Abstract
Epidermal growth factor receptor (EGFR) is frequently overexpressed in head and neck squamous cell carcinoma (HNSCC) and cetuximab, a monoclonal antibody targeting this receptor, is widely used to treat these patients. In the following investigation, we examined the role of SMAD4 down-regulation in mediating epithelial-to-mesenchymal transition (EMT) and cetuximab resistance in HNSCC. We determined that SMAD4 downregulation was significantly associated with increased cell motility, increased expression of vimentin, and cetuximab resistance in HNSCC cell lines. In the HNSCC genomic dataset obtained from The Cancer Genome Atlas, SMAD4 was altered in 20/279 (7%) of HNSCC via homozygous deletion, and nonsense, missense, and silent mutations. When SMAD4 expression was compared with respect to human papillomavirus (HPV) status, HPV-positive tumors had higher expression compared to HPV-negative tumors. Furthermore, higher SMAD4 expression also correlated with higher CDKN2A (p16) expression. Our data suggest that SMAD4 down-regulation plays an important role in the induction of EMT and cetuximab resistance. Patients with higher SMAD4 expression may benefit from cetuximab use in the clinic.
Keywords: SMAD4, epithelial-to-mesenchymal transition, cetuximab, head and neck squamous cell carcinoma
Abbreviations
- CDKN2A
cyclin-dependent kinase Inhibitor 2A
- CTX
cetuximab
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-msenchymal transition
- FDR
false delivery rate
- HB-EGF
heparin-binding EGF-like growth factor
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- KD
knocked-down
- mIR
microRNA
- qRT-PCR
quantitative real-time polymerase chain reaction
- RPPA
reverse phase protein arrays
- RSEM
RNA-Seq by Expectation Maximization
- shRNA
small hairpin RNA
- SMAD4
mothers against decapentaplegic homolog 4
- TCGA
The Cancer Genome Atlas
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease associated with several anatomical sites including the oral cavity, pharynx, and larynx. Risk factors for HNSCC development include chronic tobacco exposure, heavy alcohol use, and human papillomavirus (HPV) infection.1-4 Treatment intensification using multi-modality approaches can be effective for managing newly diagnosed HNSCC patients; however, recurrent and metastatic disease are often not curable and therapeutic options are limited.
Epidermal growth factor receptor (EGFR) is known to be an important prognostic marker and a therapeutic target in HNSCC. It has long been understood that EGFR overexpression as determined by immunohistochemistry is associated with poor survival in HNSCC patients.5-8 Furthermore, recent comprehensive genomic analyses of HNSCCs have confirmed that EGFR is altered by increased copy number and mutations.9-11 Based on these data, EGFR-targeted agents have been developed and cetuximab, a monoclonal antibody directed against EGFR, is currently FDA-approved for use in HNSCC patients.12,13 Through binding the extracellular domain of EGFR, cetuximab competes with EGFR ligands and prevents downstream activation.14 In addition to receptor inhibition, cetuximab may also promote apoptosis by inhibiting DNA damage repair and inducing antibody-dependent cellular cytotoxicity (ADCC).15-17
However, it has become apparent that a significant subset of HNSCC patients exhibit EGFR inhibitor resistance.13 Several mechanisms of resistance have been investigated including activation of HER3, MET, and downstream AKT activation.17,18 Previously, our laboratory has demonstrated multiple mechanisms of acquired cetuximab resistance in HNSCC cells including therapeutic target loss (downregulation of EGFR) and up-regulation of its cognate ligands, such as heparin-binding EGF-like growth factor (HB-EGF).19 Furthermore, we reported that the deregulation of microRNA (miR) expression, such as decrease miR-212, may contribute to the observed HB-EGF upregulation.19 We also observed that the cetuximab-resistant cells exhibit a mesenchymal phenotype in vitro compared to cetuximab-sensitive cells.
In the current study, we hypothesized that epithelial-to-mesenchymal transition (EMT) is an important contributor for mediating acquired cetuximab resistance. We demonstrate that down-regulation of Smad4 is associated with an EMT phenotype and contributes to cetuximab resistance in HNSCC. In addition, we evaluated SMAD4 genomic alterations in human HNSCC tumors which revealed higher SMAD4 expression in HPV-positive tumors suggesting that patients with HPV-positive tumors may benefit from cetuximab.
Results
Cetuximab-resistant cells exhibit a mesenchymal phenotype in vitro compared to cetuximab-sensitive cells
SCC1 and 1CC8 are an isogenic cell line pair, the latter developed as an acquired cetuximab resistance model. The sensitivity of SCC1 and 1CC8 to cetuximab treatment has previously been published.18,19 To further investigate this phenomenon, we developed a second isogenic cell line panel of acquired cetuximab resistance using cetuximab-sensitive SCC25. Twelve cetuximab-resistant clones (CTX-R1-12) were generated by chronic exposure to cetuximab in vitro (Fig. 1A). After treatment selection, we observed the cell lines with acquired cetuximab resistance exhibited a mesenchymal morphology upon visual inspection and displayed increased migratory potential compared to the sensitive parent cell lines. These features are consistent with previous reports of EMT (Fig. 1B).17,19
Figure 1.
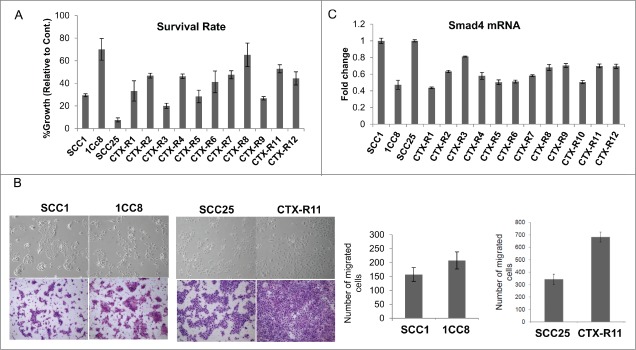
(A) Characterization of cetuximab (CTX)-resistant clones generated from SCC1 and SCC25 after chronic exposure to CTX in vitro. Cell survival was determined by Matrigel colony formation, (B) Increased cell mobility in CTX-resistant clones, 1CC8 and CTX-R11, compared to SCC1 and SCC25, respectively, determined by cell migration assay, and (C) SMAD4 mRNA levels in CTX-resistant clones compared to the isogenic parent cell lines, SCC1/1CC8 and SCC25/CTX-Rs.
To further evaluate the induction of EMT and subsequent cetuximab resistance, 218 probes representing 83 EMT-related genes (Gene Ontology set GO:0001837)20 were analyzed for coordinated differential expression between SCC1/1CC8 using previously published global gene expression data (GSE21483, File S1). These genes associated with EMT were differentially expressed between SCC1 and 1CC8 with a statistical significance (p-value = 9 × 10−5). More specifically, 31 probes representing 21 genes were significantly associated with cetuximab resistance seen in 1CC8 (Fig. 2). Among these, SMAD4 was chosen for further evaluation because SMAD4 knockout mice develop spontaneous HNSCCs that histologically resemble the human disease.21 Furthermore, with respect to our isogenic cell line pair, SMAD4 was substantially down-regulated in 1CC8 compared to SCC1 (p-value of 8 × 10−9). Lower 1CC8 SMAD4 mRNA expression was also confirmed by qRT-PCR (Fig. 1C). To expand on this result, we evaluated the newly generated SCC25-derived cetuximab-resistant cell lines (CTX-R1-12) for SMAD4 expression. While SMAD4 expression was variable across the panel, qRT-PCR analysis determined that all 12 daughter cell lines expressed lower levels of SMAD4 mRNA compared to the parental SCC25 cell line with an average decrease of 41% ± 3 (Fig. 1C).
Figure 2.
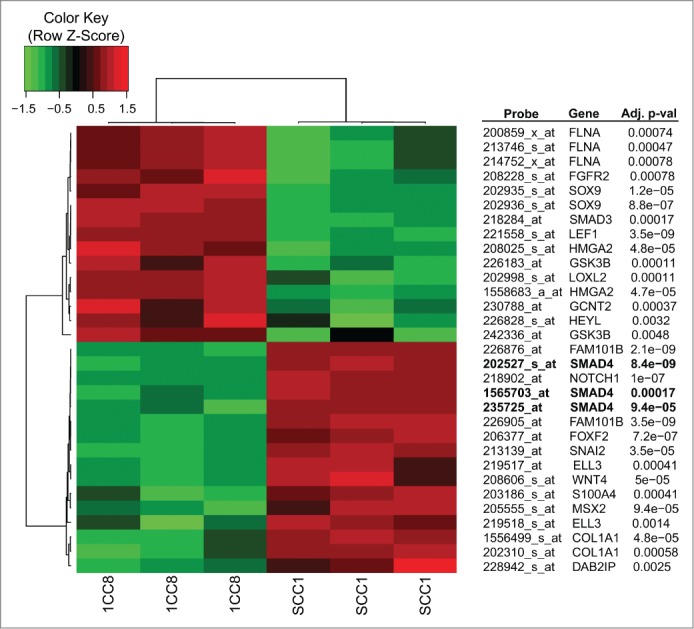
Heatmap of expression values for probes that are differentially expressed between SCC1 and 1CC8 with a statistical significance and are annotated to the GO EMT pathway.
SMAD4 knock-down increases EMT phenotype and induces cetuximab resistance in HNSCC
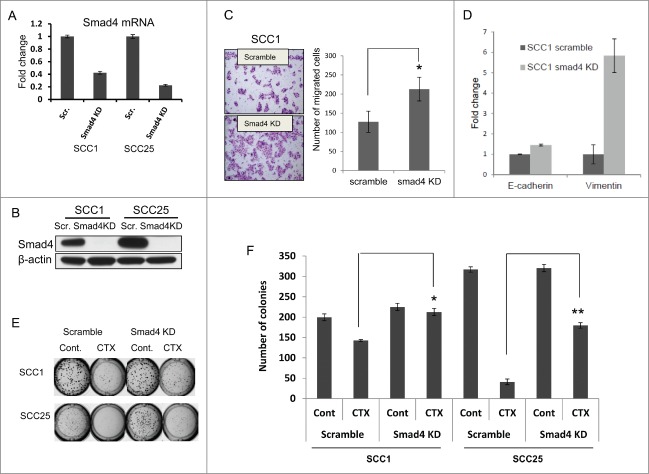
To determine the functional consequences of SMAD4 downregulation in cetuximab-sensitive HNSCC cell lines, SMAD4 was stably knocked-down (KD) in SCC1 and SCC25 using shRNAs (Fig. 3A–B). In the SMAD4 KD groups, the number of migrating cells increased nearly 2-fold compared to scrambled shRNA controls (p < 0.05). These cells also demonstrated the EMT phenotype observed in the cetuximab-resistant cell lines (Fig. 3C). In SCC1, SMAD4 KD also caused a significant increase in vimentin mRNA, supporting our hypothesis that SMAD4 down-regulation contributes to EMT in HNSCC (Fig. 3D). Additionally, downregulation of SMAD4 decreased cetuximab sensitivity of both SCC1 and SCC25, as determined by matrigel colony formation (Fig. 3E–F). These results suggest that decreased SMAD4 expression is associated with EMT in HNSCC and the functional consequence of this transformation is increased motility and cetuximab resistance.
Figure 3.
Smad4 knockdown (KD) induces cetuximab (CTX) resistance in HNSCC cell lines. (A) Lower expression of SMAD4 mRNA in CTX-sensitive cell lines, SCC1 and SCC25, after Smad4 shRNA KD, (B) Lower expression of Smad4 protein in CTX-sensitive cell lines, SCC1 and SCC25, after Smad4 shRNA KD, (C) SCC1- SMAD4 KD cells are more migratory, (D) SCC1- SMAD4 KD cells have increased expression of vimentin, and (E–F) SMAD4 KD cells are less sensitive to CTX treatment (100nM) than control (Cont) as measured by Matrigel colony formation assay. *statistically significant.
SMAD4 expression in human HNSCC
To further determine the relevance of SMAD4 down-regulation in tumors obtained from HNSCC patients, we analyzed the genomic alteration of SMAD4 in The Cancer Genome Atlas (TCGA) using DNA and RNA sequencing data obtained from 279 HNSCC samples.9 There were 36 HPV-positive and 243 HPV-negative HNSCC with both DNA and RNA data within TCGA (Table 1). While HPV-positive samples did not demonstrate any homozygous deletions, 13 of 243 (5%) HPV-negative HNSCCs harbored homozygous deletions of SMAD4. One (3%) HPV-positive HNSCC had a missense mutation (P298S) but MutationAssessor found its probability of functional significance to be low.22,23 Six of the HPV-negative HNSCCs (2%) had nonsense, missense, and silent mutations. MutationAssessor assigned the 3 missense mutations (P544L, R361H, and W99C) to have medium probability of functionally significance.22,23 We found that samples with deletions and nonsense mutations of SMAD4 had significantly reduced SMAD4 mRNA expression (p = 0.0002, Fig. 4A). We also categorized the tumors with wild-type SMAD4 into high and low expression cohorts relative to the expression levels in samples with homozygous deletion and nonsense mutations, and found 125 of the 279 samples (44.8%; Table 1) to have low SMAD4 expression. Moreover, SMAD4 mRNA expression was positively correlated with Smad4 protein expression (correlation coefficient 0.19, p-value 0.0057, Fig. 4B). In addition, we found SMAD4 mRNA expression is significantly lower in HPV-negative HNSCC tumors relative to HPV-positive tumors (p-value of 1.5 × 10−12, Fig. 4C). SMAD4 mRNA expression was also significantly correlated with CDKN2A (p16) RNA expression which is commonly used as a surrogate marker of HPV status in HNSCC24,25 (correlation coefficient 0.19, p-value 0.0018, Fig. 4D).
Table 1.
Genomic alterations of SMAD4 in tumors obtained from HNSCC patients in TCGA. Samples are called SMAD4 low when their log transformed RSEM expression for SMAD4 from RNA-sequencing data is below the maximum value of SMAD4 expression for homozygous deletions or nonsense mutations
| HPV | SMAD4 | SMAD4.Low | SMAD4.High |
|---|---|---|---|
| HPV+ | Homozygous Deletion | 0 | 0 |
| Nonsense Mutation | 0 | 0 | |
| Missense / Silent Mutation | 0 | 1 (p.P298S) | |
| WT | 3 | 32 | |
| HPV- | Homozygous Deletion | 13 | 0 |
| Nonsense Mutation | 2 (p.S242*; p.Q461*) | 0 | |
| Missense / Silent Mutation | 2 (p.W99C; p.R361H) | 2 (p.P544L; p.K46K) | |
| WT | 122 | 102 |
Figure 4.
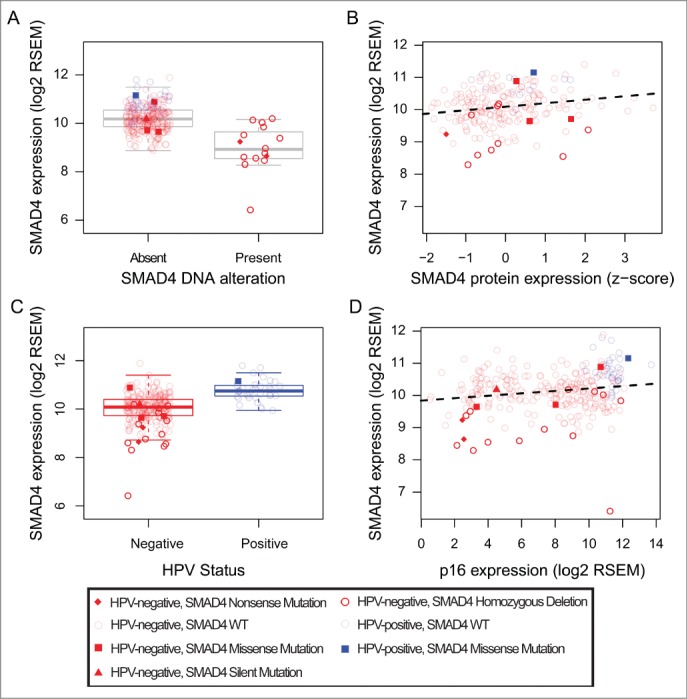
SMAD4 expression by SMAD4 alterations (homozygous deletions and nonsense mutations). Points are shaped and shaded as indicated in the figure and colored by HPV-status (red for HPV-negative, blue for HPV-positive). (A) HNSCCs with SMAD4 alterations have lower SMAD4 mRNA expression, (B) SMAD4 mRNA expression is positively correlated with Smad4 protein expression, (C) SMAD4 mRNA expression is higher in the HPV-positive HNSCCs compared to HPV-negative HNSCCs, and (D) SMAD4 mRNA expression is positively correlated with CDKN2A (p16) mRNA expression.
Discussion
Although the use of cetuximab has demonstrated positive clinical results in HNSCC treatment, de novo and acquired resistance to this targeted agent represent a significant challenges for improving patient outcome. Previous reports from our laboratory established ErbB-associated ligand upregulation and receptor crosstalk are correlated with acquired cetuximab resistance in HNSCC.19 However, additional HNSCC-specific modifications associated with this resistance required further investigation.
EMT has been well studied in developmental biology where this process leads to a migratory mesenchyme that later establishes mesodermal and endodermal tissues.26,27 In carcinogenesis, the migratory phenotype and increased plasticity associated with EMT provides many advantages for a developing malignancy to evolve and evade traditional therapies leading to distant metastasis. Thus, it is important to elucidate the molecular determinants of EMT in a disease-specific manner to improve therapeutic outcomes in HNSCC. Prior gene expression analysis of HNSCC from our laboratory demonstrated that a subset of this disease is associated with an upregulation of EMT-related genes, which then leads to poor clinical outcomes.28 Supporting this hypothesis, changes in EMT downstream markers (i.e., E-cadherin, vimentin) are associated with the development of metastasis in HNSCC.29 Additionally, putative HNSCC stem cells isolated from cell lines exhibit lower E-cadherin and higher vimentin levels, while also demonstrating increased invasiveness and EMT phenotypes.30
Smad4 is an established tumor suppressor in many cancer subtypes and its loss in HNSCC is associated with inactivating mutations or loss of heterozygosity at 18q21.9,31-35 The significance of Smad4 in the development of HNSCC was recently strengthened by Bornstein, et al. as they demonstrated conditional Smad4 loss (Smad4−/−) in the oral cavity of mice causes spontaneous tumors. The histology ranged from moderately to poorly differentiated squamous cell carcinomas and closely mirrored that of analogous human disease.21 The authors also showed that Smad4 loss resulted in genomic instability through the downregulation of DNA repair-related genes including Brca1, FancAm, FancD2 and Rad51, and subsequent abnormal centrosome amplification. In addition, Smad4 loss caused increased inflammation of the surrounding stroma characterized by numerous infiltrating leukocytes and increased inflammatory cytokines suggesting an upregulation of TGF-β1. Our current investigation adds to the existing data that SMAD4 downregulation contributes to the induction of EMT and is a common feature of acquired cetuximab resistance in HNSCC cell lines supporting important therapeutic and prognostic consequences of SMAD4 deregulation.
Lastly, we report that SMAD4 expression is low in a significant number of HNSCC, especially in HPV-negative tumors, while the genomic alteration is relatively rare (7%). Also, tumors with lower CDKN2A (p16) expression are likely to have lower SMAD4 expression. Considering that patients with HPV-negative HNSCC have worse survival compared to the HPV-positive tumors,24,36 low SMAD4 expression may contribute to poor outcome in HPV-negative patients. In addition, a retrospective analysis of a randomized phase III clinical trial comparing radiation to radiation plus cetuximab in locally advanced HNSCC patients demonstrated that p16 expression was strongly prognostic and HPV-positive patients benefited more from the addition of cetuximab to standard radiation therapy compared to the HPV-negative patients.37 We are still waiting for the phase III clinical trial data comparing radiation plus cisplatin versus radiation plus cetuximab in HPV-positive patients to determine the efficacy of cetuximab in this patient setting; however, high expression of SMAD4 in our data may suggest that HPV-positive patients may benefit from cetuximab.
To conclude, we have demonstrated that down-regulation of SMAD4 is an important mediator of EMT and cetuximab resistance. Further investigation is warranted to translate the findings to therapeutic development of cetuximab in HNSCC patients.
Materials and Methods
Cell lines and materials
SCC1 and 1CC8 represent an isogenic cell line pair, in which 1CC8 was selected in vitro as a model for acquired cetuximab resistance.18 SCC25 was purchased from American Type Culture Collection and additional cetuximab resistant clones of SCC25 were generated as previously published.18 Each cell line was authenticated using short tandem repeat (STR) analysis kit GenePrint 10 (Promega, Madison, WI) through the Johns Hopkins University Genetic Resources Core Facility. Cetuximab (Bristol-Myers Squibb, Princeton, NJ) was purchased from the Johns Hopkins Pharmacy.
Matrigel colony formation and cell migration assays
Matrigel colony formation assays were performed to assess the survival rate as previously described.19 For the cell migration assay, cells were seeded in a Transwell chamber (8-µM pore) coated with Collagen I. Migration was initiated across the membrane with a 0–10% FBS chemoattractant gradient. After 36 hrs, remaining cells were removed with a cotton swab, while migratory cells were fixed, stained, and quantified from 6 fields of view at 200X magnification. Data are presented as the average number of cells per field of view.
Quantitative real-time PCR
Total RNA was extracted as previously described.19 For real-time PCR, Taqman gene expression assays (Applied Biosystems, Foster City, CA) were performed per manufacturer's instructions. The assays utilized were SMAD4 (Hs00929647-m1) and β-actin (ACTB, Hs00357333-g1). Relative gene expression was normalized to β-actin by comparative Ct.
Stable knockdown of SMAD4
Cells were seeded in 6-well plates 24 hours prior to transduction. Per manufacturer's instructions, an appropriate amount of lentiviral transduction particles of SMAD4 (Sigma, NY) were added into the media in the presence with hexadimethrine bromide. After puromycin selection, cell pools were collected and knockdown was confirmed with qRT-PCR and Western blot.
Statistical analysis of HNSCC cell line microarrays
Raw array data was obtained for SCC1/1CC8 from GSE21483 and normalized with fRMA version 1.16.0.38 Differential gene expression associated with cetuximab resistance was assessed for SCC1/1CC8 using t-statistics moderated with empirical Bayes computed with the LIMMA Bioconductor package version 3.20.9. Gene set enrichment was inferred by applying a one-sided gene set test and implemented in the LIMMA package to the moderated t-statistics for SCC1/1CC8. The p-values reported for gene set enrichment of EMT were based upon the set of GO EMT probes from GO:0001837 identified in version 2.14.0 of the annotation for Affymetrix hgu133plus2.0.db package in Bioconductor. All reported probe-level p-values were adjusted for false discovery rate (FDR) using the Benjamini-Hotchberg correction. Probes with FDR adjusted p-value below 0.05 and absolute log fold-change greater than 0.5 were deemed significantly related to cetuximab resistance. This adjustment was performed only for the set of probes annotated to the EMT GO set, GO:0001837, hypothesized a priori to relate to cetuximab resistance. R code for all microarray preprocessing and analyses is provided in File S1.
Statistical analysis of TCGA data
Processed Level 3 mRNA expression, protein expression, copy number, and mutation data was obtained for the 279 TCGA HNSCC freeze-set samples used for the TCGA publication.9 HPV status was assessed by integrating viral detection in sequencing data and p16 status, as described in the TCGA publication.9 Samples were called to have a homozygous deletion in SMAD4 when its GISTIC2.039 score estimated from the copy number data was −2. The MutationAssessor functional impact scores 23 for reported SMAD4 mutations were obtained from cBioPortal.40 Gene expression values were reported as log transformed RSEM values 41 using version 2 of data preprocessing for TCGA RNA-sequencing data. Protein expression values were reported as z-scores measured from reverse phase protein arrays (RPPA) and were obtained from cBioPortal.40 The RPPA data were only available for 200 of the 279 samples (14 HPV-Positive and 186 HPV-Negative). SMAD4 was called significantly differentially expressed when the p-value for t-statistics comparing log transformed RSEM values were below 0.05. Samples were deemed as having DNA alterations in SMAD4 when they had either homozygous deletions or nonsense mutations. The maximum log transformed RSEM value of SMAD4 expression for these altered samples (10.2) was used to establish a threshold for low SMAD4 expression.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This project was supported by RO1 DE017982 and R21 DE023430 to CH Chung, and K25 CA141053 and R01 CA177669 to EJ Fertig.
References
- 1.Maier H, Dietz A, Gewelke U, Heller WD, Weidauer H. Tobacco and alcohol and the risk of head and neck cancer. Clin Investig 1992; 70:320-7; PMID:1521046 [DOI] [PubMed] [Google Scholar]
- 2.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et al.. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000; 92:709-20; PMID:10793107; http://dx.doi.org/ 10.1093/jnci/92.9.709 [DOI] [PubMed] [Google Scholar]
- 3.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med 2001; 345:1890-900; PMID:11756581; http://dx.doi.org/ 10.1056/NEJMra001375 [DOI] [PubMed] [Google Scholar]
- 4.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 2007; 356:1944-56; PMID:17494927; http://dx.doi.org/ 10.1056/NEJMoa065497 [DOI] [PubMed] [Google Scholar]
- 5.Grandis J, Melhem M, Gooding W, Day R, Holst V, Wagener M, Drenning S, Tweardy D. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst 1998; 90:824-832; PMID:9625170; http://dx.doi.org/ 10.1093/jnci/90.11.824 [DOI] [PubMed] [Google Scholar]
- 6.Grandis JR, Tweardy DJ. TGF-alpha and EGFR in head and neck cancer. J Cell Biochem Suppl 1993; 17F:188-91; PMID:8412192; http://dx.doi.org/ 10.1002/jcb.240531027 [DOI] [PubMed] [Google Scholar]
- 7.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, Jarrett C, Carter J, Murphy BA, Netterville J, et al.. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol 2006; 24:4170-6; PMID:16943533; http://dx.doi.org/ 10.1200/JCO.2006.07.2587 [DOI] [PubMed] [Google Scholar]
- 8.Chung CH, Zhang Q, Hammond EM, Trotti AM 3rd, Wang H, Spencer S, Zhang HZ, Cooper J, Jordan R, Rotman MH, et al.. Integrating epidermal growth factor receptor assay with clinical parameters improves risk classification for relapse and survival in head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2011; 81:331-8; PMID:20732768; http://dx.doi.org/ 10.1016/j.ijrobp.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015; 517:576-82; PMID:25631445; http://dx.doi.org/ 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker TP, Brown CD, Pugh TJ, Stojanov P, Cho J, et al.. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res 2015; 21:632-41; PMID:25056374; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, Agrawal N, Fakhry C, Quon H, Subramaniam RM, et al.. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol 2015; 26:; PMID:25712460; http://dx.doi.org/ 10.1093/annonc/mdv109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al.. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med 2006; 354:567-78; PMID:16467544; http://dx.doi.org/ 10.1056/NEJMoa053422 [DOI] [PubMed] [Google Scholar]
- 13.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, et al.. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med 2008; 359:1116-27; PMID:18784101; http://dx.doi.org/ 10.1056/NEJMoa0802656 [DOI] [PubMed] [Google Scholar]
- 14.Harris R, Chung E, Coffey R. EGF receptor ligands. Experimental Cell Research 2003; 2003:2-13; PMID:12648462; http://dx.doi.org/ 10.1016/S0014-4827(02)00105-2 [DOI] [PubMed] [Google Scholar]
- 15.Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol 2005; 76:157-61; PMID:16024112; http://dx.doi.org/ 10.1016/j.radonc.2005.06.022 [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Albaitero A, Ferris RL. Immune activation by epidermal growth factor receptor specific monoclonal antibody therapy for head and neck cancer. Arch Otolaryngol Head Neck Surg 2007; 133:1277-81; PMID:18086972; http://dx.doi.org/ 10.1001/archotol.133.12.1277 [DOI] [PubMed] [Google Scholar]
- 17.Bedi A, Chang X, Noonan K, Pham V, Bedi R, Fertig EJ, Considine M, Califano JA, Borrello I, Chung CH, et al.. Inhibition of TGF-beta enhances the in vivo antitumor efficacy of EGF receptor-targeted therapy. Mol Cancer Ther 2012; 11:2429-39; PMID:22927667; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0101-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, Gondi V, Hsu KT, Harari PM. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene 2008; 27:3944-56; PMID:18297114; http://dx.doi.org/ 10.1038/onc.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatakeyama H, Cheng H, Wirth P, Counsell A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough WG, et al.. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One 2010; 5:e12702; PMID:20856931; http://dx.doi.org/ 10.1371/journal.pone.0012702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. http://www.geneontology.org [Google Scholar]
- 21.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S, et al.. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest 2009; 119:3408-19; PMID:19841536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. www.cbioportal.org [Google Scholar]
- 23.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 2011; 39:e118; PMID:21727090; http://dx.doi.org/ 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, et al.. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010; 363:24-35; PMID:20530316; http://dx.doi.org/ 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung CH, Zhang Q, Kong CS, Harris J, Fertig EJ, Harari PM, Wang D, Redmond KP, Shenouda G, Trotti A, et al.. p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol 2014; 32:3930-8; PMID:25267748; http://dx.doi.org/ 10.1200/JCO.2013.54.5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakaya Y, Sukowati EW, Alev C, Nakazawa F, Sheng G. Involvement of dystroglycan in epithelial-mesenchymal transition during chick gastrulation. Cells Tissues Organs 2011; 193:64-73; PMID:21051858; http://dx.doi.org/ 10.1159/000320165 [DOI] [PubMed] [Google Scholar]
- 27.Massague J. TGFbeta in cancer. Cell 2008; 134:215-30; PMID:18662538; http://dx.doi.org/ 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, Ang KK, El-Naggar AK, Zanation AM, Cmelak AJ, et al.. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-{kappa}B signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res 2006; 66:8210-8218; PMID:16912200; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-1213 [DOI] [PubMed] [Google Scholar]
- 29.Nijkamp MM, Span PN, Hoogsteen IJ, van der Kogel AJ, Kaanders JH, Bussink J. Expression of E-cadherin and vimentin correlates with metastasis formation in head and neck squamous cell carcinoma patients. Radiother Oncol 2011; 99:344-8; PMID:21684617; http://dx.doi.org/ 10.1016/j.radonc.2011.05.066 [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Wei Y, Hummel M, Hoffmann TK, Gross M, Kaufmann AM, Albers AE. Evidence for epithelial-mesenchymal transition in cancer stem cells of head and neck squamous cell carcinoma. PLoS One 2011; 6:e16466; PMID:21304586; http://dx.doi.org/ 10.1371/journal.pone.0016466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al.. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996; 271:350-3; PMID:8553070; http://dx.doi.org/ 10.1126/science.271.5247.350 [DOI] [PubMed] [Google Scholar]
- 32.Kim SK, Fan Y, Papadimitrakopoulou V, Clayman G, Hittelman WN, Hong WK, Lotan R, Mao L. DPC4, a candidate tumor suppressor gene, is altered infrequently in head and neck squamous cell carcinoma. Cancer Res 1996; 56:2519-21; PMID:8653689 [PubMed] [Google Scholar]
- 33.Takebayashi S, Ogawa T, Jung KY, Muallem A, Mineta H, Fisher SG, Grenman R, Carey TE. Identification of new minimally lost regions on 18q in head and neck squamous cell carcinoma. Cancer Res 2000; 60:3397-403; PMID:10910046 [PubMed] [Google Scholar]
- 34.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al.. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011; 333:1154-7; PMID:21798897; http://dx.doi.org/ 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al.. The mutational landscape of head and neck squamous cell carcinoma. Science 2011; 333:1157-60; PMID:21798893; http://dx.doi.org/ 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rischin D, Young RJ, Fisher R, Fox SB, Le QT, Peters LJ, Solomon B, Choi J, O'Sullivan B, Kenny LM, et al.. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol 2010; 28:4142-8; PMID:20697079; http://dx.doi.org/ 10.1200/JCO.2010.29.2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenthal D, Harari P, Giralt J, Bell D, Raben D, Liu J, Schulten J, Ang K, Bonner J. Impact of p16 status on the results of the phase III cetuximab (cet)/radiotherapy (RT). J Clin Oncol 2014; 32:suppl; abstr 6001 [Google Scholar]
- 38.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics 2010; 11:242-53; PMID:20097884; http://dx.doi.org/ 10.1093/biostatistics/kxp059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011; 12:R41; PMID:21527027; http://dx.doi.org/ 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12:323; PMID:21816040; http://dx.doi.org/ 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.