord blood samples.
Statistical analysis
DNA methylation β-values were logit transformed to M-values as previously recommended for statistical analysis.57 To deconvolute the most prevalent sources of variability in DNA methylation across the array, we performed a Principal Component Analysis (PCA) on both the (a) normalized only and (b) normalized and ComBat adjusted DNA methylation data. The top 3 principle components (PCs) estimated for each data set were then examined in terms of their association with sample plate using a series of linear regression models. The top PCs computed from the normalized only DNA methylation data were significantly associated with plate; however, these associations were attenuated in ComBat adjusted DNA methylation data (Figure S1 and Table S1) and suggest that ComBat was effective in reducing plate-associated variation in DNA methylation. Using the normalized and ComBat adjusted plate methylation data, we implemented a statistical deconvolution method to estimate the proportion of white blood cell composition in cord-blood.58 This method estimates the relative proportion of 6 major cell types: CD8 T-cells, CD4 T-cells, natural killer (NK) cells, B-cells, monocytes, and granulocytes. This was achieved by using discriminatory differentially methylated regions in flow-sorted purified leukocytes from 6 adult samples found in the minfi package of R.59 Subsequently, multivariate linear regression models were used to evaluate the association between the estimated leukocyte proportion and log2-transformed toenail Hg exposure.
We next implemented a locus-by-locus analysis aimed toward identifying differentially methylated CpG loci based on toenail Hg concentration using limma models.60 Briefly, linear models were used to estimate the dose-response relationship between log2-transformed values of toenail Hg concentration and M-values. A second model that included the main effect of maternal log2-transformed toenail Hg and log2-transformed urinary arsenic levels along with their multiplicative interaction was evaluated while also adjusting for urinary creatinine to account for urine dilution. All models were adjusted for maternal age at delivery, infant sex, and the imputed white blood cell distribution from the Houseman projection.58 Although our epigenome-wide approach was exploratory in nature, P-values were adjusted for multiple comparisons by comparing results to a Bonferroni corrected threshold for statistical significance. Lastly, using the top 100 differentially methylated CpG loci based on the lowest P-values, we compared their distribution based on CpG island location to the rest of the array. All analyses were carried out using the R statistical package, version 3.1.1 (www.r-project.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the US National Institute of Environmental Health Sciences (NIEHS) grants P42 ES016465 (KC Donnelly Fellowship), P01 ES022832, US EPA grant RD83544201, and by a CTSA grant from NCATS awarded to the University of Kansas Medical Center for Frontiers: The Heartland Institute for Clinical and Translational Research # KL2TR000119. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the presentation.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Lamborg CH, Hammerschmidt CR, Bowman KL, Swarr GJ, Munson KM, Ohnemus DC, Lam PJ, Heimbürger L-E, Rijkenberg MJ, Saito MA. A global ocean inventory of anthropogenic mercury based on water column measurements. Nature 2014; 512:65-8; PMID:25100482; http://dx.doi.org/ 10.1038/nature13563 [DOI] [PubMed] [Google Scholar]
- 2.Tewalt SJ, Bragg LJ, Finkelman RB. Mercury in US Coal: abundance, distribution, and modes of occurrence. USGS Online Publication Directory: USGS, 2001. http://pubs.usgs.gov/fs/fs095-01/fs095-01.pdf. [Google Scholar]
- 3.Vahter M, Åkesson A, Lind B, Björs U, Schütz A, Berglund M. Longitudinal study of methylmercury and inorganic mercury in blood and urine of pregnant and lactating women, as well as in umbilical cord blood. Environ Res 2000; 84:186-94; PMID:11068932; http://dx.doi.org/ 10.1006/enrs.2000.4098 [DOI] [PubMed] [Google Scholar]
- 4.Oken E, Choi AL, Karagas MR, Mariën K, Rheinberger CM, Schoeny R, Sunderland E, Korrick S. Which fish should I eat? Perspectives influencing fish consumption choices. Environ Health Perspect 2012; 120:790; PMID:22534056; http://dx.doi.org/ 10.1289/ehp.1104500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freire C, Ramos R, Lopez-Espinosa M-J, Díez S, Vioque J, Ballester F, Fernández M-F. Hair mercury levels, fish consumption, and cognitive development in preschool children from Granada, Spain. Environ Res 2010; 110:96-104; PMID:19909946; http://dx.doi.org/ 10.1016/j.envres.2009.10.005 [DOI] [PubMed] [Google Scholar]
- 6.Grandjean P, Weihe P, White RF, Debes F, Araki S, Yokoyama K, Murata K, Sørensen N, Dahl R, Jørgensen PJ. Cognitive deficit in 7-year-old children with prenatal exposure to methylmercury. Neurotoxicol Teratol 1997; 19:417-28; PMID:9392777; http://dx.doi.org/ 10.1016/S0892-0362(97)00097-4 [DOI] [PubMed] [Google Scholar]
- 7.Boucher O, Muckle G, Jacobson JL, Carter RC, Kaplan-Estrin M, Ayotte P, Dewailly É, Jacobson SW. Domain-specific effects of prenatal exposure to PCBs, mercury, and lead on infant cognition: results from the Environmental Contaminants and Child Development Study in Nunavik. Environ Health Perspect 2014; 122:310; PMID:24441767; http://dx.doi.org/ 10.1289/ehp.122-A310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagiv SK, Thurston SW, Bellinger DC, Amarasiriwardena C, Korrick SA. Prenatal exposure to mercury and fish consumption during pregnancy and attention-deficit/hyperactivity disorder–related behavior in children. Arch Pediat Adolesc Med 2012; 166:1123-31; PMID:23044994; http://dx.doi.org/ 10.1001/archpediatrics.2012.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagas M, Choi AL, Oken E, Horvat M, Schoeny R, Kamai E, Grandjean P, Korrick S. Evidence on the human health effects of low level methylmercury exposure. Environ Health Perspect 2012; 120:799-806; PMID:22275730; http://dx.doi.org/ 10.1289/ehp.1104494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somers EC, Ganser MA, Warren JS, Basu N, Wang L, Zick SM, Park SK. Mercury exposure and antinuclear antibodies among females of reproductive age in the United States: NHANES. Environ Health Perspect 2015; Advanced Publication; PMID: 25665152; http://dx.doi.org/ 10.1289/ehp.1408751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maccani JZ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, Marsit CJ. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ Health Perspect 2015; Advanced Publication; PMID:25748564: http://dx.doi.org/ 10.1289/ehp.1408561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu N, Goodrich JM, Head J. Ecogenetics of mercury: from genetic polymorphisms and epigenetics to risk assessment and decision‐making. Environ Toxicol Chem 2014; 33:1248-58; PMID:24038486; http://dx.doi.org/ 10.1002/etc.2375 [DOI] [PubMed] [Google Scholar]
- 13.Miklavčič A, Casetta A, Tratnik JS, Mazej D, Krsnik M, Mariuz M, Sofianou K, Špirić Z, Barbone F, Horvat M. Mercury, arsenic and selenium exposure levels in relation to fish consumption in the Mediterranean area. Environ Res 2013; 120:7-17; PMID:22999706; http://dx.doi.org/ 10.1016/j.envres.2012.08.010 [DOI] [PubMed] [Google Scholar]
- 14.Sanders AP, Flood K, Chiang S, Herring AH, Wolf L, Fry RC. Towards prenatal biomonitoring in North Carolina: assessing arsenic, cadmium, mercury, and lead levels in pregnant women. PloS One 2012; 7:e31354; PMID:22427803; http://dx.doi.org/ 10.1371/journal.pone.0031354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vahter M. Health effects of early life exposure to arsenic. Basic Clin Pharmacol Toxicol 2008; 102:204-11; PMID:18226075; http://dx.doi.org/ 10.1111/j.1742-7843.2007.00168.x [DOI] [PubMed] [Google Scholar]
- 16.Tolins M, Ruchirawat M, Landrigan P. The developmental neurotoxicity of arsenic: Cognitive and behavioral consequences of early life exposure. Ann Global Health 2014; 80:303-14; PMID:25459332; http://dx.doi.org/ 10.1016/j.aogh.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 17.Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, Paul-Brutus R, Gamble MV, Harper KN, Parvez F. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect 2015; 123:64-71; PMID:25325195; http://dx.doi.org/ 10.1289/ehp.123-A64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect 2013; 121:971-7; PMID:23757598; http://dx.doi.org/ 10.1289/ehp.1205925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kile ML, Houseman EA, Baccarelli A, Quamruzzaman Q, Rahman M, Mostofa G, Cardenas A, Wright RO, Christiani DC. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics 2014; 9:0-1; PMID:24525453; http://dx.doi.org/ 10.4161/epi.28153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, Mahiuddin G, Mostofa G, Hsueh Y-M, Wright RO. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect 2012; 120:1061-6; PMID:22466225; http://dx.doi.org/ 10.1289/ehp.1104173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol 2015; 218:71-9; PMID:25568453; http://dx.doi.org/ 10.1242/jeb.106971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol 2011; 31:363-73; PMID:21256208; http://dx.doi.org/ 10.1016/j.reprotox.2010.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanfeliu C, Sebastià J, Kim SU. Methylmercury neurotoxicity in cultures of human neurons, astrocytes, neuroblastoma cells. Neurotoxicology 2001; 22:317-27; PMID:11456333; http://dx.doi.org/ 10.1016/S0161-813X(01)00015-8 [DOI] [PubMed] [Google Scholar]
- 24.Farina M, Rocha JB, Aschner M. Mechanisms of methylmercury-induced neurotoxicity: evidence from experimental studies. Life Sci 2011; 89:555-63; PMID:21683713; http://dx.doi.org/ 10.1016/j.lfs.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediat 2009; 21:243; PMID:19663042; http://dx.doi.org/ 10.1097/MOP.0b013e32832925cc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turk PW, Laayoun A, Smith SS, Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis 1995; 16:1253-5; PMID:7767994; http://dx.doi.org/ 10.1093/carcin/16.5.1253 [DOI] [PubMed] [Google Scholar]
- 27.Basu N, Head J, Nam D-H, Pilsner JR, Carvan MJ, Chan HM, Goetz FW, Murphy CA, Rouvinen-Watt K, Scheuhammer AM. Effects of methylmercury on epigenetic markers in three model species: mink, chicken and yellow perch. Comp Biochem Physiol Part C: Toxicol Pharmacol 2013; 157:322-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics 2009; 1:222-8; PMID:20461219; http://dx.doi.org/ 10.1039/b903049b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howe CG, Niedzwiecki MM, Hall MN, Liu X, Ilievski V, Slavkovich V, Alam S, Siddique AB, Graziano JH, Gamble MV. Folate and cobalamin modify associations between S-adenosylmethionine and methylated arsenic metabolites in arsenic-exposed Bangladeshi adults. J Nutr 2014; 144:690-7; PMID:24598884; http://dx.doi.org/ 10.3945/jn.113.188789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waly M, Olteanu H, Banerjee R, Choi S, Mason J, Parker B, Sukumar S, Shim S, Sharma A, Benzecry J. Activation of methionine synthase by insulin-like growth factor-1 and dopamine: a target for neurodevelopmental toxins and thimerosal. Mol Psychiat 2004; 9:358-70; PMID:14745455; http://dx.doi.org/ 10.1038/sj.mp.4001476 [DOI] [PubMed] [Google Scholar]
- 31.Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Front Genet 2014; 5; PMID:25076963; http://dx.doi.org/ 10.3389/fgene.2014.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodrich JM, Basu N, Franzblau A, Dolinoy DC. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environ Mol Mutagen 2013; 54:195-203; PMID:23444121; http://dx.doi.org/ 10.1002/em.21763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rees JR, Sturup S, Chen C, Folt C, Karagas MR. Toenail mercury and dietary fish consumption. J Expo Sci Environ Epidemiol 2006; 17:25-30; PMID:16912698; http://dx.doi.org/ 10.1038/sj.jes.7500516 [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto M, Chan HM, Domingo JL, Oliveira RB, Kawakami S, Murata K. Significance of fingernail and toenail mercury concentrations as biomarkers for prenatal methylmercury exposure in relation to segmental hair mercury concentrations. Environ Res 2015; 136:289-94; PMID:25460648; http://dx.doi.org/ 10.1016/j.envres.2014.09.034 [DOI] [PubMed] [Google Scholar]
- 35.Oken E, Kleinman KP, Berland WE, Simon SR, Rich-Edwards JW, Gillman MW. Decline in fish consumption among pregnant women after a national mercury advisory. Obstet Gyn 2003; 102:346; PMID:12907111; http://dx.doi.org/ 10.1016/S0029-7844(03)00484-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bose-O'Reilly S, McCarty KM, Steckling N, Lettmeier B. Mercury exposure and children's health. Curr Probl Pediat Adol Health Care 2010; 40:186-215; PMID:20816346; http://dx.doi.org/ 10.1016/j.cppeds.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shenker BJ, Maserejian NN, Zhang A, McKinlay S. Immune function effects of dental amalgam in children: a randomized clinical trial. J Am Dent Assoc (1939) 2008; 139:1496; PMID:18978388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abedi‐Valugerdi M. Mercury and silver induce B cell activation and anti‐nucleolar autoantibody production in outbred mouse stocks: are environmental factors more important than the susceptibility genes in connection with autoimmunity? Clin Exp Immunol 2009; 155:117-24; PMID:19076835; http://dx.doi.org/ 10.1111/j.1365-2249.2008.03801.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed SA, Hissong B, Verthelyi D, Donner K, Becker K, Karpuzoglu-Sahin E. Gender and risk of autoimmune diseases: possible role of estrogenic compounds. Environ Health Perspect 1999; 107:681; PMID:10502531; http://dx.doi.org/ 10.1289/ehp.99107s5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanna CW, Bloom MS, Robinson WP, Kim D, Parsons PJ, vom Saal FS, Taylor JA, Steuerwald AJ, Fujimoto VY. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod 2012; 27:1401-10; des038; PMID:22381621; http://dx.doi.org/ 10.1093/humrep/des038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onishchenko N, Karpova N, Sabri F, Castrén E, Ceccatelli S. Long‐lasting depression‐like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem 2008; 106:1378-87; PMID:18485098; http://dx.doi.org/ 10.1111/j.1471-4159.2008.05484.x [DOI] [PubMed] [Google Scholar]
- 42.Lee B-E, Hong Y-C, Park H, Ha M, Koo BS, Chang N, Roh Y-M, Kim B-N, Kim Y-J, Kim B-M. Interaction between GSTM1/GSTT1 polymorphism and blood mercury on birth weight. Environ Health Perspect 2010; 118:437; PMID:20194072; http://dx.doi.org/ 10.1289/0900731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33:245-54; PMID:12610534; http://dx.doi.org/ 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]
- 44.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. Genome-wide methylation analysis of human colon cancer reveals similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009; 41:178; PMID:19151715; http://dx.doi.org/ 10.1038/ng.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao X, Evans J, Chae H, Pilrose J, Kim S, Yan P, Huang R, Lai H, Lin H, Liu Y. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene 2012; 32:4519-28; PMID:23128390; http://dx.doi.org/ 10.1038/onc.2012.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader–Willi/Angelman region. Am J Med Genet Part A 2008; 146:2041-52; PMID:18627066; http://dx.doi.org/ 10.1002/ajmg.a.32364 [DOI] [PubMed] [Google Scholar]
- 47.Kaur P, Aschner M, Syversen T. Glutathione modulation influences methyl mercury induced neurotoxicity in primary cell cultures of neurons and astrocytes. Neurotoxicology 2006; 27:492-500; PMID:16513172; http://dx.doi.org/ 10.1016/j.neuro.2006.01.010 [DOI] [PubMed] [Google Scholar]
- 48.Ceccatelli S, Daré E, Moors M. Methylmercury-induced neurotoxicity and apoptosis. Chemico-Biol Interact 2010; 188:301-8; PMID:20399200; http://dx.doi.org/ 10.1016/j.cbi.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 49.Koga M, Serritella AV, Messmer MM, Hayashi-Takagi A, Hester LD, Snyder SH, Sawa A, Sedlak TW. Glutathione is a physiologic reservoir of neuronal glutamate. Biochem Biophys Res Commun 2011; 409:596-602; PMID:21539809; http://dx.doi.org/ 10.1016/j.bbrc.2011.04.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steegenga WT, Boekschoten MV, Lute C, Hooiveld GJ, de Groot PJ, Morris TJ, Teschendorff AE, Butcher LM, Beck S, Müller M. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age 2014; 36:1523-40; PMID:24789080; http://dx.doi.org/ 10.1007/s11357-014-9648-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol 2012; 13:R43; PMID:22703893; http://dx.doi.org/ 10.1186/gb-2012-13-6-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert-Diamond D, Cottingham KL, Gruber JF, Punshon T, Sayarath V, Gandolfi AJ, Baker ER, Jackson BP, Folt CL, Karagas MR. Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci U S A 2011; 108:20656-60; PMID:22143778; http://dx.doi.org/ 10.1073/pnas.1109127108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchiset-Ferlay N, Savanovitch C, Sauvant-Rochat M-P. What is the best biomarker to assess arsenic exposure via drinking water? Environ Int 2012; 39:150-71; PMID:22208756; http://dx.doi.org/ 10.1016/j.envint.2011.07.015 [DOI] [PubMed] [Google Scholar]
- 54.Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD. Functional normalization of 450k methylation array data improves replication in large cancer studies. bioRxiv 2014; 15:503; PMID:25599564; http://dx.doi.org/ 10.1186/s13059-014-0503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8:118-27; PMID:16632515; http://dx.doi.org/ 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 56.Chen Y-A, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the illumina infinium HumanMethylation450 microarray. Epigenetics 2013; 8:203; PMID:23314698; http://dx.doi.org/ 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010; 11:587; PMID:21118553; http://dx.doi.org/ 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13:86; PMID:22568884; http://dx.doi.org/ 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, Söderhäll C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS One 2012; 7:e41361; PMID:22848472; http://dx.doi.org/ 10.1371/journal.pone.0041361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smyth GK. Limma: linear models for microarray data. Bioinformatics Comput Biol Solut Using R and Bioconductor: Springer, 2005:397-420; http://dx.doi.org/ 10.1007/0-387-29362-0_23 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


