Abstract
Soybean GmWRKY53 functions in both biotic and abiotic stress signaling. Using GmWRKY53 as a bait yeast 2-hybrid library screening to saturation isolated multiple independent fragments for many interacting proteins, enabling delineation of minimal interacting domains and computation of a confidence score. Multiple independent clones coding for the LATE ELONGATED HYPOCOTYL clock protein GmLCL2 (MYB114) were isolated and the binding site for GmWRKY53 was mapped to 90 amino acids separate from the MYB domain. This suggests a direct input from the clock on GmWRKY53 activity. The GmWRKY53-interacting proteins also included 3 water stress-inducible AP2/ERF transcription factors. One of these (Glyma03g26310) is one of the most strongly water stress induced genes in soybean roots, suggesting that GmWRKY53/ERF complexes regulate water stress responses.
Keywords: clock protein, drought, ERF transcription factor, protein-protein interaction, soybean, WRKY transcription factor
Abbreviations
- transcription factor
TF
- resistance proteins
R proteins
- yeast two-hybrid
Y2H
- LATE ELONGATED HYPOCOTYL
LHY
- bright yellow 2
BY-2
- guanine nucleotide exchange factor
GEF
- GTPase accelerating proteins
GAP
Introduction
WRKY transcription factors (TFs) are one of the major families of plant transcription factors and play roles in regulating the responses to both biotic and abiotic stresses. Although transcription factor families are classified according to their DNA-binding domains, in general it is the other protein domains that influence transcription. These protein-protein interactions are less well understood than the process of DNA binding which involves well-defined interactions of amino acids in the WRKY domain with the W box binding site on target promoters. Interactions of WRKY TFs with proteins activate or repress transcription. By studying these protein interaction networks new insights can be obtained into the molecular mechanisms of signal transduction.
Several different types of proteins have been shown to interact with WRKY transcription factors. For example, some WRKY proteins interact with calmodulin, which binds to the C-motif (DxxVxKFKx-VISLLxxxR) that is present in some Group IId WRKY proteins.1 This suggests that the activity of these Group IId WRKY proteins is regulated by Ca2+. WRKY TFs are also a major target of MAP kinase signaling cascades and both DNA-binding and transcription activating activities can be altered by phosphorylation.2 Other interacting partners of WRKY proteins include 14–3–3 proteins,3 chromatin remodeling proteins such as Arabidopsis histone deacetylase 19,4 VQ proteins,5 CC-NBR-LRR-type R proteins,6 and E3 ubiquitin ligases.7
WRKY TFs are some of their own interacting partners and homo- and heterodimerization are features of some signaling webs involving WRKY TFs.9 It is known that some WRKY TFs contain dimerization domains such as leucine zippers.10 Recently, a class of proteins that contain a VQ motif (FxxxVQxLTG) have been shown to be interacting partners of WRKY transcription factors from Group IIc and Group I.11 VQ proteins are important regulators of DNA binding and other molecular activities of some WRKY TFs.12 Yeast 2-hybrid (Y2H) assays with mutated versions of the Arabidopsis VQ protein VQ10 identified structural features unique to Group I and IIC WRKY proteins that are critical for their interaction with VQ proteins. These include the presence of 4 amino acid residues between the conserved Cys residues involved in zinc coordination and 2 charged amino acid residues in the region that precede the WRKYGQK signature sequence. VQ proteins are involved in abiotic and biotic stress responses as well as other functions such as seed size.11,17
GmWRKY53 is a group IIc WRKY transcription factor that is up-regulated at the mRNA level in both soybean roots and leaves as a response to water stress. It was originally described as being salt and water stress inducible in 2-week-old soybean seedlings.18 Using transgenic tobacco (Nicotiana tabacum L.) Bright Yellow 2 (BY-2) cells we have previously shown that the GmWRKY53 promoter directs salt and jasmonate inducible expression and to a lesser degree ABA inducible expression.19 This suggests that the GmWRKY53 transcription factor is part of several stress-related signaling webs. Consistent with this, transcriptome analysis of the soybean root response to Fusarium virguliforme revealed that GmWRKY53 is the most strongly upregulated WRKY transcription factor gene during both resistant and susceptible interactions.20 These data suggest that GmWRKY53 is involved in regulating responses to both biotic and abiotic stresses.
Here we determine components of the interactome of GmWRKY53 using Yeast 2-Hybrid libraries screened to saturation. Twenty seven in frame GmWRKY53-interacting proteins were isolated. Multiple, independent fragments were isolated for many interacting proteins, enabling delineation of minimal interacting domains and the computation of a confidence score. These interactors included 3 water stress-inducible AP2/ERF proteins, 19 VQ proteins, and multiple independent clones coding for the LATE ELONGATED HYPOCOTYL (LHY) clock gene GmLCL2 (MYB114). This emerging GmWRKY53 interaction network provides novel insights into the molecular mechanisms of signal transduction in both stress responses and circadian rhythms.
Results
A domain-based strategy to construct highly complex, random primed cDNA libraries from a time course of water stressed soybean tissues (root and leaf) plus control tissues was employed to isolate GmWRKY53 interacting proteins. The complexity of the soybean library was greater than 10 million independent fragments in yeast. To ensure reproducible and exhaustive Y2H results the library was screened to saturation allowing testing of about 83 million interactions (fold8- coverage of the library). This strategy enabled the isolation of multiple independent fragments for many of the interacting proteins. For all of the interacting proteins a confidence score could therefore be computed and in some cases a minimal interacting domain could be defined.
For each interaction, a Predicted Biological Score (PBS) was computed to assess the interaction reliability.21 This e-value score represents the probability of an interaction being non-specific and is primarily based on the comparison between the number of independent prey fragments found for an interaction and the chance of finding them at random. Four categories were defined from A (the highest confidence rank) to D.21 D interactions generally represent interactions identified through one unique prey fragment or multiple identical ones. There were also 4 additional interacting proteins that appeared to be out of frame but that were a similar class of protein to many other positives (VQ proteins). The total number of GmWRKY53-interacting proteins was therefore 30 2.
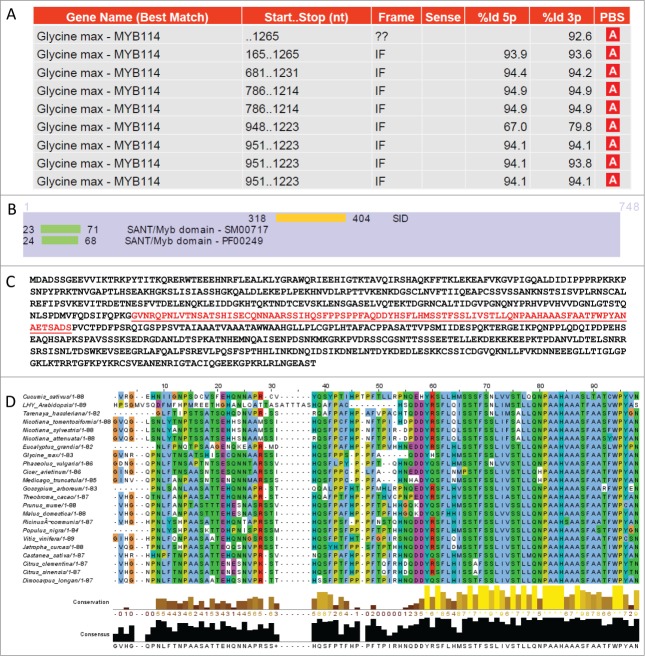
Strikingly, one A ranked interactor codes for GmLCL2 (MYB114), a soybean ortholog of the LATE ELONGATED HYPOCOTYL (LHY) clock gene. Multiple independent clones were obtained for GmLCL2 (Fig. 1A) and one of these encoded about half of the protein (nucleotides 165–1265). Other clones were much shorter and enabled a minimum GmWRKY53-interacting domain to be determined from amino acids 318 to 404 (Fig. 1B and C). Blast searches with this interacting domain revealed that it is well conserved among LHY-like genes from related species (Fig. 1D). Interestingly, Arabidopsis LHY is only similar over the C-terminal half of the GmWRKY53-interacting domain. Assuming similar interactions between Arabidopsis LHY and Group IIc/I WRKY proteins, this suggests that the amino acid sequence HNQDDYRSFLHMSSTFSSLIVSTLLQNPAAHAAASFAATFWPYAN may represent the core of the interacting domain in the GmLCL2 protein (Fig. 1D). This GmWQRKY53-GmLCL2 interaction is intriguing. Several clock proteins are known to interact with other proteins essential for their activity and at the transcriptional level several regulators of stress responses are known to be regulated by clock components such as CCA1, PRR5, PRR7, and TOC1.30 However, direct interaction between a WRKY transcription factor and a clock component would represent a different level of regulation of stress responses by the circadian system.
Figure 1.
Interaction of GmWRKY53 with the LHY-like protein GmLCL2 (MYB114). (A) Clones isolated from the yeast 2 hybrid screen. The start and stop codons are shown for each truncated protein. The percentage identity at the 5 and 3 prime ends is shown. These values are close to 100% once trimmed at the ends. (B) Cartoon of the GmLCL2 protein (lilac box) and the maximum required interaction domain (amino acids 318–404) and the SANT/Myb domain. (C) Amino acid sequence of GmLCL2 with the maximum GmWRKY53-interacting domain shown in red. (D) CLUSTALW derived multiple sequence alignment of the maximum interacting domain from LHY-like proteins from different plant species. The Jalview derived consensus sequence is shown.
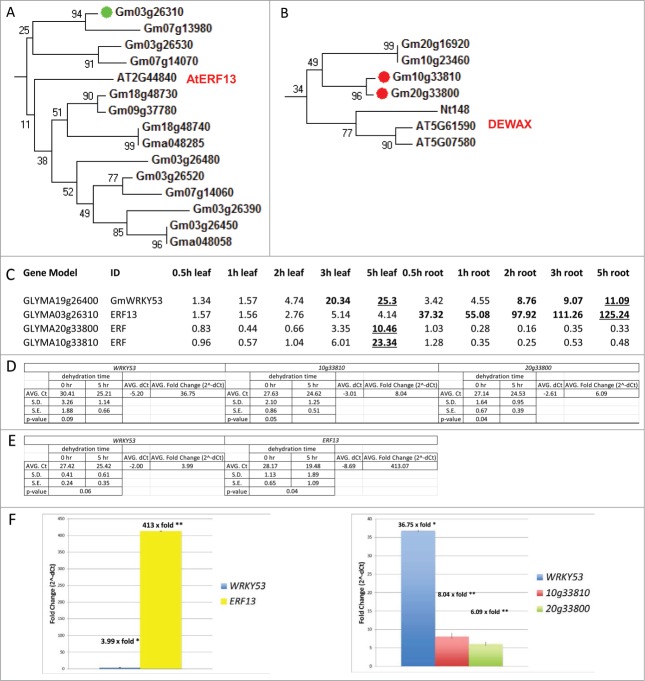
AP2/ERF transcription factors are major regulators of both biotic and abiotic stress responses in plants. The same genome wide transcriptome analyses that identified GmWRKY53 as water stress inducible also identified a large number of AP2/ERF transcription factor genes that are strongly (Rabara et al., unpublished). In roots, Glyma03g26310 is the third most strongly water stress induced soybean gene at early time points and the most strongly up-regulated AP2/ERF gene. Glyma03g26310 (ERF13-like XP_003521070) was isolated multiple times in the Y2H screen including 2 near full-length clones (Table S1). This suggests that AP2/ERF-WRKY complexes are part of stress response signaling webs in soybean. This is consistent with the identification of 2 other AP2/ERF transcription factors as GmWRKY53 interacting proteins, Glyma10g33810 with high confidence and Glyma20g33800 with a good confidence (Table S1). The soybean genome is a partially diploidized tetraploid and Glyma10g33810 and Glyma20g33800 are paralogs with 90.5% amino acid identity (182/201). The isolation of 2 paralagous AP2/ERF transcription factors supports this WRKY/ERF interaction and the 2 proteins were isolated a combined total of 7 independent times representing a total of 4 near full length clones (2 for each gene). Additionally, one of the Arabidopsis orthologues (At5g61590) is in the GO Biological Process category response to water deprivation (Fig. 2B). Additionally, both of the soybean genes are upregulated in leaves at the mRNA level by water stress, as is their GmWRKY53 interacting partner (Fig. 2C–F). These data suggest that complexes containing GmWRKY53 and these 2 AP2/ERF transcription factors form part of the response of soybean to water deprivation.
Figure 2.
AP2/ERF proteins that interact with GmWRKY53 and expression profiles of GmWRKY53, Glyma03g26310, Glyma10g33810 and Glyma20g33800 during the course of water stress. (A) Phylogenetic tree of soybean and Arabidopsis AP2/ERF proteins. Glyma03g26310 is indicated with a green circle and the closest Arabidopsis protein, AtERF13 is also indicated. (B) Phylogenetic tree of soybean and Arabidopsis AP2/ERF proteins. Glyma10g33810 and Glyma20g33800 are indicated with red circles and the closest Arabidopsis proteins, At5g61590 and At5g07580 are indicated. (C) Expression profiles using oligo array analysis. Bold values show induction of at least fold5- and underlined time points were validated by qRT-PCR using independent samples. (D) Relative expression levels of GmWRKY53, Glyma10g33810 and Glyma20g33800 during water stress in leaf and root tissue. (E) Relative expression levels of GmWRKY53 and Glyma03g26310 during water stress in root tissue. (F) Relative inducibiltities. Values denoted by * are significant at 90%; ** = significant at 95%.
One of the 2 very high confidence interactions of GmWRKY53 was with the VQ protein GmVQ61 (Glyma15g08160). GmVQ61 was isolated 11 times with 7 independent clones showing interaction with GmWRKY53. The isolation of multiple independent clones allowed a minimum GmWRKY53-interacting domain to be determined from amino acids 21 to 112. This represents just under half of the protein and includes the VQ domain that has been shown previously to be a WRKY interacting domain. Interestingly, 14 other VQ proteins were isolated (Table 1 and Table S1). The large number of VQ proteins isolated is consistent not only with the VQ domain being a WRKY (Group IIc or I) interaction domain but also with reports that WRKY proteins can typically interact with many VQ proteins.11
Table 1.
GmWRKY53 interaction and water stress inducibility of the 70 4 soybean VQ proteins. VQ proteins that interact with GmWRKY53 in the yeast 2-hybrid screen are shown in green. VQ proteins that were isolated as binding to GmWRKY53 but also found to be out of frame with the Gal4 activation domain are shown in light green. The relative fold changes in mRNA levels relative to unstressed controls in leaf and root tissues are shown and induction/repression of more than fold5- is indicated in gray
| SEQ_ID | Gene ID | 30 mins leaf | 1 h leaf | 2 h leaf | 3 h leaf | 5 h leaf | 30 mins root | 1 h root | 2 h root | 3 h root | 5 h root |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLYMA01g02280 | GmVQ1 | 0.93 | 0.85 | 0.88 | 0.97 | 1.23 | 0.99 | 0.91 | 0.91 | 0.87 | 0.81 |
| GLYMA01g12940 | GmVQ2 | 0.94 | 1.75 | 3.24 | 2.36 | 2.36 | 0.88 | 0.88 | 0.99 | 1.25 | 1.12 |
| GLYMA01g40320 | GmVQ3 | 1.1 | 2.62 | 2.52 | 2.1 | 1.08 | 11.83 | 19.47 | 27.5 | 20.42 | 31.19 |
| GLYMA02g29270/29281 | GmVQ4 | 2.21 | 1.77 | 3.38 | 2.41 | 2.93 | 0.84 | 0.5 | 0.41 | 0.81 | 0.78 |
| GLYMA02g37130/37135 | GmVQ5 | 0.93 | 1.08 | 1.01 | 0.83 | 0.51 | 0.95 | 0.89 | 0.77 | 0.92 | 0.72 |
| GLYMA03g27560 | GmVQ6 | 0.83 | 5.12 | 6.86 | 2.82 | 1.34 | 3.08 | 2.36 | 1.99 | 2.46 | 2.32 |
| GLYMA03g28360 | GmVQ7 | 1.17 | 0.77 | 0.59 | 0.37 | 0.33 | 0.82 | 0.79 | 0.68 | 0.77 | 0.74 |
| GLYMA03g36330 | GmVQ8 | 0.54 | 3.9 | 9.67 | 2.01 | 0.77 | 9.3 | 9.66 | 9.01 | 9.13 | 10.91 |
| GLYMA03g40940/40941 | GmVQ9 | 0.83 | 1.33 | 3.24 | 2.58 | 2.3 | 0.39 | 0.75 | 1.59 | 1.36 | 2.3 |
| GLYMA04g10780 | GmVQ10 | 0.83 | 0.8 | 0.62 | 0.79 | 0.82 | 0.75 | 0.83 | 0.62 | 0.79 | 0.5 |
| GLYMA04g11200/11211 | GmVQ11 | 0.89 | 0.68 | 0.65 | 0.43 | 0.44 | 0.87 | 1.36 | 1.48 | 0.75 | 0.6 |
| GLYMA04g16880 | GmVQ12 | 1.37 | 7.71 | 12.1 | 18.88 | 22.29 | 0.76 | 1.82 | 2.05 | 4.5 | 2.3 |
| GLYMA04g39270 | GmVQ13 | 1.54 | 2.06 | 2.76 | 1.74 | 1.53 | 1.65 | 1.72 | 1.44 | 1.69 | 1.22 |
| GLYMA04g41820 | GmVQ14 | 0.48 | 0.99 | 0.87 | 0.68 | 0.63 | 1.2 | 0.85 | 0.91 | 1.4 | 1.25 |
| GLYMA05g22760 | GmVQ15 | 1.18 | 2.12 | 1.76 | 2.53 | 1.23 | 3.54 | 6.01 | 6.2 | 5.17 | 7.72 |
| GLYMA05g26340 | GmVQ16 | 1.55 | 4.27 | 12.99 | 5.81 | 15.49 | 1.81 | 2.99 | 4.5 | 7.44 | 6.04 |
| GLYMA05g27220 | GmVQ17 | 1.2 | 2.75 | 5.24 | 2.5 | 1.55 | 5.88 | 4.4 | 4.44 | 2.17 | 5.65 |
| GLYMA05g31340/31341 | GmVQ18 | 0.91 | 0.88 | 1.13 | 1.14 | 1.99 | 0.76 | 0.96 | 1.23 | 0.93 | 0.79 |
| GLYMA05g32350 | GmVQ19 | 1.16 | 3.09 | 3.04 | 3.21 | 2.98 | 1.45 | 1.27 | 1.02 | 0.86 | 0.93 |
| GLYMA05g33250 | GmVQ20 | 0.82 | 0.6 | 0.45 | 0.28 | 0.22 | 1.11 | 2.09 | 2.85 | 4.15 | 6.91 |
| GLYMA06g10630 | GmVQ21 | 0.96 | 0.81 | 0.69 | 1.16 | 1.21 | 1.35 | 1.24 | 0.72 | 1.04 | 0.76 |
| GLYMA06g10950/10951 | GmVQ22 | 0.99 | 1.19 | 0.97 | 0.81 | 0.85 | 1.09 | 1.36 | 1.46 | 0.79 | 0.47 |
| GLYMA06g10960 | GmVQ23 | 0.77 | 0.6 | 0.51 | 0.58 | 0.64 | 0.9 | 1.28 | 2.02 | 1.17 | 0.75 |
| GLYMA06g12960 | GmVQ24 | 0.5 | 0.93 | 0.73 | 1.2 | 1.22 | 1.18 | 0.91 | 1 | 1.43 | 1.16 |
| GLYMA06g15650 | GmVQ25 | 1.19 | 1.87 | 3.16 | 1.89 | 1.89 | 1.16 | 1.34 | 1.09 | 1.23 | 1.2 |
| GLYMA06g36640 | GmVQ26 | 0.91 | 0.65 | 0.72 | 1.49 | 1.55 | 0.53 | 0.56 | 0.77 | 0.59 | 0.56 |
| GLYMA07g03240 | GmVQ27 | 0.82 | 0.87 | 1.01 | 0.95 | 1.03 | 0.75 | 1.04 | 1.13 | 1.2 | 0.82 |
| GLYMA07g10290 | GmVQ28 | 1.36 | 1.53 | 1.33 | 1.23 | 1.21 | 1.49 | 1.54 | 3.42 | 2.4 | 2.4 |
| GLYMA07g31780/31781 | GmVQ29 | 0.97 | 0.92 | 0.93 | 0.82 | 0.94 | 1.01 | 1.29 | 1.31 | 0.95 | 0.91 |
| GLYMA08g00850 | GmVQ30 | 1.4 | 0.85 | 0.5 | 0.11 | 0.09 | 1.26 | 3.09 | 6.35 | 7.68 | 8.85 |
| GLYMA08g04650/04651 | GmVQ31 | 1.38 | 0.85 | 0.68 | 0.99 | 0.9 | 0.3 | 0.21 | 0.17 | 0.31 | 0.32 |
| GLYMA08g09250 | GmVQ32 | 1.96 | 5.85 | 16.43 | 12.71 | 33.85 | 1.91 | 3.79 | 3.86 | 10.82 | 5.73 |
| GLYMA08g10166/no model | GmVQ33 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| GLYMA08g14580/14581 | GmVQ34 | 1.36 | 1.4 | 1.9 | 2.02 | 3.44 | 0.92 | 1.07 | 1.28 | 0.97 | 1.02 |
| GLYMA08g15620 | GmVQ35 | 1.45 | 2.67 | 4.28 | 4.10 | 4.17 | 1.65 | 1.34 | 1.14 | 1.11 | 1.26 |
| GLYMA08g16790 | GmVQ36 | 2.01 | 1.63 | 6.11 | 9.6 | 5.76 | 0.41 | 0.24 | 0.27 | 1.42 | 1.25 |
| GLYMA08g18820 | GmVQ37 | 0.87 | 1.30 | 3.47 | 7.19 | 3.98 | 1.30 | 1.38 | 2.72 | 2.73 | 3.41 |
| GLYMA08g22860 | GmVQ38 | 0.78 | 0.86 | 1.13 | 1.33 | 1.39 | 0.87 | 1 | 1.13 | 0.99 | 0.62 |
| GLYMA08g36730 | GmVQ39 | 0.88 | 1.12 | 1.17 | 1.26 | 1.43 | 1.01 | 0.98 | 0.96 | 1.02 | 0.93 |
| GLYMA08g36740 | GmVQ40 | 0.95 | 1.07 | 1.04 | 1.27 | 1.45 | 1 | 0.99 | 0.97 | 0.99 | 0.93 |
| GLYMA08g36750 | GmVQ41 | 0.9 | 0.92 | 0.91 | 1.1 | 1.37 | 0.78 | 0.84 | 0.58 | 0.67 | 0.66 |
| GLYMA08g42125/42120 | GmVQ42 | 0.50 | 0.51 | 0.30 | 0.31 | 0.20 | 1.04 | 1.32 | 1.01 | 0.58 | 0.63 |
| GLYMA09g05700 | GmVQ43 | 1.53 | 2.72 | 6.08 | 3.13 | 4.35 | 2.63 | 2.65 | 3.41 | 3.82 | 3.84 |
| GLYMA09g17151/17140 | GmVQ44 | 2.12 | 1.97 | 3.16 | 1.86 | 2.43 | 1.13 | 0.6 | 0.54 | 1.08 | 0.98 |
| GLYMA09g31600 | GmVQ45 | 1.11 | 0.91 | 1.63 | 1.29 | 1.06 | 0.89 | 1.27 | 1.47 | 2.62 | 2.91 |
| GLYMA10g41970 | GmVQ46 | 1.94 | 1.5 | 2.43 | 1.45 | 2.03 | 2.35 | 1.62 | 2.14 | 1.92 | 2.53 |
| GLYMA11g04970 | GmVQ47 | 1.95 | 1.23 | 2.17 | 2.41 | 1.48 | 10.52 | 18.55 | 28.85 | 20.17 | 27.95 |
| GLYMA11g38180 | GmVQ48 | 1.31 | 1.26 | 0.58 | 0.2 | 0.33 | 0.69 | 0.82 | 0.91 | 0.78 | 0.61 |
| GLYMA12g24250 | GmVQ49 | 0.99 | 0.76 | 0.67 | 0.65 | 0.8 | 0.74 | 0.68 | 0.88 | 0.8 | 0.89 |
| GLYMA12g35380 | GmVQ50 | 0.82 | 0.78 | 0.7 | 0.77 | 0.97 | 1.27 | 1.4 | 1.48 | 1.51 | 1.5 |
| GLYMA13g08831/08820 | GmVQ51 | 1.22 | 2.26 | 2.46 | 1.35 | 1.69 | 2.40 | 3.22 | 3.35 | 3.19 | 6.50 |
| GLYMA13g10840 | GmVQ52 | 1.15 | 5.02 | 5.34 | 3.45 | 3.95 | 3.07 | 3.34 | 2.85 | 3.24 | 3.45 |
| GLYMA13g24700 | GmVQ53 | 0.8 | 0.82 | 0.87 | 0.93 | 0.82 | 1.09 | 1.21 | 1.12 | 0.92 | 0.95 |
| GLYMA13g26290 | GmVQ54 | 1.02 | 5.35 | 18.20 | 80.00 | 113.62 | 1.65 | 3.96 | 11.47 | 18.00 | 26.91 |
| GLYMA13g29005/29000 | GmVQ55 | 0.31 | 0.66 | 1.57 | 1.18 | 0.35 | 0.84 | 1.78 | 0.84 | 0.55 | 0.64 |
| GLYMA13g31180 | GmVQ56 | 0.91 | 1.12 | 1.26 | 1.11 | 0.66 | 0.99 | 1.04 | 1.05 | 0.74 | 0.72 |
| GLYMA13g35130 | GmVQ57 | 0.9 | 0.78 | 0.69 | 0.71 | 1 | 1.42 | 1.52 | 1.95 | 1.7 | 2.08 |
| GLYMA14g00570 | GmVQ58 | 1.91 | 8.98 | 13.21 | 6.68 | 4.79 | 1.64 | 1.46 | 1.69 | 1.89 | 1.66 |
| GLYMA14g29580 | GmVQ59 | 1.28 | 1.46 | 1.79 | 3.21 | 2.54 | 2.53 | 2.22 | 3.53 | 3.3 | 6.64 |
| GLYMA14g34680/34681 | GmVQ60 | 0.74 | 0.58 | 0.71 | 1.07 | 0.81 | 1.05 | 1.57 | 1.53 | 1.26 | 1.06 |
| GLYMA15g08160 | GmVQ61 | 0.92 | 1.41 | 1.19 | 0.43 | 0.23 | 1.38 | 1.29 | 1.11 | 0.91 | 0.92 |
| GLYMA15g16990 | GmVQ62 | 1.3 | 3.89 | 8.02 | 8.95 | 7.36 | 1.96 | 2.52 | 4.07 | 4.76 | 4.62 |
| GLYMA15g37230 | GmVQ63 | 1.01 | 1.55 | 9.75 | 39.55 | 70.03 | 2.02 | 3.50 | 6.94 | 9.97 | 14.11 |
| GLYMA15g40000 | GmVQ64 | 0.94 | 1.84 | 4.55 | 8.73 | 4.35 | 1.41 | 1.60 | 3.29 | 2.74 | 3.84 |
| GLYMA15g42280 | GmVQ65 | 1.06 | 0.87 | 2 | 1.44 | 1.31 | 0.6 | 0.46 | 0.46 | 1.2 | 0.98 |
| GLYMA17g17210 | GmVQ66 | 1.28 | 1.98 | 3.64 | 5.4 | 2.93 | 5.05 | 6.21 | 6.66 | 7.33 | 7.72 |
| GLYMA18g02140 | GmVQ67 | 1.49 | 1.02 | 0.58 | 0.29 | 0.42 | 0.77 | 0.69 | 0.73 | 0.71 | 0.59 |
| GLYMA18g13001/no model | GmVQ68 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| GLYMA19g30531/30530 | GmVQ69 | 0.86 | 4.94 | 6.44 | 2.71 | 1.34 | 2.95 | 1.60 | 1.82 | 2.42 | 2.23 |
| GLYMA19g31080 | GmVQ70 | 1.02 | 0.69 | 0.40 | 0.54 | 0.50 | 0.70 | 0.52 | 0.36 | 0.56 | 0.32 |
| GLYMA19g38980 | GmVQ71 | 1.42 | 2.56 | 15.72 | 1.93 | 0.76 | 14.53 | 14.76 | 15.8 | 5.23 | 18.37 |
| GLYMA19g43590 | GmVQ72 | 0.54 | 1.21 | 0.95 | 0.77 | 0.67 | 0.52 | 0.96 | 1.48 | 0.97 | 2.03 |
| GLYMA20g15230 | GmVQ73 | 1.28 | 2.58 | 4.75 | 2.34 | 2.56 | 1.66 | 1.88 | 2.42 | 2.4 | 2.89 |
| GLYMA20g25070 | GmVQ74 | 1.14 | 1.19 | 1.72 | 1.47 | 1.99 | 3.14 | 2.57 | 3.17 | 3.76 | 4.28 |
The soybean VQ protein family has previously been studied by Wang et al.31 and 74 VQ (GmVQ 1–74) motif-containing genes were found.31 Our data illustrated that that GmWRKY53 can interact with 15 of these proteins (Table 1). Four additional VQ clones had inserts in the correct orientation but in the wrong reading frame for a fusion protein with the Gal4 activation domain, suggesting that these proteins can interact with GmWRKY53 and activate transcription in yeast without the Gal4 activation domain (Table 1). In total 19 of the 74 soybean VQ proteins interacted with GmWRKY53, consistent with the formation of multiple GmWRKY53-VQ transcriptional regulatory complexes (Fig. 3). Seventy two of the 70 4 soybean VQ genes were present on our oligo array and Table 1 shows that 28 VQ genes were differentially regulated by water stress when using a conservative cut off threshold of fold5- inducibility/repressibility. Of these 28 VQ genes, 9 interacted with GmWRKY53 in the Y2H screen (Table 1) and might be involved in regulating changes in gene expression as a result of water deficit.
Figure 3.

Phylogenetic tree and GmWRKY53 interactions of 70 4 soybean VQ proteins. Alignments were constructed using MUSCLE23 and a Neighbor Joining tree was constructed using MEGA6,27 the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were determined.25 The evolutionary distances were computed using the Poisson correction method.26 Red circles indicate VQ proteins that interact with GmWRKY53 in the yeast 2-hybrid screen. Orange circles indicate interacting proteins that were out of frame with respect to the Gal4 activation domain.
In addition to the proteins already mentioned, several other signaling proteins were found to interact with GmWRKY53 in the Y2H screen (Table S1). Glyma14g11260 codes for an F-box family protein that is the apparent soybean ortholog of the Arabidopsis protein SLOMO (identities = 563/880 (63%), positives = 672/880 (76%)). SLOMO is an F-box protein required for auxin homeostasis and normal timing of lateral organ initiation at the shoot meristem. SLOMO has ubiquitin-protein ligase activity and this suggest that under specific conditions GmWRKY53 is targeted for degradation by the proteasome. Glyma08g07260 codes for a MADS Box transcription factor and was isolated as an apparent full length clone in the screen. The most similar Arabidopsis protein is AGAMOUS-LIKE 22/FLOWERING ARABIDOPSIS QTL1/SHORT VEGETATIVE PHASE which is a floral repressor that functions within the thermosensory pathway. It is implicated in regulating jasmonic acid mediated signaling pathways, defense responses, hypersensitive responses, responses to bacteria, responses to temperature stimulus, systemic acquired resistance, and salicylic acid mediated signaling pathways (www.arabidopsis.org). Our data suggest that GmWRKY53/Glyma08g07260 complexes are a component of stress-responsive signaling webs.
Another interacting protein Glyma14g00230 encodes a guanine nucleotide exchange factor (GEF). GEFs are involved in the activation of small GTPases. Small GTPases act as molecular switches in intracellular signaling pathways. Glyma14g00230 is a SEC7-like GEF and similar proteins function in cytokinesis, gravitropism, microtubule cytoskeleton organization, regulation of ARF protein signal transduction, and vesicle-mediated transport. A possible link between WRKY proteins and G protein signaling has not previously been suggested and it may represent another signaling input for GmWRKY53.
Discussion
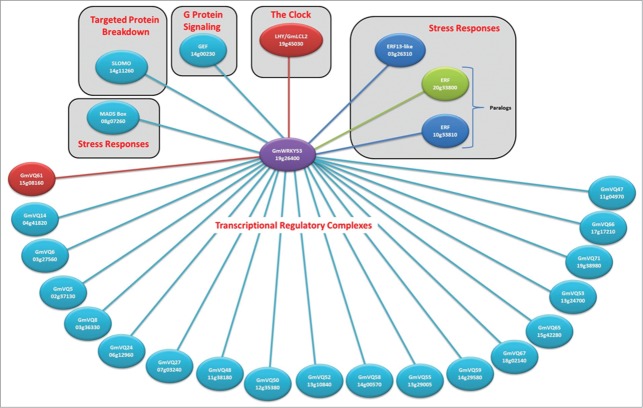
GmWRKY53 is up-regulated at the mRNA level as a response to water stress,19 salt,18 and Fusarium virguliforme infection.20 This suggests that GmWRKY53 is part of several stress-related signaling webs including biotic and abiotic stresses. Protein interaction networks provide insights into the molecular mechanisms of signal transduction. We therefore used Y2H screening with a highly complex random primed cDNA library from control and water stressed soybean tissues and screened to saturation. As a consequence, we were able to not only discover interacting proteins but also to assign a confidence value to these interactions and map interacting domains (Fig. 4).
It is usual to verify the interactions from a yeast 2 hybrid screen using independent methods. Our future work will focus on further defining the interactions that we have identified using GmWRKY53 as a bait, but in the meantime, we have good confidence in the interactions because of the multiple independent fragments for many interacting proteins that enabled delineation of minimal interacting domains and computation of a confidence score (MYB114, Glyma10g33810, Glyma20g33800, and Glyma03g26310, and also Glyma15g08160). Additionally, there are over 200 ERF genes in soybean and so the isolation of 2 paralogs (Glyma20g33800, and Glyma03g26310) is therefore very unlikely by chance alone when there are only 3 total ERF proteins isolated as interacting with GmWRKY53. Finally, there are now numerous publications that show that multiple VQ proteins typically interact with Group IIc and Group I WRKY transcription factors. Our data on VQ proteins interacting with GmWRKY53 is fully consistent with not only the identity of the interacting proteins (VQ) but also the large number of interactors from this family (19).
One of the highest confidence interactors was a soybean orthologs of the LHY clock gene, GmLCL2 (MYB114). Multiple independent clones were obtained for GmLCL2 (Fig. 1A) and we mapped the interaction domain on the GmLCL2 protein to a region of 80 6 amino acids. Comparisons with the Arabidopsis LHY protein suggest that the C-terminal half of this region may represent the core of the interaction domain (Fig. 1).
This raises interesting possibilities concerning the role of this interaction. Several clock components are known to interact with other proteins but these proteins tend to be essential for their regulatory activity.30 There are also interactions between clock components and outputs. Usually this involves regulation of transcription or targeted protein breakdown.30 For example, in abiotic and biotic stress responses it has been shown that CBF genes are influenced by the clock. Also disruption of the clock gene CCA1 in Arabidopsis results in reduced resistance to downy mildew at dawn. Conversely, overexpression enhances resistance.32 It appears that not only does the clock affect immune responses, but immune responses can also affect the clock. Mutants in LHY itself affect basal and resistance gene-mediated defense and treatment with the elicitor flg22 can feedback-regulate clock activity demonstrating crosstalk between the 2 pathways.33 Our results indicate that one possible mechanism of crosstalk might involve direct interaction of the GmWRKY53 TF with LHY (Fig. 1). In such a mechanism, altered levels or modifications of the GmWRKY53 protein due to stress would crosstalk with the clock by directly interacting with LHY. The importance of disease resistance, abiotic stress, and the clock make further analysis of the GmWRKY53/LHY interaction a priority.
GmWRKY53 formed complexes with 2 different types of water stress-inducible ERF proteins (with Glyma03g26310 or Glyma10g33810/Glyma20g33800) suggesting these complexes recognize promoters with both W and GCC-like boxes. Examples of such promoters are known. For example the Gst1 box from the potato GST1 gene34 contains a GCC-like box and a W box separated by just 4 bp. Synthetic promoters made from this promoter region consisting of both a W and GCC box direct strong pathogen and wound-inducible expression in transgenic Arabidopsis.35 It is therefore possible that WRKY/ERF complexes are formed at the sites of these composite promoter elements and this may make for a stronger targeting of stress inducible gene promoters by stress responsive transcription factors.
Soybean contains at least 74 VQ proteins31 and 19 of these were isolated as GmWRKY53-interacting proteins in the Y2H screen (Table 1). Similar data with the Arabidopsis AtWRKY51 protein showed that about half of the 34 Arabidopsis VQ proteins are interactors.11 It is therefore clear that there is not a one-to-one relationship between VQ proteins and WRKY transcription factors and a single WRKY protein can have multiple VQ protein partners. Interaction with a VQ protein can alter the DNA-binding specificity of a WRKY transcription factor and the 19 VQ proteins that we have shown to interact with GmWRKY53 may alter the DNA-binding specificity of the WRKY protein and therefore its target gene preference and biological role.
Other GmWRKY53 interacting proteins suggest that GmWRKY53 may be targeted for degradation by the proteasome and that GmWRKY53 could be involved in G protein signaling. The GEF, Glyma14g00230, is likely involved in the activation of small GTPases. Blast searches with the amino acid sequences of some of the VQ proteins that were isolated in the screen revealed that NCBI annotation predicted by automated computational analysis suggests that 15g08160 is also a GEF and that 13g24700 and 12g35380 are GTPase accelerating proteins (GAPS). GEFS and GAPs are the “on” and “off” switches in G protein signaling, respectively but support for the NCBI automatic annotation is lacking for all except Glyma14g00230. Nevertheless, the possibility of a link between WRKY proteins and G protein signaling has not previously been suggested and it may represent another signaling input for GmWRKY53.
In conclusion, it has recently been shown that the circadian clock affects immune responses and immune responses can also affect the clock.36 Our results indicate that one possible mechanism of this crosstalk might involve direct interaction of the GmWRKY53 transcription factor with the LHY transcription factor, possibly with increased levels or modification of the GmWRKY53 protein under stress promoting interaction with LHY. GmWRKY53 interactions with water stress inducible AP2/ERF transcription factors may represent a component of water stress responses where promoters are targeted by both WRKY and ERF transcription factors in a complex that may increase binding affinity for a promoter and also place a single promoter at the end of 2 separate parts of the signaling web, one regulated by WRKY TFs and the other by AP2/ERF TFs. It is likely that multiple VQ proteins are also part of these signaling networks as 19 different VQ proteins interacted with GmWRKY53 in the Y2H screen.
Materials and Methods
Soybean cv. ‘Williams 82’ seeds were grown in hydroponics using 0.5× Hoagland's solution, pH 5.8 in a growth chamber with a 16 hour/8 hour day/night cycle at 25°C and 50% relative humidity. After 30 days, plants were subjected to water stress by transferring them to empty boxes. Leaves and roots samples were harvested after 0, 0.5, 1, 2, 3 and 5 hr of dehydration and immediately frozen in liquid nitrogen and stored in −80°C. Nine plants were utilized for each time-point (3 replicates per time-point and 3 plants per replicate). PolyA + RNA extraction was performed with pooled samples of roots and leaves from all time points and the control. GmWRKY53 (Glyma19g26400) was used as bait. cDNA synthesis and the yeast-2 hybrid screen (Y2H) was performed at Hybrigenics services, Paris, France as previously described.21 The screen was performed with 50 mM of 3-Aminotriazol to avoid auto-activation. Results were analyzed using BLAST searches against the soybean genome V1.1 at Phytozome (www.phytozome.net),22 the NR database at NCBI (http://blast.ncbi.nlm.nih.gov) as well as against the TAIR10 proteins (www.arabidopsis.org). Alignments were constructed in MEGA6 using MUSCLE23 and the default parameters. For each Neighbor Joining tree,24 the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were determined.25 The evolutionary distances were computed using the Poisson correction method.26 Evolutionary analyses were conducted in MEGA6.27 All positions containing alignment gaps and missing data were eliminated in pairwise sequence. Multiple sequence alignments and consensus sequences were produced using ClustalW2 (http://hmmer.janelia.org)28 using the default settings and visualized using Jalview (www.jalview.org).29 RNA was isolated using QIAGEN© RNeasy-MIDI. 10μg total RNA from each sample was used for micro-array analysis. A custom made 12 × plex array was designed by Roche NimbleGen, Inc.. containing multiple 60mer oligomers to all high and low confidence genes from the GLYMAv1.0 release of the soybean genome. Oligoarray experiments were performed at MOgene, LC (St Louis, MO). Data analysis was performed using ArrayStar v4. Validation of expression of the candidate genes was done by qRT-PCR using Qiagen QuantiTect SYBR Green. The PCR cycle was set as following: step 1: 95°C 15 min; step 2: 95°C 15 sec, 50°C 30 sec, and 72°C 30 sec, × 40; Step 3: melt curve: 95°C 15 sec, 50°C 30 sec, and 95°C 15 sec. The qRT-PCR primers used were as follows: Glyma03g26310 F aagtatgcggcggagattag, R ctcagcggtttcgtaggttc; Glyma20g33800 F ggtcataaagccgtgttgaa, R gaacttgtggcagctgttgt; Glyma10g33810 F tggcacatttgacactgaga, R attcaacacggctttgtgac; GmWRKY53 F tccaattcctcaagctaccc, R catcaccaccaccttgtctc.
Figure 4.
The interactome of GmWRKY53 as determined by Yeast 2-Hybrid library screening to saturation. Red circles and connecting lines indicate very high confidence interactions, dark blue indicates high confidence, green indicates good confidence, and light blue indicates moderate confidence. The names and Glyma IDs of each protein are shown and red lettering indicates possible functions of the interactions..
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank Naveen Kumar, Malini Rao, and Nikhil Kesarla in the Rushton lab.
Funding
This project was supported by National Research Initiative grants 2008-35100-04519 and 2008-35100-05969 from the USDA National Institute of Food and Agriculture.
References
- 1.Park CY, Lee JH, Yoo JH, Moon BC, Choi MS, Kang YH, Lee SM, Kim HS, Kang KY, Chung WS, et al.. WRKY group IId transcription factors interact with calmodulin. FEBS Lett 2005; 579:1545-50; PMID:15733871; http://dx.doi.org/ 10.1016/j.febslet.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 2.Ishihama N, Yamada R, Yoshioka M, Katou S, Yoshioka H. Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 2011; 23:1153-70; http://dx.doi.org/ 10.1105/tpc.110.081794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang I-F, Curran A, Woolsey R, Quilici D, Cushman JC, Mittler R, Harmon A, Harper JF. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 2009; 9:2967-85; PMID:19452453; http://dx.doi.org/ 10.1002/pmic.200800445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J-M, To TK, Nishioka T, Seki M. Chromatin regulation functions in plant abiotic stress responses. Plant, Cell Environ 2010; 33:604-11; http://dx.doi.org/ 10.1111/j.1365-3040.2009.02076.x. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 2013; 74:730-45; PMID:23451802; http://dx.doi.org/ 10.1111/tpj.12159. [DOI] [PubMed] [Google Scholar]
- 6.Shen Q-H, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear Activity of MLA Immune Receptors Links Isolate-Specific and Basal Disease-Resistance Responses. Science 2007; 315:1098-103; PMID:17185563; http://dx.doi.org/ 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 7.Miao Y, Zentgraf U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 2010; 63:179-88; PMID:20409006; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04233.x. [DOI] [PubMed] [Google Scholar]
- 8.Shang Y, Yan L, Liu Z-Q, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al.. The Mg-Chelatase H Subunit of Arabidopsis Antagonizes a Group of WRKY Transcription Repressors to Relieve ABA-Responsive Genes of Inhibition. Plant Cell 2010; 22:1909-35; http://dx.doi.org/ 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z. Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 2013; 6:287-300; PMID:23455420; http://dx.doi.org/ 10.1093/mp/sst026. [DOI] [PubMed] [Google Scholar]
- 10.Cormack RS, Eulgem T, Rushton PJ, Köchner P, Hahlbrock K, Somssich IE. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochimica Biophys Acta 2002; 1576:92-100; http://dx.doi.org/ 10.1016/S0167-4781(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y, Zhou Y, Yang Y, Chi YJ, Zhou J, Chen JY, Wang F, Fan B, Shi K, Zhou YH, et al.. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol 2012; 159:810-25; PMID:22535423; http://dx.doi.org/ 10.1104/pp.112.196816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 2011; 23:3824-41; PMID:21990940; http://dx.doi.org/ 10.1105/tpc.111.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NHT, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al.. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 2005; 24:2579-89; PMID:15990873; http://dx.doi.org/ 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perruc E, Charpenteau M, Ramirez BC, Jauneau A, Galaud J-P, Ranjeva R, Ranty B. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J 2004; 38:410-20; PMID:15086802; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02062.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang A, Garcia D, Zhang H, Feng K, Chaudhury A, Berger F, Peacock WJ, Dennis ES, Luo M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 2010; 63:670-9; PMID:20545893; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04271.x. [DOI] [PubMed] [Google Scholar]
- 16.Hu P, Zhou W, Cheng Z, Fan M, Wang L, Xie D. JAV1 Controls Jasmonate-Regulated Plant Defense. Mol Cell 2013; 50:504-15; PMID:23706819; http://dx.doi.org/ 10.1016/j.molcel.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 17.Pecher P, Eschen-Lippold L, Herklotz S, Kuhle K, Naumann K, Bethke G, Uhrig J, Weyhe M, Scheel D, Lee J. The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of 'VQ-motif'-containing proteins to regulate immune responses. New Phytol 2014; 203:592-606; PMID:24750137; http://dx.doi.org/ 10.1111/nph.12817. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q-Y, Tian A-G, Zou H-F, Xie Z-M, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 2008; 6:486-503; PMID:18384508; http://dx.doi.org/ 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 19.Tripathi P, Rabara RC, Lin J, Rushton PJ. GmWRKY53, a water- and salt-inducible soybean gene for rapid dissection of regulatory elements in BY-2 cell culture. Plant Signal Behav 2013; 8:e24097; PMID:23511199; http://dx.doi.org/ 10.4161/psb.24097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radwan O, Liu Y, Clough SJ. Transcriptional Analysis of Soybean Root Response to Fusarium virguliforme, the Causal Agent of Sudden Death Syndrome. Mol Plant Microbe Interact 2011; 24:958-72; PMID:21751852; http://dx.doi.org/ 10.1094/MPMI-11-10-0271. [DOI] [PubMed] [Google Scholar]
- 21.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al.. Protein interaction mapping: a Drosophila case study. Genome Res 2005; 15:376-84; PMID:15710747; http://dx.doi.org/ 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 2012; 40:D1178-86; PMID:22110026; http://dx.doi.org/ 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-7; PMID:15034147; http://dx.doi.org/ 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evolut 1987; 4:406-25; PMID:3447015. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985; 39:783-91; http://dx.doi.org/ 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 26.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol 1965; 8:357-66; PMID:5876245; http://dx.doi.org/ 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evolut 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/ 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 2011; 39:W29-37; PMID:21593126; http://dx.doi.org/ 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics 2004; 20:426-7; PMID:14960472; http://dx.doi.org/ 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 30.Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends Plant Sci 2014; 19:240-9; PMID:24373845; http://dx.doi.org/ 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Zhang H, Sun G, Jin Y, Qiu L. Identification of active VQ motif-containing genes and the expression patterns under low nitrogen treatment in soybean. Gene 2014; 543:237-43; PMID:24727126; http://dx.doi.org/ 10.1016/j.gene.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, Lee DU, Fu XD, Dong X. Timing of plant immune responses by a central circadian regulator. Nature 2011; 470:110-4; PMID:21293378; http://dx.doi.org/ 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Xie Q, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong D, Kwak JM, et al.. Crosstalk between the Circadian Clock and Innate Immunity in Arabidopsis. PLoS Pathog 2013; 9:e1003370; PMID:23754942; http://dx.doi.org/ 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strittmatter G, Gheysen G, Gianinazzi-Pearson V, Hahn K, Niebel A, Rohde W, Tacke E. Infections with various types of organisms stimulate transcription from a short promoter fragment of the potato gst1 gene. Mol Plant Microbe Interact 1996; 9:68-73; PMID:8589425; http://dx.doi.org/ 10.1094/MPMI-9-0068. [DOI] [PubMed] [Google Scholar]
- 35.Rushton PJ, Reinstadler A, Lipka V, Lippok B, Somssich IE. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 2002; 14:749-62; PMID:11971132; http://dx.doi.org/ 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roden LC, Ingle RA. Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 2009; 21:2546-52; PMID:19789275; http://dx.doi.org/ 10.1105/tpc.109.069922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.