Abstract
Background: NCCN states that chemotherapies for advanced esophageal and gastric cancers may be used interchangeably. Biomarkers from gastroesophageal cancer patients were interrogated to identify actionable alterations with therapeutic implications. Methods: 666 gastric and 640 esophageal cancer cases referred to Caris Life Sciences between 2009 thru 2013 were evaluated. Specific testing was performed, which included a combination of sequencing (Sanger, NGS) and protein expression (IHC). Results: In the complete cohort (n = 1306), 30 of 45 genes tested harbored mutations; highest rates were seen in TP53 (54%), APC (10%), SMAD4 (5.9%), KRAS (5.9%), and PIK3CA (5.1%). IHC of TOP2A was high in 76% of cases, TOPO1 in 51% and SPARC in 25%; low IHC of ERCC1 was seen in 65%, RRM1 in 62%, TS in 61% and MGMT in 45%, indicating potential benefit from epirubicin, irinotecan, nab-paclitaxel, platinum-based agents, gemcitabine, 5FU/capecitabine and temozolomide, respectively. In the HER2+ cohort (n = 88), 50% of patients demonstrated possible benefit from a combination of trastuzumab with 5FU/capecitabine based on concurrent low TS, 53% with irinotecan (high TOPO1), 63% with cisplatin (low ERCC1) and 55% with gemcitabine (low RRM1). Subgroup analysis by tumor origin demonstrated significant differences in actionable biomarker profiles with HER2 (13% vs. 4.6%), SPARC (34% vs. 15%), TOP2A (86% vs. 67%), and TOPO1 (55% vs. 46%) in esophageal and gastric adenocarcinoma cases respectively (P < 0.05). Conclusion: A comprehensive multiplatform biomarker analysis suggested significant biomarker differences between gastric and esophageal cancers. These results can assist in the development of future clinical trials.
Keywords: biomarker, esophageal cancer, gastric cancer, molecular profiling, personalized medicine
Abbreviations
- GC
Gastric cancer
- EC
Esophageal cancer
- HER2
Human epidermal growth factor receptor 2
- IHC
Immunohistochemistry
- FISH
Fluorescent and chromogenic in-situ hybridization
Introduction
Gastric (GC) and esophageal (EC) cancer are currently the second and eighth leading causes of cancer related mortality worldwide, with 5 y survival rates of less than 25%.1 The overall poor outcomes associated with these cancers have been attributed to the aggressive underlying tumor biology, resulting in most patients presenting with advanced disease at the time of diagnosis. Currently, surgical resection for early stage disease offers the best chance for a cure. However, a high rate of tumor recurrence has prompted the development of treatment strategies that are dependent on the delivery of systemic therapies in either the neoadjuvant or adjuvant setting.2 Unfortunately, identifying effective cytotoxic agents has proven to be more difficult with current response rates in the range of 40–50%.
According to the NCCN compendium, chemotherapeutic regimens for GC and EC are mostly interchangeable, especially in the advanced and metastatic setting.3,4 Combination therapies consisting of anthracycline, fluoropyrimidine, taxane, and platinum based agents are current first line therapy recommendations. However, the absence of a single, unified standard of care cytotoxic regimen has created complexities in the treatment approach.
The recent advances of tumor profiling have provided the opportunity to better understand the molecular signature of various cancers. Over the last decade, advancements in the understanding of critical tumorigeneis mechanisms has led to a shift from empiric approaches based on clinicopathologic features to biomarker driven algorithms. Although still in its early phases, an example highlighting the potential role of molecular profiling to guide treatment in GC and EC is the human epidermal growth factor receptor (HER2; also known as ERBB2) overexpression in gastric cancer. The pivotal ToGA trial was the first well powered randomized control trial that demonstrated a survival benefit when Trastuzumab, a monoclonal antibody against HER2, was combined with chemotherapy for advanced gastric cancers.5 Evaluation of HER-2 expression has since become standard for advanced gastric and esophageal cancers. However, in spite of GC and EC both sharing aggressive biological behaviors, genomic profiling has shown that the molecular mechanisms leading to tumor formation are unique for each cancer type.6
In the present study, high throughput multiplatform assays were used to interrogate biomarkers in a large cohort of gastric and esophageal cancers in order to identify molecular alterations with established therapeutic implications. Improved understanding of the unique molecular alterations has the potential to augment and refine the effectiveness of current chemotherapy treatment strategies.
Results
From 2009 to 2013, a total of 1306 gastroesophageal tumor samples were evaluated; 666 gastric adenocarcinoma and 640 esophageal carcinoma (553 esophageal adenocarcinoma, 37 esophageal squamous carcinoma, 50 esophageal carcinoma NOS). The median age of the collective cohort was 61 (Interquartile Range [IQR]: 53–71), with the majority being male (n = 929, 71.1%).
Assessment of actionable targets by IHC and FISH/CISH
Standard-of-care for gastroesophageal adenocarcinoma includes first line cytotoxic regimens consisting of 2 or 3 drug combinations of an anthracycline, fluoropyrimidine, taxane, irinotecan, or platinum based agent. Associated biomarkers with potential sensitivity to first line agents include TOP2A (epirubicin/anthracycline), TUBB3 (taxane), PGP (taxane), TS (fluoropyrimidine), TOPO1 (irinotecan), and ERCC1 (platinum agent). IHC biomarker analysis of the collective cohort demonstrated TOP2A (76.2% High) and TUBB3 (75.1% negative) to have the greatest percentage of independent actionable targets among first line agents, suggesting sensitivity to anthracyline and taxane agents respectively (Table 1). Potential sensitivity to trastuzumab by HER2 IHC positivity was also present in 8% of the total cohort (92 out of 1155). When considering tumors that are HER2 2+ and FISH amplified (HER2/CEP17 > 2.0), additional 8 tumors were identified as potential responders to trastuzumab, bringing HER2 positivity rate to 9%.
Table 1.
Overall frequency of actionable targets among all tumors tested by immunohistochemistry along with associated therapies (n = 1306)
| Target Biomarker | N | % Actionable | Associated Agents |
|---|---|---|---|
| TOP2A (+)* | 947 | 76.2 | Epirubicin |
| TUBB3 (−)* | 261 | 75.1 | Taxane |
| ERCC1 (−)* | 837 | 65.1 | Cisplatin, Oxaliplatin |
| RRM1 (−) | 1055 | 61.9 | Gemcitabine |
| TS (−)* | 1050 | 61 | Capecitabine, 5-FU |
| PTEN (−) | 1134 | 57.9 | PI3K/AKT/mTor Inb |
| PGP (−)* | 918 | 52.6 | Taxane |
| TOPO1 (+)* | 1046 | 51.3 | Irinotecan |
| MGMT (−) | 1124 | 45.3 | Temozolomide |
| SPARC (+) | 1140 | 25 | Nab-paclitaxel |
| PDGFR (+) | 237 | 24.5 | Multi-kinase Inb |
| cMET (+) | 444 | 23.9 | cMET Inb |
| TLE3 (+) | 458 | 16.6 | Taxane |
| HER2 (+)* | 1155 | 8 | Trastuzumab |
| cKIT (+) | 675 | 4 | cKIT Inb |
| PR (+) | 1084 | 3 | Hormonal Therapy |
| ER (+) | 1087 | 0.8 | Hormonal Therapy |
| AR (+) | 1081 | 0.4 | Hormonal Therapy |
(−) Negative expression; (+) high expression; * Biomarker with associated agent on NCCN compendium; Inb Inhibitor; 5-FU 5-Fluorouracil.
Additional chemotherapeutic agents with associated biomarkers were assessed. Overall, the greatest percentage of non NCCN compendium actionable targets included RRM1 (61.9% negative, gemcitabine), PTEN (57.9% negative, PI3K/AKT/mTOR Inhibitors), MGMT (45.3% negative, temozolomide) and SPARC (24.5%, nab-paclitaxel).
Genotype analysis
Of the 45 genes that were evaluated by sequencing, only 5 genes were found to harbor mutations with a frequency of greater than 5% (Fig. 1). Tumor suppressor gene, TP53, had the highest relative gene mutation frequency of 54.2%. This is in comparison to other established tumor associated genes, which include APC (10.2%), SMAD4 (5.9%), KRAS (5.9%), and PIK3CA (5.1%).Various additional targetable genes including PTEN (2.5%), ERBB2 (1.5%), cMET (1.5%), BRAF (1.3%) and EGFR (1.3%) were mutated in 1–5% of patient samples. The specific mutations for each gene can be found in the supplementary materials (Table S1 and S2). A total of 15 genes that underwent analysis were found to harbor no underlying genomic alteration.
Figure 1.
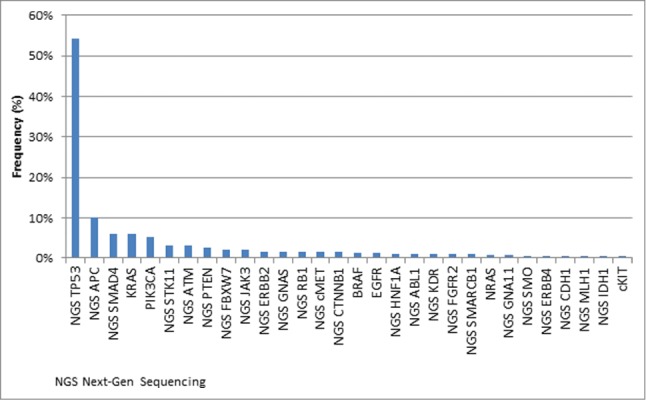
Frequency of cancer gene mutations identified by a combination of Sanger and Next-Gen Sequencing of all tumors that were tested (n = 1306). Genes tested without alterations: AKT1, ALK, CSF1R, FGFR1, FLT3, GNAQ, HRAS, JAK2, MPL, NOTCH1, NPM1, PDGFRA, PTPN11, RET, VHL.
Actionable targets among HER2 positive cohort
A total of 100 tumors were defined as HER2 positive in our cohort, among which 28 were gastric cancer and 72 were esophageal cancer cases. These tumors were either HER2 IHC 3+ or HER2 IHC2+ and FISH amplified. Tumors with FISH amplification but no evidence of HER2 protein overexpression were not considered HER2 positive5. Multiple actionable targets were identified in the HER2 positive cohort and are shown in Figure 2. Coexpression of TOP2A among HER2 positive patients occurred most frequently (93%), suggesting potential sensitivity to a combined anthracycline/ trastuzumab treatment approach. Additional actionable targets that were noted to have drug-amendable alterations in greater than 50% of the HER2 positive tumors included TUBB3 (69% negative, taxanes), ERCC1 (63% negative, platinums), RRM1 (60% negative, gemcitabine), PgP (58% negative, taxanes and anthracyclines), TOPO1 (56% positive, Irinotecan), PTEN (53%, negative, mTor inhibitors), and TS (52% low, fluoropyrimidine).
Figure 2.
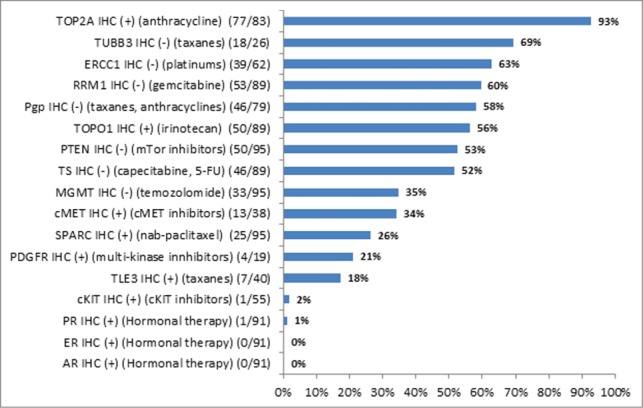
Actionable biomarker targets by immunohistochemistry among HER2+ gastroesophageal tumors (n = 100).
Comparison of actionable targets between esophageal and gastric adenocarcinoma
The molecular makeup of the 666 gastric adenocarcinoma tumors was compared to that of the 553 esophageal adenocarcinoma and a differential expression pattern was revealed (Table 2). Esophageal tumors with squamous features or labeled as carcinoma NOS (not otherwise specified) were excluded from the analysis because of a different or unclear histology. By IHC, significant differences in actionable biomarker expression profiles were noted including TOP2A (86% vs 67%, P < 0.01), MGMT (50% vs 60%, P < 0.01), TOPO1 (55% vs. 46%, p = 0.01) and TUBB3 (68% vs 84.1%, P < 0.01) in gastric and esophageal adenocarcinomas, respectively. Interestingly, HER2 (12.5% vs 4.6%, P < 0.01), SPARC (34% vs 15%, P < 0.01), and TLE3 (20.4% vs 9.4%, P < 0.01) were more than twice overexpressed in esophageal than in gastric adenocarcinoma tumors. Additionally, genetic profiling demonstrated KRAS (8.4% vs 3.8%) and PIK3CA (7.8% vs 2.4%) to be more frequently mutated in gastric cancers as compared to esophageal cancers (P < 0.05). In contrast, TP53 was mutated in 63% of esophageal cancers versus 40% of gastric cancers (P < 0.01).
Table 2.
Biomarker comparison of gastric and esophageal adenocarcinoma by Immunohistochemistry
| Gastric adenocarcinoma |
Esophageal adenocarcinoma |
||||
|---|---|---|---|---|---|
| Target Biomarker | Total | % Actionable | Total | % Actionable | P value |
| TOP2A (+) | 507 | 67 | 377 | 86 | <0.01 |
| TUBB3 (−) | 126 | 84.1 | 106 | 67.9 | <0.01 |
| ERCC1 (−) | 447 | 64.2 | 349 | 67.1 | |
| RRM1 (−) | 553 | 63.8 | 434 | 61.8 | |
| TS (−) | 548 | 62.4 | 433 | 60.7 | |
| PTEN (−) | 577 | 58.4 | 478 | 57.9 | |
| PGP (−) | 479 | 54.1 | 378 | 47.1 | |
| TOPO1 (+) | 543 | 46 | 433 | 54.7 | <0.01 |
| MGMT (−) | 576 | 40 | 471 | 51 | <0.01 |
| SPARC (+) | 584 | 15.4 | 477 | 34.4 | <0.01 |
| PDGFR (+) | 157 | 22.3 | 73 | 28.8 | |
| cMET (+) | 208 | 22.1 | 192 | 27.1 | |
| TLE3 (+) | 213 | 9.4 | 201 | 20.4 | <0.01 |
| HER2 (+) | 586 | 4.6 | 488 | 12.5 | <0.01 |
| cKIT (+) | 368 | 4.3 | 273 | 3.3 | |
| PR (+) | 559 | 3.2 | 451 | 3.1 | |
| ER (+) | 560 | 1.6 | 452 | 0 | |
| AR (+) | 559 | 0.7 | 449 | 0 | |
Discussion
The identification of the unique molecular mechanisms driving tumorigenesis for EC and GC has resulted in an emerging interest in biomarker analysis to provide theranostic information to assist in treatment delivery.7,8 In the present study, a multiplatform approach was utilized that confirmed a high frequency of biomarker expression profiles that would suggest potential sensitivity to first line chemotherapeutic drugs. Interestingly, biomarkers such as RRM1, MGMT, PTEN, and SPARC were identified by IHC with increasing frequency, along with mutations including KRAS, PIK3CA, PTEN, ERBB2, BRAF, and EGFR, highlighting additional areas for targeted therapies. However, subgroup analysis by cancer type demonstrated significant differences in biomarker profiles between EC and GC, which further add to the complexity in treatment and underscores the need to approach each cancer type independently.
A study by Von Hoff et al. was one of the earlier trials to first demonstrate the potential efficacy of a treatment regimen selected by molecular profiling.9 Of the 66 refractory metastatic cancer patients treated with a cytotoxic regimen determined by the molecular profiling results, 18 (27%) experienced a longer progression free survival. Similarly, the TOGA trial was the first study to demonstrate the benefit of targeted therapies (trastuzumab) integrated with standard cytotoxic agents for gastric or gastroesophageal junction adenocarcinoma that overexpressed HER-2.5 Patients treated with trastuzumab plus chemotherapy group experienced a median overall survival of 13.8 months compared with 11.1 months in those assigned to chemotherapy alone (HR 0.74, p = 0.0046). However studies have shown amplification of HER2 in only 7–34% of gastric tumors,10,11 which is similar to what was found in the present study (8%). Therefore, detection of additional targetable molecular alterations are in great need.
Our study shows that there was no individual biomarker suggesting uniform sensitivity to an individual chemotherapy agent in all gastroesophageal tumors. At most, 75% of patients possessed a molecular profile that would predict sensitivity to either epirubicin (76.2% TOP2A high) or taxane (75.1% TUBB3 negative) therapy. To increase the likelihood of tumor responsiveness, 2 drug and 3-drug chemotherapy combinations have been introduced, which has resulted in an approximate 17% reduction in the risk of death (HR 0.83, 95%CI 0.74–0.93).12 However, up to 50% of patients will not respond to current multi-drug regimens. Molecular profiling has the potential to identify patients with biomarker features that predict resistance to one or more chemotherapies, and thus helping to devise a more effective treatment plan.
For patients who are potential responders to trastuzumab, further decisions have to be made by the clinician as of which chemotherapeutic agent to use in combination. While fluorpyrimidines, platinums as well as taxanes are common choices, the biomarker profile in the HER2 positive cohort suggest that not all patients carry a profile associated with sensitivity to these agents. However, other potential options exist which include epirubicin, mTor inhibitors, gemcitabine, irinotecan, and temozolomide, based on the expression level of their associated biomarkers. Anthracyclines are not commonly considered in combination with trastuzumab based on observed cardiotoxicity of doxorubicin in breast; however in gastroesophageal cancer, epirubicin carries a much milder cardiotoxcicity and has shown preliminary but promising clinical activity.13 Based on an overexpression of TOP2A in over 90% of HER2 positive patients, it's logical to hypothesize that adding epirubicin to trastuzumab containing regimen would be effective on select patients.
Interestingly, IHC and genomic analysis identified additional actionable targets beyond those that are associated with cytotoxic drugs on the NCCN compendium for gastric or esophageal cancer. Three biomarkers, RRM1 (62%), MGMT (45%), and PTEN (58%), had negative expression profiles by IHC in greater than 40% of the gastroesophageal tumors, suggesting potential sensitivity to gemcitabine, temozolomide, or PI3K inhibitors respectively. While the overall efficacy of gemcitabine as a monotherapy or when delivered in combination for advanced gastroesophageal cancers has resulted in equivocal results, recent studies have demonstrated that negative RRM1 expression is associated with higher response rates to gemcitabine based chemotherapy regimens.14-16 Therefore, stratifying patients based on RRM1 expression may increase the likelihood of gemcitabine efficacy and further expand the types of cytotoxic agents currently available. Similar principles can also be applied to other biomarkers when attempting to identify additional cytotoxic agents that have not yet been considered.
Mutations in TP53 (∼50%) remains a common event in most major types of solid tumors. However, it was not until recently that targeted therapies (e.g. PRIMA-1, p28) to TP53 have been investigated in phase I/II clinical trials with promising results.17,18 In the present study, a high frequency of TP53 mutations (55%) were identified, which is consistent with previous studies. In contrast, the vast majority of genes were not found to harbor alterations among the collective cohort. APC (10.1%), SMAD4 (5.85%), KRAS (5.85%), and PIK3A (5.1%) were the next most frequently mutated genes. Potential targeted therapies to the aforementioned genes would include inhibitors of the Wnt/β-Catenin Pathway (APC), downstream signaling targets of KRAS, such a MEK/ERK, as well as PI3K/AKT/mTOR inhibitors, which are actively being investigated in clinical trials.19–21 However, the low frequency of somatic mutations in gastroesophageal tumors underscores the genetic heterogeneity of these cancers, supporting the need for a broad based multiplatform profiling in order to identify potential targets for therapy.
Lastly, subgroup analysis by cancer site revealed a differential biomarker expression pattern between gastric and esophageal tumors. Recognizing that certain biomarkers such as HER2, SPARC and TLE3 are overexpressed more than twice as frequently in esophageal cancer as compared to gastric cancer, underscores the need to tailor therapy to the underlying molecular signature rather than relying heavily on empiric strategies.
The current study has several limitations. Despite being one of the largest series that applied molecular profiling to a series of gastric and esophageal cancers, prospective data capturing the potential efficacy of the targeted agents is lacking. Instead, clinical trials, which compare the efficacy of established therapies to those determined through molecular profiling are needed. Furthermore, it was unclear as to the extent of treatment each tumor sample had received prior to analysis. Heavily pretreated tumors that were exposed to first and second line therapies may reflect more advanced gastroesophageal tumors, and thus are not representative of other patients with the same type of cancer. Lastly, HER-2 overexpression in both gastric (4.6%) and esophageal (12.5%) cancers occurred at a lower frequency when compared to published series. This finding potentially highlights the paradigm shift in how gastroesophageal cancers are being treated. Assessment of HER-2 expression has become standard of care. Therefore it is possible that the majority of tumors in the present study underwent prior HER-2 screening, resulting in a greater proportion of HER-2 negative tumors at the time of analysis. Nonetheless, as more effective targeted agents are identified, the adoption of molecular profiling will continue to expand and will likely have a larger role in treatment algorithms.
In conclusion, molecular profiling has the capacity to identify targets to which established therapies are currently available. Whether the application of treatment regimens influenced by biomarker analysis can be effective remains ill defined. In the present study, a multiplatform approach that included IHC, FISH/CISH and mutation profiling identified multiple targets both on and off NCCN compendium for gastroesophageal cancer treatment. More importantly, molecular profiling identified a differential pattern in biomarker expression between gastric and esophageal tumors, highlighting the unique mechanisms driving tumorigenesis for each respective cancer. Further studies that apply the results of these biomarker analyses have the potential to refine and enhance current treatment strategies.
Materials and Methods
Approval was granted from the Institutional Review Board at the Medical College of Wisconsin. A retrospective biomarker analysis was performed on pathologically confirmed gastroesophageal tumor samples that were submitted to a commercial referral diagnostic laboratory (Caris Life Sciences, Phoenix, AZ) for molecular profiling aimed to provide theranostic information based on tumor biomarkers. A multiplatform approach was taken that included sequencing, immunohistochemistry (IHC) as well as fluorescent and chromogenic in situ hybridization (FISH/CISH) to investigate targetable biomarker aberrations. Those specimens labeled gastroesophageal junction were excluded from analysis.
Immunohistochemistry (IHC)
IHC analysis was performed on formalin fixed paraffin embedded tumor samples using commercially available detection kits, automated staining techniques (Benchmark XT, Ventana, Tucson, AZ; and AutostainerLink 48, Dako, Carpinteria, CA) in a CLIA/CAP certified, ISO validated lab (Caris Life Sciences, Phoenix, AZ). The primary antibody clones used are as follows: AR (AR441/ AR318), cKIT (polyclonal), ER (SP1), PDGFR (polyclonal), HER2 (4B5), ERCC1 (8F1), MGMT (MT23.3), PGP (C494), PR (1E2/100), PTEN (6H2.1), RRM1 (polyclonal), SPARC monoclonal (122511), SPARC (polyclonal), TOPO1 (1D6), TOP2A (3F6) and TS (TS106/4H4B1), cMET(SP44), TUBB3(Neuronal Class III Beta-Tubulin Polyclonal) and TLE3(M-201). The intensity of the staining was scored as 0, 1+, 2+ or 3+, and the percentage of staining cells was scored as 0 to 100% by board- certified pathologists.
The thresholds to categorize the immunohistochemical staining to high and low are as follows: TOP2A: 1+ and 10%; TUBB3: 1+ and 10%; ERCC1: 2+ and 50%; RRM1: 2+ and 50%; TS: 1+ and 10%; PTEN: 1+, 50%; PGP: 1+ and 10%; TOPO1: 2+ and 30%; MGMT: 1+ and 35%; SPARC: 2+ and 30%; cMET: 2+ and 50%; TLE3: 2+ and 30%; HER2: 3+ and 10%; PR: 1+ and 10%; ER: 1+ and 10% and AR: 1+ and 10%.
Fluorescent and chromogenic In Situ hybridization (FISH)
HER2 gene amplification was evaluated by FISH (HER2/CEP17 probe) and CISH (INFORM HER2 Dual ISH DNA probe cocktail). HER-2/CEP17 ratio > 2.0 was considered amplified.5,22
Next-gen sequencing
Direct sequence analysis was performed on genomic DNA isolated from formalin-fixed paraffin-embedded tumor samples using the Illumina MiSeq platform. Specific regions of the genome were amplified using the Illumina TruSeq Amplicon Cancer Hotspot panel. All variants that are reported here are detected with >99% confidence based on the frequency of the mutation present and the amplicon coverage. The sequencing included hotspot regions of 45 genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR (VEGFR2), KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFR, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, STK11, TP53, and VHL.
Sanger sequencing
Sanger sequencing included selected regions of BRAF, KRAS, c-KIT, EGFR, NRAS and PIK3CA genes and was performed by using M13-linked PCR primers designed to flank and amplify targeted sequences. PCR products were bi-directionally cycle sequenced by using the BigDye Terminator v1.1 chemistry (Applied Biosystems) and analyzed using the 3730 DNA Analyzer (Applied Biosystems). Sequence traces were analyzed using Mutation Surveyor software v3.25 (Soft Genetics). A sample was deemed mutated if the same nucleotide change was identified in both the forward and reverse traces as well as being absent in the control sample traces.
Statistical analysis
Summary statistics were obtained using established methods. Continuous variables were described as medians and IQR. Categorical variables were described as totals and frequencies. Non-parametric comparisons between groups were analyzed using the Kruskal-Wallis and Fischer exact test as appropriate. Alpha was set at 0.05 and all tests were 2 sided.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127:2893-917; PMID:21351269; http://dx.doi.org/ 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2. Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ, Falk SJ, Iveson TJ, et al. . Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med 2006; 355:11-20; PMID:16822992; http://dx.doi.org/ 10.1056/NEJMoa055531 [DOI] [PubMed] [Google Scholar]
- 3. National Comprehensive Cancer Network Gastric Cancer (Including cancer in the proximal 5cm of the stomach). [Google Scholar]
- 4. National Comprehensive Cancer Network Esophageal and Esophagogastric Junction Cancers. [DOI] [PubMed] [Google Scholar]
- 5. Bang Y, Van Cutsem E, Feyereislova A, Chung H, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. . Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010; 376:687-97; PMID:20728210; http://dx.doi.org/ 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 6. Chen Z, Saad R, Jia P, Peng D, Zhu S, Washington MK, Zhao Z, Xu Z, El-Rifai W. Gastric adenocarcinoma has a unique microRNA signature not present in esophageal adenocarcinoma. Cancer 2013; 119:1985-93; PMID:23456798; http://dx.doi.org/ 10.1002/cncr.28002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boland P, Burtness B. Esophageal carcinoma: are modern targeted therapies shaking the rock? Curr Opin Oncol 2013; 25:417-24; PMID:23680713; http://dx.doi.org/ 10.1097/CCO.0b013e328362105e [DOI] [PubMed] [Google Scholar]
- 8. Miura JT, Johnston FM, Thomas J, George B, Eastwood D, Tsai S, Christians KK, Turaga KK, Gamblin TC. Molecular profiling in gastric cancer: examining potential targets for chemotherapy. J Surg Oncol 2014; 110:302-6; PMID:24844210; http://dx.doi.org/ 10.1002/jso.23639 [DOI] [PubMed] [Google Scholar]
- 9. Von Hoff DD, Stephenson JJ, Jr., Rosen P, Loesch DM, Borad MJ, Anthony S, Jameson G, Brown S, Cantafio N, Richards DA, et al. . Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010; 28:4877-82; PMID:20921468; http://dx.doi.org/ 10.1200/JCO.2009.26.5983 [DOI] [PubMed] [Google Scholar]
- 10. Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol 2008; 19:1523-9; PMID:18441328; http://dx.doi.org/ 10.1093/annonc/mdn169 [DOI] [PubMed] [Google Scholar]
- 11. Tanner M, Hollmén M, Junttila TT, Kapanen AI, Tommola S, Soini Y, Helin H, Salo J, Joensuu H, Sihvo E, et al. . Amplification of HER-2 in gastric carcinoma: association with Topoisomerase IIalpha gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann Oncol 2005; 16:273-8; PMID:15668283; http://dx.doi.org/ 10.1093/annonc/mdi064 [DOI] [PubMed] [Google Scholar]
- 12. Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE. Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol 2006; 24:2903-9; PMID:16782930; http://dx.doi.org/ 10.1200/JCO.2005.05.0245 [DOI] [PubMed] [Google Scholar]
- 13. Palacio S, Loaiza-Bonilla A, Kittaneh M, Kyriakopoulos C, Ochoa RE, Escobar M, Arango B, Restrepo MH, Merchan JR, Rocha Lima CM, et al. . Successful use of Trastuzumab with anthracycline-based chemotherapy followed by trastuzumab maintenance in patients with advanced HER2-positive gastric cancer. Anticancer Res 2014; 34:301-6; PMID:24403478 [PubMed] [Google Scholar]
- 14. Christman K, Kelsen D, Saltz L, Tarassoff PG. Phase II trial of gemcitabine in patients with advanced gastric cancer. Cancer 1994; 73:5-7; PMID:8275437; http://dx.doi.org/ 10.1002/1097-0142(19940101)73:1%3c5::AID-CNCR2820730103%3e3.0.CO;2- [DOI] [PubMed] [Google Scholar]
- 15. De Lange SM, van Groeningen CJ, Kroep JR, Van Bochove A, Snijders JF, Peters GJ, Pinedo HM, Giaccone G. Phase II trial of cisplatin and gemcitabine in patients with advanced gastric cancer. Ann Oncol 2004; 15:484-8; PMID:14998853; http://dx.doi.org/ 10.1093/annonc/mdh109 [DOI] [PubMed] [Google Scholar]
- 16. Dong X, Hao Y, Wei Y, Yin Q, Du J, Zhao X. Response to first-line chemotherapy in patients with non-small cell lung cancer according to RRM1 expression. PLoS One 2014; 9:e92320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lehmann S, Bykov VJ, Ali D, Andrén O, Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A, et al. . Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol 2012; 30:3633-9; PMID:22965953; http://dx.doi.org/ 10.1200/JCO.2011.40.7783 [DOI] [PubMed] [Google Scholar]
- 18. Warso MA, Richards JM, Mehta D, Christov K, Schaeffer C, Rae Bressler L, Yamada T, Majumdar D, Kennedy SA, Beattie CW, et al. . A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br J Cancer 2013; 108:1061-70; PMID:23449360; http://dx.doi.org/ 10.1038/bjc.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lesko AC, Goss KH, Prosperi JR. Exploiting APC function as a novel cancer therapy. Curr Drug Targets 2014; 15:90-102; PMID:24200292; http://dx.doi.org/ 10.2174/1389450114666131108155418 [DOI] [PubMed] [Google Scholar]
- 20. Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA, Grinsted L, Zazulina V, Smith P, et al. . Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013; 14:38-47; http://dx.doi.org/ 10.1016/S1470-2045(12)70489-8 [DOI] [PubMed] [Google Scholar]
- 21. Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, Naing A, Falchook GS, Moroney JW, Piha-Paul SA, et al. . PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther 2011; 10:558-65; PMID:21216929; http://dx.doi.org/ 10.1158/1535-7163.MCT-10-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rüschoff J, Dietel M, Baretton G, Arbogast S, Walch A, Monges G, Chenard MP, Penault-Llorca F, Nagelmeier I, Schlake W, et al. . HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch 2010; 457:299-307; PMID:20665045; http://dx.doi.org/ 10.1007/s00428-010-0952-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


