Abstract
Ubiquitin-specific protease 22 (USP22) is closely related with poor prognosis of cancer patients. However, the role of USP22 expression in nasopharyngeal carcinoma (NPC) has not been determined. The main aim of this study was to determine the role of USP22 in the pathologic processes of NPC. Immunohistochemistry (IHC), western blot (WB), and real-time polymerase chain reaction (RT-PCR) were used to measure the expression of USP22 in cell lines and tissues of NPC in comparison with expression in non-cancerous cells and tissues. USP22-specific short hairpin RNA (shRNA) was used to knock down USP22 expression in the NPC cell line CNE-1 and CNE-2. Furthermore, the impact of USP22 in cellular proliferation, growth, and cell cycle were detected respectively. WB was used to determine the role of USP22 in the AKT/GSK-3/Cyclin signaling pathway. The expression levels of USP22 were remarkably higher in NPC cell lines and tissues. With cell counting and the MTS assay, cellular growth and proliferation progression of USP22 knockdown cell line was shown to be effectively restrained. The USP22 silencing both in CNE-1 and CNE-2 cells caused them to accumulate in the G0/G1 phase of the cell cycle. USP22 knockdown was also found to modulate the AKT/GSK-3/Cyclin pathway, resulting in downregulation of p-AKT, p-GSK-3β, and cyclinD1. This study suggests that USP22 plays a critical regulatory role in the pathologic processes of NPC, and that it may be a potential biological treatment target in the future.
Keywords: Nasopharyngeal carcinoma, USP22, cell growth, cell proliferation, cell cycle, AKT/GSK-3/Cyclin pathway
Abbreviations
- DUBs
Deubiquitinating Enzymes
- EB
Epstein-Barr
- hSAGA
human Spt-Ada-Gcn5 acetyltransferase
- IHC
Immunohistochemistry
- NC
Negative Control
- NPC
Nasopharyngeal carcinoma
- ORF
Open Reading Frame
- RT-PCR
real-time polymerase chain reaction
- shRNA
short hairpin RNA
- shUSP22
small hairpin RNA of USP22
- USP22
Ubiquitin-specific Protease 22
- WB
Western Blot
Introduction
Nasopharyngeal carcinoma (NPC) is a head and neck malignant tumor derived from the nasopharyngeal epithelium with remarkable geographic and ethnic worldwide distribution. NPC is a rare malignant tumor with an annual incidence well under 1 per 100,000 persons in Caucasians from North America and other Western countries. Conversely, the highest incidence is found among Southern Chinese (the annual incidence is approximately 25–30 per 100,000 persons per year), especially those of Cantonese descent.1,2 In recent years, significant advances have been made in diagnostic techniques and therapy for NPC, however the survival rate for patients with NPC has not experienced a proportional increase, due to recurrence and distant metastases.3 Except for the obvious differences in geographic and racial distribution, Epstein-Barr (EB) virus infection, genetic susceptibility, and environmental factors have been considered to be the main risk factors for NPC.4 Up to now, knowledge on the accurate molecular mechanism underlying the onset and progression of NPC has been extremely limited. The molecular pathogenesis of NPC includes abnormal expression and alteration of dominant oncogenes, tumor-suppressor genes, and signaling pathways, including the Akt pathway, mitogen-activated protein kinases, and the Wnt signaling pathway.5 Therefore, more in-depth studies on the molecular mechanisms of NPC are imperative for the exploration of novel effective therapeutic strategies.
Previous studies have found that USP22 is one of 11-genes in the Polycomb group with a cancer stem cell signature, which can encode various proteins that promote invasive tumor growth. The degree of expression of these proteins is associated with short-term tumor recurrence, distant metastatic potential, and chemotherapy-resistance. The level of expression can also powerfully predict the therapeutic outcome of patients with a wide variety of cancers.6–8 Sequence analysis reveals that USP22 is a newly discovered ubiquitin specific peptidase gene, which belongs to the deubiquitinating enzymes (DUBs) protease family, which have ubiquitin hydrolase activity.9,10 In humans, genomic USP22 is located on chromosome 17 and is composed of 14 exons; its cDNA includes an open reading frame (ORF) of 1578 bp, encoding 525 amino acids and a highly conserved ubiquitin enzyme family area.9 Furthermore, previous analysis showed that USP22 is an enzymatic subunit of the human Spt-Ada-Gcn5 acetyltransferase (hSAGA) cofactor complex, which is required for activator-driven transcription, and can function as an activator for nuclear receptor-mediated transactivation, regulating the expression of genes related to the progression of oncogenesis.11,12 Within hSAGA, USP22 causes deconjugation of ubiquitin from both the H2A and H2B histones and regulates transcription associated with epigenetic alteration and cancer progression.11 In addition, USP22 has been shown to regulate cellular growth and proliferation; most importantly, studies have found that knock-down of USP22 reduces transformation of c-Myc and plays an important role in cell cycle progression and anchorage-independent growth, which encourages cancerous growth. Additionally, the down-regulation of USP22 results in tumor cells remaining arrested in the G1 phase.10,13,14, Liu, et al., identified that USP22 may act as an oncogene modulating the cell cycle of the colorectal cancer cell line HCT116, both through the BMI-1-mediated INK4a/ARF pathway and the Akt pathway. The downregulation of USP22 expression in HCT116 cells resulted in cells accumulating in the G1 phase of the cell cycle via the BMI-1-mediated Akt pathway.15 Therefore, the role of USP22 in cell growth and proliferation, hSAGA-mediated transcription, and the cell cycle suggests that USP22 may be a significant regulator of biological processes and a potential therapeutic target in cancers.
Recent experimental and clinical studies have shown that overexpression of USP22 is highly correlated with invasion, metastasis, and poor prognosis of certain malignant tumors, such as salivary duct and esophageal carcinomas, breast, gastric, colorectal, and cervical cancers, among others.16–21 Scholars have also found that silencing USP22 expression can significantly induce inhibition of cell proliferation and arrest of the cell cycle in bladder cancer, colorectal cancer, and human brain glioma cell lines.22–24 These results strongly support the idea that carcinogenic activation of USP22 may lead to the invasive progression and poor prognosis of cancers. However, until now, the expression of USP22 in NPC cells and its role in the accurate molecular mechanisms underlying the pathogenesis and progression of NPC has not yet been elucidated. In this study, we investigated the aberrant expression of USP22 in NPC, as well as the effect of the knockdown of the USP22 gene on NPC cellular growth, proliferation, and cell cycle progression. We also sought to illuminate its molecular mechanism in the Akt signaling pathway. Finally, we are committed to exploring both molecular diagnostics and novel treatments of NPC in the future.
Results
Elevated USP22 expression levels were seen in cell lines and tissues of NPC, compared with levels in non-cancerous nasopharyngeal mucosa and cells
In order to evaluate whether USP22 activation may be due to NPC carcinogenesis, immunohistochemistry analysis was used to examine expression of USP22 in the paired NPC and non-cancerous nasopharyngeal mucosa tissues. Immunohistochemistry analysis showed that more USP22 staining appearing as brown particles was localized within the cell of NPC tissues, which means that higher expression levels of USP22 in NPC were observed, when compared to corresponding adjacent non-cancerous mucosal tissues (Fig. 1A). Many studies have shown that the increased expression of an oncogene is often involved in gene amplification.16-24 In agreement with the envisaged carcinogenic effects of USP22, RT-PCR analysis showed that the expression levels of USP22 mRNA in NPC tissues were higher than in normal nasopharyngeal mucosa (Fig. 1B), which was consistent with the results of USP22 protein immunohistochemistry analyses. The results indicate that USP22 mRNA and proteins are overexpressed in NPC tissues.
Figure 1.
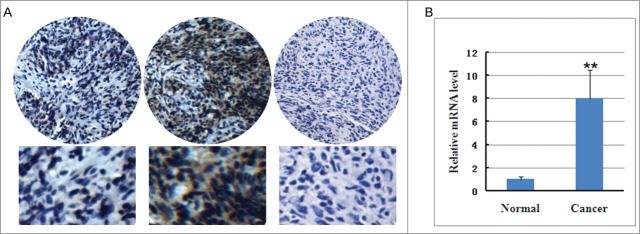
USP22 protein and mRNA expression in nasopharyngeal carcinoma tissues and non-cancerous nasopharyngeal mucosa. (A) The expression of USP22 in nasopharyngeal carcinoma tissue and non-cancerous nasopharyngeal mucosa via immunohistochemistry analysis. lefe pannel: non-cancerous nasopharyngeal mucosa; middle pannel: nasopharyngeal carcinoma tissues; right pannel: negative control without USP22 antibody. (B) USP22 mRNA expression in nasopharyngeal carcinoma tissue and non-cancerous nasopharyngeal mucosa via quantitative RT-PCR analysis. All experiments were performed in triplicate and results are expressed as means ± SD analyzed with t-test. **: P < 0.01.
We also studied the expression level of USP22 in NPC cell lines compared with normal nasopharyngeal cells. To obtain the expression data of USP22 protein, we performed a WB analysis, which revealed that protein levels in nasopharyngeal cell lines, including CNE-1, CNE-2, Sune-5–8F, and Sune-6–10B cells, were higher than in non-cancerous nasopharyngeal cells (Fig. 2A). In addition, RT-PCR analysis was utilized to detect the expression level of USP22 mRNA. USP22 mRNA were highly expressed in CNE-1, CNE-2, Sune-5–8F, and Sune-6–10B NPC cell lines, and the expression levels were markedly higher than in normal nasopharyngeal cells. Additionally, the quantification analysis showed that USP22 mRNA expression in the CNE-2 and CNE-1 cell lines were the highest (Fig. 2B). Therefore, the CNE-2 and CNE-1 cell lines were selected for further research. The results indicate that USP22 protein and mRNA are over expressed in the CNE-1, CNE-2, Sune-5–8F, and Sune-6–10B cell lines of nasopharyngeal carcinoma.
Figure 2.
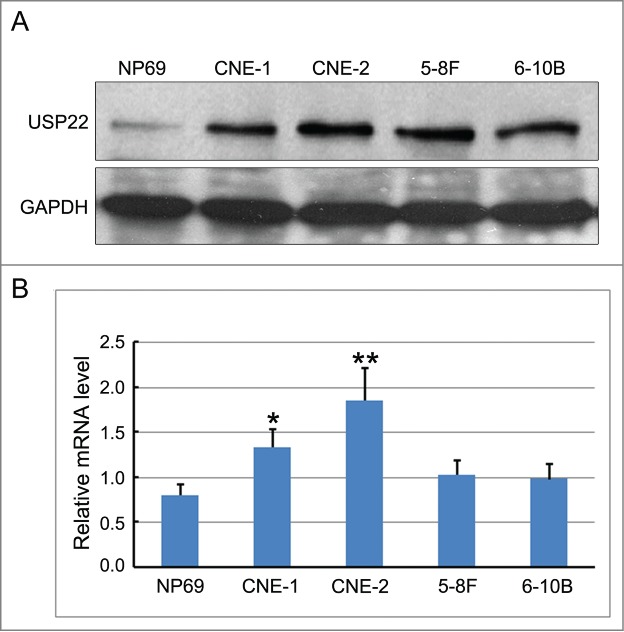
USP22 protein and mRNA expression in nasopharyngeal carcinoma cell lines and non-cancerous nasopharyngeal cells. (A) USP22 protein expression in CNE-1, CNE-2, Sune-5–8F, and Sune-6–10B cell lines of nasopharyngeal carcinoma and non-cancerous nasopharyngeal cells via Western blot analysis. (B) USP22 mRNA expression in CNE-1, CNE-2, Sune-5–8F, and Sune-6–10B cell lines of nasopharyngeal carcinoma and non-cancerous nasopharyngeal cells via RT-PCR analysis. All experiments were performed in triplicate and results are expressed as means ± SD analyzed with ANOVA. *: P < 0.01.
Inhibition of USP22 expression by shRNA and establishing the CNE-2-shUSP22 and CNE-1-shUSP22 stable cell lines
Lentiviral vector mediated RNA interference technology was used to infect the NPC CNE-2 and CNE-1 cells with negative control (NC) and USP22-special shRNA lentivirus, generating CNE-2-shUSP22 and CNE-1-shUSP22 stable cell lines (Fig. 3A). Transfection of CNE-2 and CNE-1 cells with USP22-special shRNA resulted in a marked down-regulation of USP22 protein levels, compared with NC cells, as determined by western blot (Fig. 3B). RT–PCR identified that infection of NPC CNE-2 and CNE-1 cells with USP22-special shRNA resulted in an obvious downregulation of USP22 mRNA expression compared with NC cells (Fig. 3C). All of these results indicate that USP22-special shRNA infection can effectively inhibit the USP22 gene and protein expression in NPC CNE-2 and CNE-1 cells.
Figure 3.
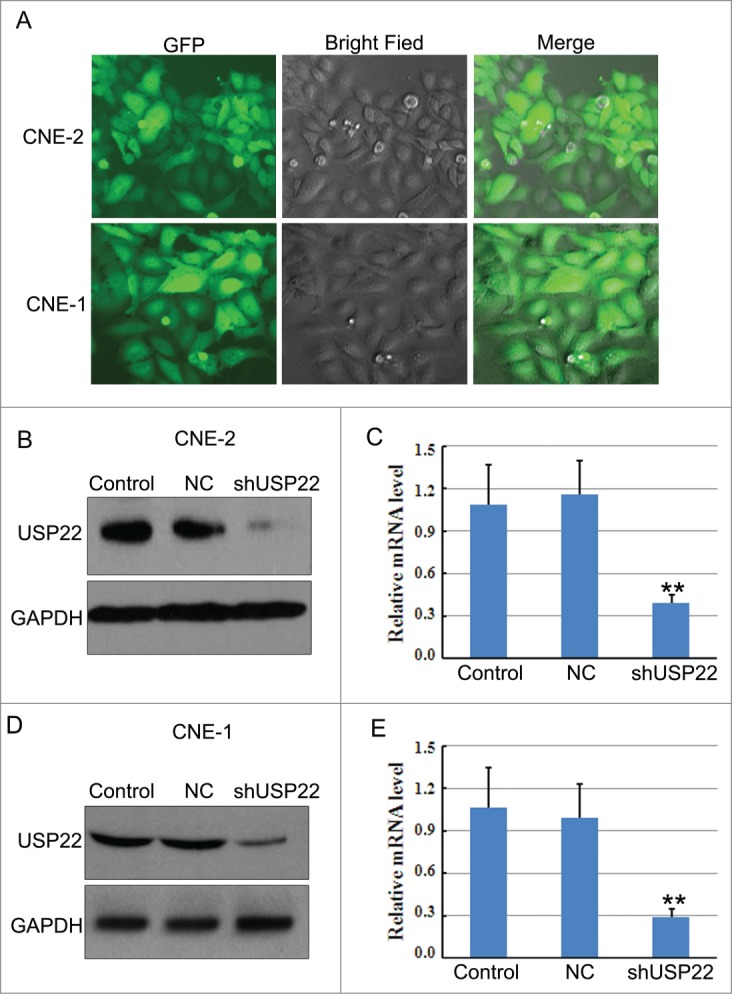
RNA interference to knock down USP22 gene expression in NPC cells. (A) CNE-1 and CNE-2 cells were transfected with USP22-special and control shRNA. (B) Western blot and (C) quantitative RT-PCR analysis were used to determine interference efficiency following transfection in CNE-1 and CNE-2 cells. All experiments were performed in triplicate and results are expressed as means ± SD analyzed with ANOVA. **: P < 0.01
Down-regulation of USP22 expression reduces cellular viability and induces G0/G1 cell cycle arrest
In order to investigate whether downregulation of USP22 expression alters cellular proliferation of NPC cells, the MTS assay was used to assess the viability of CNE-2-NC and CNE-2-shUSP22 stable cells at 24, 48, and 72 h. In comparison to negative control cells, CNE-2-shUSP22 cells significantly reduced cellular proliferation (Fig. 4A). The examination of whether RNA interference-mediated down-regulation of USP22 expression affects the growth of NPC cells was conducted through the analysis of the viability of CNE-2-NC and CNE-2-shUSP22 cells at 24, 48, and 72 h with a cell counting assay. Similar results were obtained from CNE-1-NC and CNE-1-shUSP22 cells (Fig. 4B). The results indicate that knockdown USP22 expression with USP22 shRNA significantly reduced the cell growth of human nasopharyngeal carcinoma cell lines.
Figure 4.
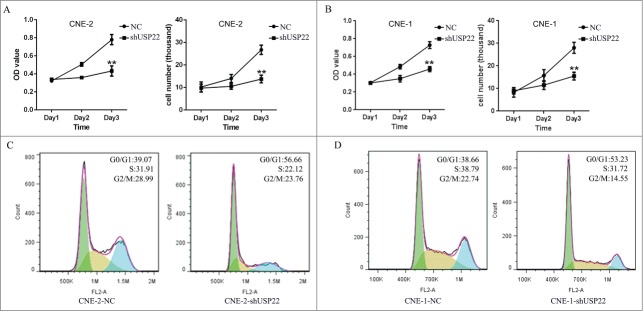
Down-regulation of USP22 expression reduces cellular viability and induces G0/G1 cell cycle arrest in NPC cells. (A) MTS assay and cell counting assay were performed to analyze proliferation viability and growth viability of CNE-2 and CNE-1 stable cell lines (B), respectively. The result showed that downregulation of USP22 expression inhibits cellular cell growth and. Proliferation. (C) USP22 expression silencing arrested CNE-2 cells and CNE-1 cells (D) in the G0/G1 phase, as determined by flow cytometric analysis. Each independent experiment was repeated 3 times. All experiments were performed in triplicate and results are expressed as means ± SD analyzed with ANOVA. **: P < 0.01
Simpson, et al.25, and Liang, et al.,26 identified that the cell cycle is linked to proliferation, and that its arrest can result in inhibition of proliferation. We measured whether the alterations of cell viability mediated by downregulation of USP22 expression were associated with cell cycle arrest. Flow cytometric analysis was used to analyze the cell cycle. Under down-regulation of USP22, there were 17.59% and 14.57% increase in the number of CNE-2 and CNE-1 cells in the G0/G1 phase (56.66% and 53.23%), with a concomitant 9.79% and 7.07% decrease of those found in the S phase (22.12% and 31.72%), compared with the control cell lines (Fig. 4C, D), suggesting that the majority of the cells were accumulated in the G0/G1 phase. These results indicate that USP22 may play a potential role in nasopharyngeal tumorigenesis by modulating cell viability and cell cycle transition.
USP22 acts as an oncogene by activating the AKT/GSK-3/Cyclin signaling pathway
Alterations of Akt pathway also play a key role in G1 activation, which lead to sustained G1/S transition.27 Recently, Kida et al.28 reported that cell-cycle progression is regulated by Akt activation through phosphorylation and consequent inhibition of glycogen synthase kinase 3β to avert cyclin D1 degradation.29,30 Since we have found that downregulation of USP22 cause significant G1 arrest, we turned to check the changes of Akt pathway to further investigate the molecule mechanism affecting the cell cycle by USP22. Western blot was used to detect the key protein of the AKT/GSK-3/Cyclin signaling pathway, including the cycle-related proteins p-AKT, p-GSK-3β, cyclinD1, GSK-3β, and AKT. As can be seen in Figure 5, the knockdown of USP22 resulted in down-regulation of p-AKT, p-GSK-3β, and cyclinD1 and was accompanied with an up-regulation of GSK-3β and AKT. As not a protein kinase, USP22 is unable to activate the PI3K/Akt pathway directly. The polycomb group protein Bmi-1, a well-known USP22 target, induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells, through repressing the expression of tumor suppressor PTEN, which negatively regulates the PI3K/Akt pathway.31 We further investigated the level of Bmi-1 and PTEN when knockdown USP22, WB results shown an obviously decrease of Bmi-1 and a significant upregulation of PTEN (Fig.5). Therefore, the results suggest that USP22 regulates the cell cycle in NPC, via affecting the crucial protein in the Bmi-1/PTEN/AKT/GSK-3/Cyclin signaling pathway.
Figure 5.
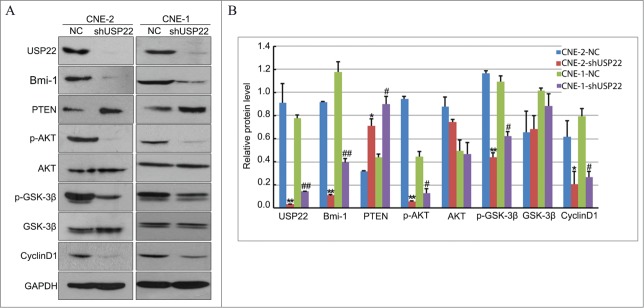
Konckdown of USP22 expression inactivates AKT/GSK-3/Cyclin signaling pathway. (A) Western blot was used to analyze the expression of USP22, Bmi-1, PTEN, p-AKT, AKT, p-GSK-3β, GSK-3β, and cyclinD1 in the CNE-2 and CNE-1 cell lines. GAPDH serve as a loading control. (B) Analysis the gray level of western blot results using ImageJ. The relative protein levels were indicated as bars. All experiments were performed in triplicate and results are expressed as means ± SD analyzed with ANOVA. **: P < 0.01 vs CNE-2 NC, *: P < 0.05 vs CNE-2 NC, ##: P < 0.01 vs CNE-1 NC, #: P < 0.01 vs CNE-1 NC.
Discussion
USP22, a novel deubiquitinating enzyme that regulates numerous cellular mechanisms, including pre-implantation, growth and differentiation, cell cycle progression, transcriptional activation, signal transduction, and oncogenesis, is the core of a multitude of physiological and pathological processes.32 USP22 was recently identified as one of a small set of 11 biological markers capable of predicting potential metastatic-enabling and poor prognosis in human malignant tumors.6-8 Glinsky, et al., recently reported that the highly expressed gene signature in primary tumors was associated with shorter intervals of tumor recurrence, more distant metastases, and poorer prognosis in patients diagnosed with 11 distinct types of cancer, including five epithelial malignancies (prostate, breast, lung, ovarian, and bladder cancers) and five non-epithelial (lymphoma, mesothelioma, medulloblastoma, glioma, and acute myeloid leukemia) malignancies.7 Numerous studies have identified that knockdown of USP22 can significantly inhibit proliferation, promote apoptosis, and arrest the cell cycle of various tumor cells.22-24 However, little is known about the effects of the downregulation of the USP22 gene in NPC cells. Therefore, in this study, we investigated the role of USP22 in NPC and its molecular mechanisms.
Immunohistochemistry analysis and quantitative RT-PCR were performed, showing a higher USP22 protein and mRNA expression in NPC tissues and cell lines compared with non-cancerous nasopharyngeal mucosa tissues and cells. This suggests that over-expression of USP22 may represent a novel biological marker and treatment target for NPC.
Down-regulation of USP22 expression was achieved by small RNA interfering technology, and was assessed by western blot and RT–PCR. The biological status of the NPC cells was then analyzed. The knockdown of the USP22 gene resulted in a reduced replication rate of NPC cells, revealing that the USP22 gene plays a regulatory role in NPC cell growth and proliferation. Previous studies have shown that the USP22 gene is highly expressed in salivary duct carcinoma and esophageal, breast, gastric, colorectal, and cervical cancer cells.16–21 USP22 also has been shown to promote the proliferation of those malignant tumors cells. Consistent with these experimental results and analyses, in our study, USP22 was demonstrated to modulate oncogenic progression of NPC cells, including growth and proliferation.
Moreover, we observed that downregulation of USP22 leads to cellular growth and proliferation inhibition, but also induces cell cycle arrest. Flow cytometry revealed that down-regulation of USP22 gene expression resulted in the accumulation of cells in the G0/G1 phase of the cell cycle, with a concomitant decrease in the S and G2/M phases. This is consistent with previous studies that have demonstrated that knockdown of USP22 gene expression can significantly induce cell cycle arrest of human brain gliomas, as well as bladder, and colorectal cancers22–24, suggesting the requirement for USP22 in the cell cycle.
AKT/GSK-3 pathway played an important role in cell cycle regulation.33 Bmi-1 is a well-known target of USP22.34 Previous report show that Bmi-1 plays an important role in the pathogenesis of NPC partially by targeting PTEN, thus activating the PI3K/Akt pathway.35 Thus we propose that USP22 may activate AKT/GSK-3 pathway by targeting Bmi-1 and subsequently PTEN. In the current study, results confirm our hypothesis. We detected the key protein of the AKT/GSK-3/Cyclin signaling pathway. Western blot analyses showed the knockdown of USP22 resulted in downregulation of p-AKT, p-GSK-3β, and cyclinD1, as showed in Figure 5. Decrease in expression of Bmi-1 and a significant upregulation of PTEN were observed in USP22-depleted cells, which consistent with previous study showing an inverse relationship between Bmi-1 and PTEN expression in NPC cells and human tumors.35 Our results suggest that the knockdown of the USP22 gene results in a reduction in the number of NPCs undergoing cell division, which leads to a subsequent inhibition of cell replication via a Bmi-1/PTEN/AKT/GSK-3 pathway. Since Bmi-1 is a target of USP22 and function in the pathogenesis of NPC, our result also indicated a role of USP22 in NPC progression and metastasis.
In summary, over expression of the USP22 gene in NPC pathologic tissues and NPC cells was identified for the first time and USP22-special shRNA-mediated down-regulation of the USP22 gene was found to restrain NPC cell growth and proliferation via cell cycle arrest by downregulation of Bmi-1/PTEN, p-AKT, p-GSK-3β, and cyclinD1, and was accompanied with an upregulation of GSK-3β and AKT. We demonstrated that USP22 plays a significant but unknown role in NPC progression within the NPC pathologic tissue and the NPC cell line. The established correlation among USP22, cell-cycle progression, and the Akt signaling pathway in NPC cells indicates the possibility that the loci regulated by USP22 may encode attractive therapeutic targets. Therefore, the specific enzymatic inhibitor to USP22 may have therapeutic significance to treat NPC.
Materials and methods
Cell culture
Human nasopharyngeal carcinoma cell line CNE-1, CNE-2, SUNE-5–8F, SUNE-6–10B and Control cell line NP69 were cultured in RPMI-1640 medium (GIBCO), containing 10% fetal bovine serum,incubated in an incubator (Thermo) including 5% CO2 and 95% O2 at 37°C. When the cells reached the logarithmic growth phase, the next experimental analyses were performed.
Generation of stable cell lines
The vector LV-008 (from Forevergen Biosciences) with an U6 promoter was used to generate small hairpin RNAs. The following oligonucleotides were subcloned into the HpaI/XhoI sites: small hairpin RNA of USP22 (shUSP22), The USP22 shRNA sequence is as follows: sense 5′-AACTCACGGACAGTCTC-AACAATTTCAAGAGAATTGTTGAGACTGTCCGTGTTTTTTC-3′,and antisense 3′-TTGA-GTGCCTGTCAGAGTTGTTAAAGTTCTCTTAACAACTCTGACAGGCACAAAAA-AGAGCT-5′. Negative Control (NC) were used as the control group, sense 5′-AACTTTCTCCGAACGTGTCAC-GTTTCA-AGAGAACGTGACACGTTCGGAGAATTTTTTC-3′; antisense 3′-TTGAAAGAGGCTTGCACAGTGCAAAG-TTCTCTTGCACTGTGCAAGCCTCTTAAAAAAGAGCT-5'.
Lentiviral production was performed as described. Briefly, HEK 293T cells were cotransfected with LV-008 and packaging vectors, and the resulting supernatant was collected after 48 and 72 h. Lentiviruses were recovered after ultracentrifugation for 1.5 h at 25,000 rpm in a Beckman Instruments (Fullerton) SW28 rotor and resuspended in PBS. Infections were carried out in the presence of 5–10 μg/ml of polybrene. After 48 hours, the cells were cultured in the medium with 2 μg/ml puromycin for 10–15 days to generate CNE-2-NC, CNE-2-shUSP22, CNE-1-NC and CNE-1-shUSP22 stable cell lines.
RNA extraction and RT-PCR determination
Total RNA was isolated from tissue samples with Trizol (Invitrogen), according to the manufacturer's instructions. RNA was reverse transcribed with the ReverTra Ace qPCR RT Kit (Toyobo Biochemicals), also according to the manufacturer's protocol. Sequence-specific primers for USP22 and GAPDH were, respectively, as follows:
USP22, 5′-CCATTGATCTGATGTACGGAGG-3′ (forward)
and 5′-TCCTTGGCGATTATTTCCATGTC-3′ (reverse); GAPDH,
5′ -GAGTCAACGGATTTGGTCGT-3′ (forward) and
5′ -GACAAGCTTCCCGTTCTCAG-3′ (reverse). Real-time PCR was performed with GoTaq® qPCR Master Mix (Promega) on a MiniOpticon™ Real-Time PCR detection instrument (Bio-Rad) using the SyBr Green detection protocol, as outlined by the manufacturer. Briefly, the amplification mixture consisted of 0.5 μM primers, 25 μl of GoTaq® qPCR Master Mix, and 1 μl template DNA in a total volume of 40 μl. Samples were amplified as follows: initial denaturation at 98°C for 30 seconds, followed by 40 cycles of denaturation for 15 seconds at 98°C and annealing/elongation for 60 seconds at 60°C. All PCRs were run in triplicate, and control reactions without template were included.
Western blot analysis
Cells of all experimental groups were collected using a cell scraper and proteins were extracted by cell lysis buffer. Protein concentration was assessed with a Bradford assay and equal amounts of total protein were separated by SDS-PAGE electrophoresis, then transferred to PVDF membranes, and blocked with 5% skimmed milk powder for 1 hour at room temperature. Next, PVDF membranes were washed with TBST (containing Nacl, Tris-Hcl and tween-20) and incubated overnight with primary antibodies against target proteins at 4°C, followed by twice TBST wash. USP22 antibody was from Abcam; Bmi-1, PTEN, AKT, p-AKT Ser473,GSK-3β, p-GSK-3β Ser9 antibodies were from Cell Signaling Technology, CyclinD1 antibody was from Santa Cluz. Membranes were incubated with appropriate special secondary antibodies for 1 hour at room temperature and washed three times with TBST. Proteins were visualized by chemiluminescence (ECL, Forevergen).
Immunohistochemistry
Ten pairs of paraffin-embedded tissue blocks were collected from the NPC and adjacent nasopharyngeal mucosa tissue specimens, then all blocks were cut into 4 mm using microtome and pasted onto the slide. The tissue sections were dewaxed in xylene and rehydrated through graded alcohol concentrations according to standard procedures. The sections were subsequently submerged in EDTA (pH 8.0) and autoclaved at 121°C for 5 min to retrieve the antigenicity. After washing in phosphate-buffered saline (PBS, 0.1 M, PH 7.4, 3 times for 5 min), endogenous peroxidase was blocked by incubation in 3% hydrogen peroxide for 15 min at room temperature. Then, incubation with the USP22 antibody (Abcam) diluted at 1:400 in PBS containing 0.5% BSA was carried out overnight at 4°C in a moist chamber, followed by further washing with buffer to remove the unbound antibody. The sections were incubated with the biotin-labeled secondary antibody, followed by peroxidase-conjugated streptavidin for 30 min. 3, 30-diaminobenzidine tetrahydrochloride (Dako) was added in order to visualize the reaction, followed by counterstaining with commercial hematoxylin, whereupon it was sequentially dehydrated in alcohol and xylene and mounted. Negative controls with PBS were obtained by omitting the primary antibodies.
Cell count assay
Cells without activity can be stained with Trypan blue solution, while viable cells will not be stained. 10μl of cell suspension was added to 30μl PBS(GIBCO)containing 0.6% Trypan blue. After mixing, the cell suspension was added to a Freshney counting plate. Next, the number of viable cells on each corner of one side of the counting plate surrounded by three lines was calculated (C1). Then cells on the other side of the plate were also calculated (C2); cells on the pressure line were not counted. The following equation was using to calculate the number of living cells: C = (C1+C2)×1 × 104 (note: 104 is the conversion coefficient for the Freshney counting plate and C is the original cell concentration per milliliter.)
Cell growth curves
Cells were digested into a single cell suspension by 0.25% Trypsin-EDTA(1X)(GIBCO). A Freshney counting plate was used to count the number of living cells, then the cell concentration was adjusted to 1 × 105 / ml. 1ml cells, approximately 1 × 105 cells, were seeded in three wells of a 12-well plate. After digestion by trypsin, the living cells were counting at 24 h, 48 h, and 72 h. The experiments were repeated three times and averages were used to draw the cell growth curves.
MTS assay
Cell viability at different time points was detected with an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. An MTS assay was used to quantitate cell viability of CNE-2 and CNE-1 stable cell lines in 96-well flat-bottom plates. Cells were plated at a density of 1 × 103 per well in 96-well plates. After 24 h of culture, test drugs were added and the cultures were incubated at 37°C for different time periods (0 h, 24 h, 48 h, and 72 h). The treated cells were then stained with MTS reagent in complete medium with a ratio of 1:10 for 4 h. The absorbance values of each well were measured using a microplate reader (Diatek) at a wavelength of 490 nm. Each experiment was performed in triplicate and repeated three times. The mean value was calculated and the proliferation curves were constructed.
Cell cycle analysis with flow cytometry
Cells were collected by trypsinization and washed 3 times with PBS. Cells were then fixed with 1 ml of 70% ice-cold ethanol overnight at 4°C. After washing twice with PBS and centrifugation for 10 min at 1,000 rpm, using a centrifugal machine (Eppendorf), the supernatant was discarded and the cell pellets were stained with 50 μg/ml of propidium iodide and 100 U/ml of RNase A, and incubated in PBS for 30 min at room temperature in the dark. The percentage of cells in the different phases of the cell cycle was determined by evaluating DNA content. A total of 10,000 cells per sample were counted. Cytometric analysis was performed on a flow cytometer (BD) with Cell Quest software. Each assay was performed in triplicate.
Ethical standards
All human studies have been approved by The Institute Research Medical Ethics Committee of Guangzhou Medical University. All human studies have been performed in accordance with the Helsinki Declaration of 1975. All persons gave their informed consent prior to their inclusion in the study.
Statistical analyses
SPSS 18.0 statistical software was used for statistical analysis. Values are presented as mean ± SD. Statistical analysis was performed using the Student's t-test or ANOVA. P
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
ZWL and TCZ are responsible for the study design. YJZ and ZWL performed the experiments and drafted the manuscript.HWY and YJZ collected the data. XLS, YL, XYS, and XDL participated in the data analysis and interpretation. All authors read and approved the final manuscript.
Funding
This study was supported by a grant of the Program for Young Talents of Guangzhou Medical University (No.2012A15 to ZWL) and a key grant from the Cancer Center of Guangzhou Medical University (No.2011-yz-06 to TCZ).
References
- 1. Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol 2002;12:421-429; PMID:12450728; http://dx.doi.org/ 10.1016/S1044579X02000858 [DOI] [PubMed] [Google Scholar]
- 2. Titcomb CJ. High incidence of nasopharyngeal carcinoma in Asia. J Insur Med 2001;33:235-238; PMID:11558403 [PubMed] [Google Scholar]
- 3. Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu LZ, Sun Y, Lin AH, Liu MZ, Ma J. How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? Int J Radiat Oncol Biol Phys 2011;80:661-668; PMID:20643517; http://dx.doi.org/ 10.1016/j.ijrobp.2010.03.024 [DOI] [PubMed] [Google Scholar]
- 4. McDermott AL, Dutt SN, Watkinson JC. The aetiology of nasopharyngeal carcinoma. Clin Otolaryngol Allied Sci 2001;26:82-92; PMID:11309046; http://dx.doi.org/ 10.1046/j.1365-2273.2001.00449.x [DOI] [PubMed] [Google Scholar]
- 5. Chou J, Lin YC, Kim J, You L, Xu Z, He B, Jablons DM. Nasopharyngeal carcinoma–review of the molecular mechanisms of tumorigenesis. Head Neck 2008;30:946-963; PMID:18446839; http://dx.doi.org/ 10.1002/hed.20833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle 2005;4:1171-1175; PMID:16082216; http://dx.doi.org/ 10.4161/cc.4.9.2001 [DOI] [PubMed] [Google Scholar]
- 7. Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503-1521; PMID:15931389; http://dx.doi.org/ 10.1172/JCI23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet 2007;39:157-158; PMID:17200673; http://dx.doi.org/ 10.1038/ng1941 [DOI] [PubMed] [Google Scholar]
- 9. Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr Patterns 2006;6:277-284; PMID:16378762; http://dx.doi.org/ 10.1016/j.modgep.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 10. Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 2008;29:102-111; PMID:18206973; http://dx.doi.org/ 10.1016/j.molcel.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 2008;7:1522-1524; PMID:18469533; http://dx.doi.org/ 10.4161/cc.7.11.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol 2005;25:1173-1182; PMID:15657442; http://dx.doi.org/ 10.1128/MCB.25.3.1173-1182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 2008;29:92-101; PMID:18206972; http://dx.doi.org/ 10.1016/j.molcel.2007.12.011 [DOI] [PubMed] [Google Scholar]
- 14. Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev 2001;15:2042-2047; PMID:11511535http://dx.doi.org/ 10.1101/gad.907901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu YL, Jiang SX, Yang YM, Xu H, Liu JL, Wang XS. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys 2012;62:229-235; PMID:21928107; http://dx.doi.org/ 10.1007/s12013-011-9287-0 [DOI] [PubMed] [Google Scholar]
- 16. Piao S, Ma J, Wang W, Liu Y, Zhang M, Chen H, Guo F, Zhang B, Guo F. Increased expression of USP22 is associated with disease progression and patient prognosis of salivary duct carcinoma. Oral Oncol 2013;49:796-801; PMID:23664741; http://dx.doi.org/ 10.1016/j.oraloncology.2013.03.454 [DOI] [PubMed] [Google Scholar]
- 17. Li J, Wang Z, Li Y. USP22 nuclear expression is significantly associated with progression and unfavorable clinical outcome in human esophageal squamous cell carcinoma. J Cancer Res Clin Oncol 2012;138:1291-1297; PMID:22447106; http://dx.doi.org/ 10.1007/s00432-012-1191-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J Cancer Res Clin Oncol 2011;137:1245-1253; PMID:21691749; http://dx.doi.org/ 10.1007/s00432-011-0998-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang DD, Cui BB, Sun LY, Zheng HQ, Huang Q, Tong JX, Zhang QF. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell Biochem Biophys 2011;61:703-710; PMID:21735131; http://dx.doi.org/ 10.1007/s12013-011-9229-x [DOI] [PubMed] [Google Scholar]
- 20. Liu YL, Yang YM, Xu H, Dong XS. Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J Gastroenterol Hepatol 2010;25:1800-1805; PMID:21039844; http://dx.doi.org/ 10.1111/j.1440-1746.2010.06352.x [DOI] [PubMed] [Google Scholar]
- 21. Yang M, Liu YD, Wang YY, Liu TB, Ge TT, Lou G. Ubiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeutics. Tumour Biol 2014;35:929-934; PMID:23979981; http://dx.doi.org/ 10.1007/s13277-013-1121-4 [DOI] [PubMed] [Google Scholar]
- 22. Li ZH, Yu Y, DU C, Fu H, Wang J, Tian Y. RNA interference-mediated USP22 gene silencing promotes human brain glioma apoptosis and induces cell cycle arrest. Oncol Lett 2013;5:1290-1294; PMID:23599781 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Lv L, Xiao XY, Gu ZH, Zeng FQ, Huang LQ, Jiang GS. Silencing USP22 by asymmetric structure of interfering RNA inhibits proliferation and induces cell cycle arrest in bladder cancer cells. Mol Cell Biochem 2011;346:11-21; PMID:20824490; http://dx.doi.org/ 10.1007/s11010-010-0585-4 [DOI] [PubMed] [Google Scholar]
- 24. Xu H, Liu YL, Yang YM, Dong XS. Knock-down of ubiquitin-specific protease 22 by micro-RNA interference inhibits colorectal cancer growth. Int J Colorectal Dis 2012;27:21-30; PMID:21773699; http://dx.doi.org/ 10.1007/s00384-011-1275-8 [DOI] [PubMed] [Google Scholar]
- 25. Simpson PJ, Moon C, Kleman AM, Connolly E, Ronnett GV. Progressive and inhibitory cell cycle proteins act simultaneously to regulate neurotrophin-mediated proliferation and maturation of neuronal precursors. Cell Cycle 2007;6:1077-1089; PMID:17404514; http://dx.doi.org/ 10.4161/cc.6.9.4132 [DOI] [PubMed] [Google Scholar]
- 26. Liang QC, Xiong H, Zhao ZW, Jia D, Li WX, Qin HZ, Deng JP, Gao L, Zhang H, Gao GD. Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett 2009;273:164-171; PMID:18793823; http://dx.doi.org/ 10.1016/j.canlet.2008.08.011 [DOI] [PubMed] [Google Scholar]
- 27. Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002;2:489-501; PMID:12094235; http://dx.doi.org/ 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- 28. Kida A, Kakihana K, Kotani S, Kurosu T, Miura O. Glycogen synthase kinase-3beta and p38 phosphorylate cyclin D2 on Thr280 to trigger its ubiquitin/proteasome-dependent degradation in hematopoietic cells. Oncogene 2007;26:6630-6640; PMID:17486076; http://dx.doi.org/ 10.1038/sj.onc.1210490 [DOI] [PubMed] [Google Scholar]
- 29. Takahashi-Yanaga F, Sasaguri T. GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy. Cell Signal 2008;20:581-589; PMID:18023328; http://dx.doi.org/ 10.1016/j.cellsig.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 30. Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 1998;12:3499-3511; PMID:9832503; http://dx.doi.org/ 10.1101/gad.12.22.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 2009;119:3626-3636; PMID:19884659; http://dx.doi.org/ 10.1172/JCI39374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature 2009;458:438-444; PMID:19325623; http://dx.doi.org/ 10.1038/nature07960 [DOI] [PubMed] [Google Scholar]
- 33. Liu YL, Jiang SX, Yang YM, Xu H, Liu JL, Wang XS. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys 2012;62:229-235; PMID:21928107; http://dx.doi.org/ 10.1007/s12013-011-9287-0 [DOI] [PubMed] [Google Scholar]
- 34. Liu YL, Jiang SX, Yang YM, Xu H, Liu JL, Wang XS. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys 2012;62:229-235; PMID:21928107; http://dx.doi.org/ 10.1007/s12013-011-9287-0 [DOI] [PubMed] [Google Scholar]
- 35. Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 2009;119:3626-3636; PMID:19884659; http://dx.doi.org/ 10.1172/JCI39374 [DOI] [PMC free article] [PubMed] [Google Scholar]


