Abstract
Carcinogenesis is etiologically associated with somatic mutations of critical genes. Recently, a number of somatic mutations and key molecules have been found to be involved in functional networks affecting cancer progression. Suitable animal models are required to validate cancer-promoting or -inhibiting capacities of these mutants and molecules. Sleeping Beauty transposon system consists of a transposon that carries gene(s) of interest and a transposase that recognizes, excises, and reinserts genes in given location of the genome. It can create both gain-of-function and loss-of-function mutations, thus being frequently chosen to investigate the etiological mechanisms and gene therapy for cancers in animal models. In this review, we summarized current advances of Sleeping Beauty transposon system in revealing molecular mechanism of cancers and improving gene therapy. Understanding molecular mechanisms by which driver mutations contribute to carcinogenesis and metastasis may pave the way for the development of innovative prophylactic and therapeutic strategies against malignant diseases.
Keywords: animal model, driver, gene function, gene therapy, malignant diseases, Sleeping Beauty transposon system, somatic mutation
Abbreviations
- Alb-Cre
Albumin promoter-Cre
- CAG promoter
CMV enhancer/chicken β-actin promoter
- CAR
chimeric antigen receptor
- CIS
common insertion site
- CMV
chimeric cytomegalovirus
- Cre
cyclization recombination enzyme
- CRC
colorectal cancer
- DDE
Asp, Asp, Glu
- DMBA/TPA
12-dimethylbenzanthracene/12-O-tetradecanoylphorbol-13-acetate
- DR
direct orientation
- Fah
fumarylacetoacetate hydrolase gene
- GWAS
gnome wide analysis study
- HBV
Hepatitis B Virus
- HBx
HBV X protein
- HCC
hepatocellular carcinoma
- IRs
inverted repeat sequences
- LsL
loxP-stop-loxP
- PAI
Pro, Ala, Ile
- MPNSTs
malignant peripheral nerve sheath tumor
- MSCV
murine stem cell virus
- PBMCs
peripheral blood mononuclear cells
- RED
Arg, Glu, Asp
- Rtl1
Retrotransposon-like 1
- RosaSBaseLsL
Cre-inducible SBase allele
- SB
Sleeping Beauty
- SBase
Sleeping Beauty transposase
- sgRNA
single guide RNA
- shp53
short hairpin RNA against the Trp53 gene
- StatinAE
angiostatin-endostatin fusion gene
- Trp53
transformation related protein 53
Introduction
Cancer is the leading cause of death in developed countries and the second leading cause of mortality in developing world.1 Carcinogenesis is a long-term process as the human body is continuously exposed to physical, chemical, and biological carcinogenic factors and their complex interactions with genetic variations. It is, at least partly, attributable to the mutations in critical genes responsible for normal programming of cell proliferation, differentiation, and death. These cancer-inducing somatic mutations can be generally classified as driver and passenger mutations. Driver mutations are indispensible for cancer development. They provide pro-cancerous milieu and are positively selected in cancer evolution. It has been summarized that driver mutations in more than 120 genes contribute to the development of cancers.2 However, most somatic mutations are passenger mutations. Passenger mutations accumulated in somatic cells through DNA replication are not subject to positive selection and not directly associated with carcinogenesis. Therefore, it is important to distinguish functional driver mutations from random passenger mutations during systematic mutation screening.2 With the use of genome-wide association study (GWAS) and next-generation sequencing technologies, it is possible to study the complicated associations of genetic mutations with cancer occurrence and progression. Recently, the National Cancer Institute of USA have released the largest-ever database of cancer-related genetic variations, providing a comprehensive resource to investigate targeted treatments for cancers. This gradually enables personalized prophylaxis and treatment of malignant diseases. One challenge of novel molecular therapy is to perform replicable experiments in appropriate animal models before clinical trials. Nowadays, transposon systems are often applied to construct animal models. To choose a proper transposon system for the construction of animal models in cancer etiological research and cancer gene therapy is one of the key steps toward personalized medicine.
Sleeping Beauty Transposon System
Transposon system
Transposon system is a non-viral DNA-mediated gene transfer system. It includes a transposase that is capable of recognizing, excising, and reinserting particular DNA sequences in targeted locations of the genome. In the past decades, a number of transposable elements, including Tc1, Tol2, Minos, Himar1, Hsmar1, Mos1, Frog Prince, and Piggyback, in vertebrate cells have been developed.3-9 Of those, Sleeping Beauty (SB) has been extensively characterized.
SB transposon system and its function
SB transposon system was created from Salmonid, which was first reported in 1997.4 It was named the “Sleeping Beauty” because the transposase isolated from Salmonid fish was transpositionally inactive due to the accumulation of mutations and artificially reawaken by eliminating the inactivating mutations.4,10-12 SB transposon system is a Tc1/mariner-type delivery system that consists of 2 components: the transposase (SBase) and the integration cassette (transposon). SB transposon has 210-250 bp inverted repeats (IRs) at their termini and directly repeated DNA sequence motifs (DRs) at the ends of each IR (termed as IR/DR domain). SB transposon can sandwich a desired genetic cargo within the IR/DR domains (Fig. 1A). SBase has several conserved domains that are critical for its function. At the N-terminus of SBase, a bipartite DNA-binding domain [PAI (Pro, Ala, Ile) and RED (Arg, Glu, Asp)] can confer specific binding to IRs because it overlaps with a nuclear localization signal (NLS) sequence. A domain directing cleavage and insertion as well as targeting genome sequence is located at the C-terminal DDE (Asp, Asp, Glu) motif. It binds to the IRs of SB transposon in a substrate-specific manner, and mediates a precise “cut-and-paste” transposition in vertebrate cells (Fig. 1B).13
Figure 1.
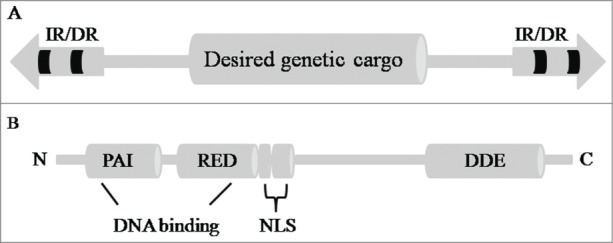
The structures of SB transposon and transposase. (A) The transposon has a desired genetic cargo, which is flanked by terminal inverted repeats (IR/DRs, 2 big arrows), each containing 2 binding sites for the transposase. (B) SB transposase has an N-terminal, bipartite, paired-like DNA-binding domain [PAI (Pro, Ala, Ile), RED (Arg, Glu, Asp)] containing a nuclear localization signal (NLS); and C-terminal, which has the DDE (Asp, Asp, Glu) catalytic domain.
SBase recognizes the IR/DRs terminals of the transposon, excises the transposon, and facilitates its insertion into targeted chromosomal DNA through NLS, a “cut-and-paste” process.10,14 This process can be divided into 5 major steps: (i) specific binding of SBsase to designated sites within the transposon IR/DRs; (ii) pairing of a synaptic complex within 2 ends of the elements and binding together by SBsase subunits; (iii) excision from the donor locus; (iv) recognition of the target sequence in genome; and (v) reintegration at the target locus (Fig. 2).
Figure 2.
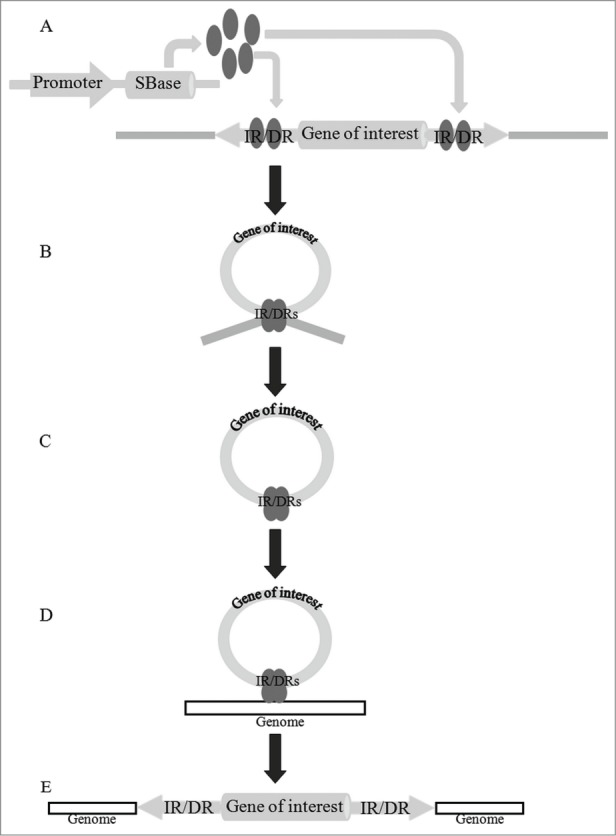
The “cut-and-paste” process of integrating gene(s) of interest into host genome. (A) SBase, whose expression driven by the promoter, recognizes the IR/DR sequence of the transposon and binds to these sites. (B) The synaptic complex formatted. (C) SBase tetramers cut the donor sequence between IR/DR sites. (D) SBase tetramers recognize target sites in the genome and bind to it. (E) The gene(s) of interest between IR/DR sites reintegrate into the genome.
Different subtypes of SB transposon system
To enhance the transposition efficiency, hyperactive versions of SB transposon system have been developed via modifying SBase or the transposon IR coding region.10-12,15,16 For example, several modified versions of SBase including SB10, SB11, SB100,17,18 and SB100X with increasing catalytic activities have been developed. SB100X is up to 100 times more active than the original one, displaying the highest efficiency (24%) compared with SB11 (1.23%) and Piggyback (3.8%).19
However, the transposition efficiency decreases sharply if the inserted sequence is more than 4 kb in length. To solve this problem, a biologically mimic SB transposon system has been developed. It contains a gene of interest flanked by 2 mutant SB transposon elements in an inverted orientation. Since each single mutant SB transposon element contains CA to GC mutations at the terminus of the right IR/DR domain, the induced mutations interfere only with the catalytic steps of transposition but not with SBase binding. As a result, the new SB transposon system has superior ability to transpose genes of >10 kb in length.11
In addition, a cyclization recombination enzyme (Cre) inducible SBase allele, RosaSBaseLsL, which allows the restriction of transposon mutagenesis to a specific tissue of interest, has been established to facilitate the insertion of genes to specific tissue(s).20 Since mammalian genome lacks high affiliative loxP sites, so Cre/loxP system can be effectively applied in mammalian system without confusing with its own system.21,22 The temporal and spatial Cre recombinase expression in mammalian has been well established.23-26 Thus, this system can be applied to transpose gene(s) of interest to specific tissue(s).
Advantages of SB transposon system
SB transposon system becomes popular in mammalian gene transfer due to following reasons. First, the ability of SBase to distinguish its own substrate from very similar sequences and the correction process during synaptic complex formation period make the SB transposon system enable to accurately recognize IR/DRs and catalyze the right substrate.13 Second, SB transposon system is relatively safe for transposition. Studies have shown that neither SBase nor SB transposon has remarkable toxicity in mice.27-30 The effective expression of SBase stops 4 days after hydrodynamic injection.31 Third, the expression of integrated genes via SB system is long-term and reliable, even pass to next generation via germline transmission.32 Fourth, SB transposon system can integrate in various tissues including human cells, one-cell mouse embryo, mouse embryonic stem cells, and mouse somatic tissues. It can theoretically integrate in more than 340 million TA sites, the target sequences for SB transposon system in mouse genome.32-41 Although both Piggyback and SB transposon systems display bias toward integration in actively transcribed loci, SB transposon system can integrate within a wide region of 4 Mb near the donor locus.42 Fifth, with the help of RosaSBaseLsL, SB system can target or integrate genes in given tissues.
Disadvantages of SB transposon system
Nevertheless, there are some disadvantages of SB transposon system. First, overall size of the transposon vector and the ratio of SBase to transposon would affect transposition efficiency, a phenomenon that occurs in SB but not observed in Piggyback.43,44 However, this limitation can be circumvented by that the SB transposon and SBase are transfected with different vectors or that SBase is provided in the form of either mRNA or protein.38,45,46 Second, SB-mediated integrated gene(s) might be transcriptionally silenced in mammalian cells.47,48 Gene-regulatory domain at the terminal of SB may produce complementary RNAs by RNA interference response against the transposon.49,50 The addition of 2 heterologous 5’-HS4 chicken β-globin insulators between genetic cargo and the IR/DR domain may prevent the transcriptional silencing of SB transposon system, thus improving the transposition efficiency.47 Third, due to the “cut-and-paste” mechanism, the SB transposon ends may not be excised, leaving a “footprint” mutation–a 5-bp insertion mutation containing a TA element.51-55
The Application of SB Transposon System for Genetic Etiological Research and Gene Therapy of Cancers
SB transposon system to construct cancer models
Animal models are frequently used in genetic etiological research of cancers. Tumorigenesis can be induced via over-expressing oncogenes and/or down-regulating tumor suppressor genes in animal models. Thus, mammalian models of cancers can be obtained by integrating or targeting specific genes in the genome of animals.56 Transposition using the SB system provides a novel method to construct cancer models.
There are several approaches for SB transposon system to induce tumorigenesis in animals. First, as SB-mediated integrated gene can be transmitted to next generation, it is relatively easy to generate and maintain whole libraries of integrated mutants in the founder animals. Tumors can be observed and analyzed via breeding the founders, skipping the process of classical embryonic stem cell–germline chimera–mutant.20,29,30,57-59 Second, combination of SBase with T2/Onc2 or T2/Onc3 transposon can randomly elicit mutations that result in different types of cancer. T2/Onc2 transposon contains the 5’ long terminal repeat (LTR) of murine stem cell virus (MSCV). T2/Onc3 is identical to T2/Onc2 other than replacing the MSCV LTR with CAG promoter, which consists of cytomegalovirus (CMV) enhancer/β-actin promoter.20 Both transposons can elicit over-expression of nearby proto-oncogenes. SBase can help T2/Onc insert into host genome to activate proto-oncogenes. The 5’LTR in T2/Onc2 usually drives the expression of proto-oncogenes at higher rates in haematopoietic cells than in cells of other histotypes.20 CAG promoter in T2/Onc3 is active in a variety of cell types, including epithelial cells.20 In addition, the RosaSBaseLsL can express SBase depending on the expression of Cre gene. A variety of carcinomas can be induced in different tissues based on different SB transposon systems.20,29,30,57-61 For example, over 20 different types of cancers have been induced using SBase with T2/Onc3 in mice.20 Triple transgenic (Rosa26-lsl-SB11; T2/Onc; albumin promoter-driven Cre) and quadruple transgenic (Rosa26-lsl-SB11; T2/Onc; albumin promoter-driven Cre; p53-lsl-R270H) mice partially generate liver tumors displaying hepatocellular carcinoma (HCC) characteristics and lung metastasis at late stage.29 Similarly, by using Villin-Cre to activate SBase expression in gastrointestinal epithelium specifically, the transgenic mice can develop intraepithelial neoplasia, adenocarcinoma and adenoma.30 With the use of bovine keratin K5 promoter to drive SB11 expression in epidermal stem cells, the transgenic mice (K5-SB11 and T2Onc2) are more likely to generate papilloma, squamous cell carcinoma, and basal cell carcinoma of the skin than the wild-type counterparts after 7,12-dimethylbenzanthracene/12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA) or only TPA treatment.57 SB transposon system harboring the T2/Onc element can facilitate tumorigenesis compared to controls without this SB transposon system.58,59 T2/Onc3 is more powerful than T2/Onc2 in inducing carcinogenesis.59 In addition, different genetic background can influence tumorigenesis in mice. On the Ptch+/− background, the transgenic mice with T2/Onc transposon and cerebellar progenitor cells-specific expressed SB11-transposase driving by Math1 promoter have an increased progression of medulloblastoma. On Tp53mut (Tp53+/− or Tp53−/−) background, this Math1-SB11/T2Onc transposon system facilitates the development of disseminated medulloblastoma.61 Third, replacement of T2/Onc with specific oncogene, SB transposon system can induce specific oncogene-driven carcinomas. For example, through hydrodynamic tail vein injection, SB transposon system containing an activated N-RAS oncogene can elicit multifocal liver cancer in p19Arf-null or heterozygous mice.62 Subcutaneous injection of SB transposon system harboring oncogenes, including c-Myc, H-RAS, and short hairpin RNA against the transformation related protein 53 (Trp53) gene (shp53), into female C57BL/6 mice can induce sarcomatoid carcinomas in skin (Table 1).63 The SB system can also be applied in other mammalians, such as rats. Single gene transgenic rats can be interbred to obtain double-transgenic rats.64 With the help of electroporation method, SB transposon system can also deliver c-Myc, H-RAS, and shp53 oncogenes into rats to produce liver tumor.65 In addition, the SB100X transposon system has been ever used for enzyme-catalyzed gene integration into the embryonic porcine genome.66 Thus, SB transposon system can be applied to construct a variety of animal models for genetic etiological research of cancers.
Table 1.
SB transposon system used for the construction of cancer mouse models
| Transposon insert element | Cancer type | Method | Animals | Ref. |
|---|---|---|---|---|
| T2/Onc3 | Squamous cell carcinoma, HCC | Pro-nuclear injection, knock in ES cell technology, ES cell electroporation, hybridization | C57BL/6J mice and C57BL/6J C3H hybrid mice | 20 |
| T2/Onc | HCC | Pro-nuclear injection, knock in ES cell technology, hybridization | Hepatocyte-specific Alb-Cre mice, Rosa26-lsl-SB11 mice and p53-lsl-R270H mice | 29 |
| T2/Onc2 | Intestinal intraepithelial neoplasia, adenocarcinoma, and adenoma | Pro-nuclear injection, knock in ES cell technology, hybridization | Rosa26-lsl-SB11 mice, Villin-Cre mice, and T2/Onc mice | 30 |
| T2/Onc2 | Skin cancers | Pro-nuclear injection, hybridization | C57BL/6J × DBA/2J F2 embryos, AC heterozygous mice | 57 |
| T2/Onc | Liver cancer | Pro-nuclear injection, knock in ES cell technology, hybridization | Tet-on-MYC mice, LAPtTA mice, Rosa26-SB11 mice, and T2/Onc mice | 58 |
| T2/Onc2 and T2/Onc3 | Pancreatic adenocarcinoma | Pro-nuclear injection, knock in ES cell technology, hybridization | LSL-KrasG12D, Pdx1-Cre, T2Onc2, T2Onc3, and Rosa26-LSL-SB11 transgenic mice | 59 |
| N-RAS | liver cancer | Hydrodynamic tail vein injection | C57BL/6J p19Arf-null mice | 62 |
| H-RAS, c-Myc, shp53 | Sarcomatoid carcinoma | Subcutaneous injection | C57BL/6 female mice | 63 |
| Fah, HBx, shp53 and N-RAS | HCC | Hydrodynamic tail vein injection | Fah−/− mice | 72 |
Alb, Albumin; c-Myc, v-myc avian myelocytomatosis viral oncogene homolog; CRC, colorectal cancer cell; Cre, a cyclization recombination enzyme; ES cell, embryonic stem cell; Fah, fumarylacetoacetate hydrolase gene; HCC, hepatocellular carcinoma; H-RAS, Harvey rat sarcoma virus oncogene; Kras, Kirsten rat sarcoma viral oncogene homolog; LAPtTA, a liver-specific tet-transactivator protein; lsl, loxP-stop-loxP; N-RAS, neuroblastoma ras oncogene; p19Arf, a positive regulator of the p53 tumor suppressor, and loss of Arf predisposes to a wide spectrum of tumors; Pdx1, pancreatic and duodenal homeobox 1; p53-lsl-R270H, R270H targeted point mutations of p53, which is considered as a conditional dominant negative p53 transgene; Rosa26-lsl-SB11, SB11 transposase cDNA preceded by lsl knocked into the Rosa26 locus; tet-o-MYC, tetracycline-repressible MYC transgene; shp53, a short hairpin RNA against tumor suppressor Trp53 encoding gene.
SB transposon system for the discovery of cancer driver mutants
GWAS, microarray, and deep sequencing for cancer gene discovery are carried out via comparing the differences between cases and controls or between cancerous tissues and non-cancerous tissues from the same individual.67-71 These designs are hard to distinguish driver mutations from passenger mutations because cross-sectional case-control studies can only indicate statistical associations between factors of interest with the diseases. If loss-of-function or gain-of-function mutations can promote carcinogenesis, these mutations are more likely to be driver mutations. SB transposon system can introduce gain-of-function mutations, such as combining with T2/Onc to cause mutations randomly or deliver some oncogenes or tumor suppressor genes. On the other hand, loss-of-function mutations can be achieved by using SB transposon system to deliver specific elements to silence genes, such as shp53.72 Thus, SB can be used to distinguish driver mutations from passenger mutations. Here, we introduce several examples. To identify genetic drivers of malignant peripheral nerve sheath tumor (MPNST), the SB transposon system has been used to characterize mutations in mice based on the following steps. First, SB expression and activity are confirmed by immunohistochemistry and PCR-excision assay, respectively. Second, common insertion site (CIS) analysis is utilized to identify potential driver-mutations by both TAPDANCE CIS and gene centric CIS analysis. Third, relevance of the CIS-associated genes to MPNST is evaluated by cross-species comparative analysis of the CISs to previously generated human array comparative genomic hybridization, SNP array, human gene expression profiling, and methylome data from normal Schwann cells, neurofibromas, and MPSNTs. Fourth, Ingenuity Pathway Analysis, Database for Annotation, Visualization and Integrated Discovery are utilized to identify significantly altered signaling pathways in CISs including Wnt/CTNNB1, PI3K/Akt/mTOR, and growth factor receptor signaling pathways. Last, further validation of novel candidate driver-mutations, like Foxr2, is performed by over expression and knockout experiments.60 The second Nebulin family member, NEBL, is involved in MLL gene rearrangement, a phenomenon frequently observed in infant acute myeloid leukemia. Stable transfection of SB transposon system harboring the expression cassettes for MLL-NEBL and NEBL-MLL has demonstrated that the fusions have oncogenic potential.73 In SB transposon system–induced mouse models, retrotransposon-like 1 (Rtl1) and PDE4D have been identified as drivers of HCC and prostate cancer, respectively.74,75 MYC is a dysregulated gene in human malignancies. Introduction of MYC through SB transposon system can generate liver cancer in a mouse model. Genetic screening and functional validation studies in this model have shown that Ncoa2/Src-2 is a tumor suppressor gene in liver cancer.58 MyoD is a well-known muscle differentiation factor. A recent SB transposon screening study has shown that its expression in cerebellum hinders the development of medulloblastoma, providing further evidence that MyoD is a tumor suppressor gene for medulloblastoma.76 The SB transposon system can also be utilized to identify genes critical in tumor dissemination. Functional genomics has demonstrated that ectopic expression of Eras, Lhx1, Ccrk, and Akt are associated with Sonic Hedgehog signaling induced dissemination process of medulloblastomas in Patched+/− mice.77
SB transposon system for cancer gene therapy
With the continual discovery of genes and genetic mutants that promote cancer development, gene therapy becomes more and more practicable options for cancer treatment. SB transposon system can be implicated in cancer gene therapy.
Viral-based gene delivery is a preferred choice for gene transfer, but it has several limitations. First, viral vectors are very likely to elicit immune/inflammatory or neurotoxic responses that are associated with contamination during bacterial extraction process. Second, viral preparations impose risks of contamination by infectious factors such as endogenous proviruses, or replication-competent viruses. Due to their tendencies to integrate near promoters or transcriptional units, viral vectors may cause unwanted cellular consequences. Third, it is relatively costly and time consuming of viral preparation. To overcome these limitations, SB transposon system is an alternative for cancer gene therapy. For example, an engineered SB transposon system coexpressing a single-chain chimeric antigen receptor (CAR) for human CD19 and CD20 has been used to integrate into the chromosome of T cells from peripheral blood mononuclear cells (PBMNCs) and umbilical cord blood. Stable dual-gene expression in T cells from PBMNCs and umbilical cord blood allows for the enrichment by positive selection with Rituxan. Both CD4+ T cells and CD8+ T cells can display the cytotoxicity against CD19+ leukemia, lymphoma, and erythroleukemia cell lines and release high-levels of antigen-dependent Th1 (but not Th2) cytokines, like granulocyte-macrophage colony-stimulating factor, TNF-α, and IFN-γ. In animal experiments, these engineered T cells significantly decrease tumor growth and increase survival time.78 Following the similar procedures, CD19-CAR-specific T cells,79-81 MART-1 and p53 targeting PBMNCs82 and IL-11Rα-CAR-specific T cells83 constructed using SB transposon system show optimal effects in treating lymphoid malignance or osteosarcoma pulmonary metastases. In addition, SB transposon system is able to directly transfer therapeutic genes in vivo for cancer treatment. A modified SB transposon system containing an angiostatin-endostatin fusion gene (StatinAE) has been applied in the CT26 mouse model of CRC metastatic to the liver. This study has demonstrated that this SB transposon system is effective in treating metastatic CRC.84 SB-mediated insertions of each of StatinAE and a soluble vascular endothelial growth factor receptor are proven to be effective for the treatment of glioma.85 The same is true for the SB-mediated insertion of suicide gene herpes simpex virus thymidine kinase controlled by human telomerase reverse transcriptase promoter and a SV40 enhancer for the treatment of HCC.86 Gene transfer mediated by SB transposon system, can improve the efficacy of immune gene therapy via sustaining cytokine secretion and direct intratumoral delivering of DNA/polyethylenimine complexes of mIFN-γ/SB construction (Table 2).87 Furthermore, the first clinical trial of SB-mediated gene therapy was initiated at MD Anderson Cancer Center in 2013. After modifying T cells collected from patients or from matched donors, CD19-CAR-specific T cells were then used to treat patients with leukemia or lymphoma. No acute and late toxicities were found and one of the first 5 treated patients remained in remission.88
Table 2.
SB transposon system used for cancer gene therapy
| Insert gene | Cancer type | Experimental animals or cells | Method | Ref. |
|---|---|---|---|---|
| CAR for human CD19 and CD20 | CD19+ lymphoid malignancies | PBMNCs and umbilical cord blood T cells; (NOD/SCID) mice irradiated and injected intraperitoneally with Daudi-LVhfflucN | Transfection; infusion in vivo | 78 |
| CAR for human CD19 and CD28 | CD19+ lymphoid malignancies | Daudi (Burkitt lymphoma), HLAnull K562 (erythroleukemia) cells and human PBMCs | Transfection | 79 |
| CD19RCD28 transgene | B-lymphoid malignancies | K562 cells and PBSC | Electroporation | 80 |
| CAR for human CD19 and CD28 | CD19+ lymphoid malignancies | PBMCs from healthy adult volunteer donors, CLL cells, MCL cells, diffuse large B-cell lymphoma cells | Electroporation | 81 |
| p53 TCR and anti-MART-1 TCR | Cancer and immunologic disease | PBMNC | Electroporation | 82 |
| IL-11Rα-CAR | OS lung metastases | Human T cells, K562, Human OS cell lines (CCH-OS-D, KRIB, SAOS-2 and LM7) | Co-electroporation | 83 |
| StatinAE | CRC metastasized to liver | BALB/c female mice with intrasplenically transplanted CT26 colorectal tumors | Hydrodynamically injected | 84 |
| sFlt-1 and statinAE | Glioma | Nude mice with GBM xenografts (subcutaneous injection of U373 or U87 cell lines) | Injection | 85 |
| Suicide gene HSV-TK | HCC | HepG2, Hep3B, Huh7, and hNHeps cell lines | Transfection | 86 |
| INF-γ | Glioblastoma | GL261 cells and C57BL/6 mice | Transfection; slow injection into the skull | 87 |
CAR, chimeric antigen receptor; CLL, chronic lymphocytic leukemia; CRC, colorectal cancer; GBM, glioblastoma multiforme; HCC, hepatocellular carcinoma; HLA, human lymphocyte antigen; HSV-TK, herps simplex virus thymidine kinase; IL-11Rα-CAR, Interleukin-11 receptor α-chain CAR; INF-γ, interferon gamma gene; MCL, mantle cell lymphoma; OS, osteosarcoma; PBMCs, peripheral blood mononuclear cells; PBSC, peripheral blood stem cell; sFlt-1, soluble vascular endothelial growth factor receptor; StatinAE, an angiostatin-endostatin fusion gene; TCR, T-cell receptors.
SB transposon system in Hepatitis B Virus (HBV) integration
SB transposon system had been applied in elucidating the mechanism of HBV-induced carcinogenesis in animal model. With the use of hydrodynamic delivery method, SB transposon system can introduce HBV X (HBx) gene into the livers of fumarylacetoacetate hydrolase (Fah) mutant mice and induce hepatic inflammation. Coexpression of Fah cDNA from the transposon vector allows for the selective repopulation of genetically corrected hepatocytes in Fah mutant mice. The subsequent selective repopulation of hepatocytes carrying the gene(s) of interest could provide useful genetic information about the mechanisms of HBV-induced neoplasm. In this mouse model, introduced HBx can activate the expression of β-catenin and HBx coinjected with shp53 accelerates the formation of liver hyperplasia. Constitutively active v-ras oncogene homolog with Gly12Val substitution (NRASG12V) alone and in combination with shp53 coinjection facilitate hepatic tumorigenesis.72 Thus, SB transposon system can be applied to investigate the oncogenic effects of viral genes.
Conclusion
SB transposon system is a reliable tool to integrate mutations into mammalian cell genome and transmit the genes (or mutants) of interest to next generations. In combination with Cre, SB transposon system allows for the activation of introduced genes (or mutants) in specific tissue(s). SB transposon system theoretically reintegrate in more than 340 million TA sites in mouse genome, which can be applied in systemically screening for cancer-related genes. It can distinguish the driver mutations from passenger mutations and also serve as a delivery toolkit in inserting therapeutic genes into host genome to adjust the imbalances between oncogenes and tumor suppressor genes. SB transposon system can combine different therapeutic approaches to improve cancer treatment. It has been applied in clinical trial for the treatment of leukemia and lymphoma. Although SB transposon system has a number of advantages, it might activate carcinogenesis by unwanted integrations. Furthermore, its safety and duration of gene expression are still uncertain. More researches are needed to improve and optimize the SB transposon system for clinical cancer gene therapy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgement
This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
Funding
This work was supported by the National Key Basic Research Program (Grant No.2015CB554006) and the National Natural Scientific Foundation of China (Grant No. 81025015, 81221061, 81302492, and 91129301). The study sponsors had no role in the study design, in the collection, analysis and interpretation of data; in the writing of the manuscript and in the decision to submit the manuscript for publication.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/ 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2. Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007; 446:153-8; PMID:17344846; http://dx.doi.org/ 10.1038/nature05610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chitilian JM, Thillainadesan G, Manias JL, Chang WY, Walker E, Isovic M, Stanford WL, Torchia J. Critical components of the pluripotency network are targets for the p300/CBP interacting protein (p/CIP) in embryonic stem cells. Stem Cells 2014; 32:204-15; PMID:24115386; http://dx.doi.org/ 10.1002/stem.1564 [DOI] [PubMed] [Google Scholar]
- 4. Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 1997; 91:501-10; PMID:9390559; http://dx.doi.org/ 10.1016/S0092-8674(00)80436-5 [DOI] [PubMed] [Google Scholar]
- 5. Miskey C, Izsvak Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res 2003; 31:6873-81; PMID:14627820; http://dx.doi.org/ 10.1093/nar/gkg910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urschitz J, Moisyadi S. Transpositional transgenesis with. Mob Genet Elements 2013; 3:e25167; PMID:23956948; http://dx.doi.org/ 10.4161/mge.25167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jursch T, Miskey C, Izsvak Z, Ivics Z. Regulation of DNA transposition by CpG methylation and chromatin structure in human cells. Mob DNA 2013; 4:15; PMID:23676100; http://dx.doi.org/ 10.1186/1759-8753-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pflieger A, Jaillet J, Petit A, Auge-Gouillou C, Renault S. Target capture during Mos1 transposition. J Biol Chem 2014; 289:100-11; PMID:24269942; http://dx.doi.org/ 10.1074/jbc.M113.523894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meir YJ, Wu SC. Transposon-based vector systems for gene therapy clinical trials: challenges and considerations. Chang Gung Med J 2011; 34:565-79; PMID:22196059 [PubMed] [Google Scholar]
- 10. Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol 2004; 24:9239-47; PMID:15456893; http://dx.doi.org/ 10.1128/MCB.24.20.9239-9247.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther 2004; 9:292-304; PMID:14759813; http://dx.doi.org/ 10.1016/j.ymthe.2003.11.024 [DOI] [PubMed] [Google Scholar]
- 12. Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 2003; 8:108-17; PMID:12842434; http://dx.doi.org/ 10.1016/S1525-0016(03)00099-6 [DOI] [PubMed] [Google Scholar]
- 13. Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem 2002; 277:34581-8; PMID:12082109; http://dx.doi.org/ 10.1074/jbc.M204001200 [DOI] [PubMed] [Google Scholar]
- 14. Ma K, Wang DD, Lin Y, Wang J, Petrenko V, Mao C. Synergetic targeted delivery of Sleeping-Beauty transposon system to mesenchymal stem cells using LPD nanoparticles modified with a phage-displayed targeting peptide. Adv Funct Mater 2013; 23:1172-81; PMID:23885226; http://dx.doi.org/ 10.1002/adfm.201102963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther 2005; 12:1148-56; PMID:16150650; http://dx.doi.org/ 10.1016/j.ymthe.2005.06.484 [DOI] [PubMed] [Google Scholar]
- 16. Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 2009; 41:753-61; PMID:19412179; http://dx.doi.org/ 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- 17. Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 2009; 47:404-8; PMID:19391106; http://dx.doi.org/ 10.1002/dvg.20508 [DOI] [PubMed] [Google Scholar]
- 18. Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A 2006; 103:15008-13; PMID:17005721; http://dx.doi.org/ 10.1073/pnas.0606979103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xue X, Huang X, Nodland SE, Mates L, Ma L, Izsvak Z, Ivics Z, LeBien TW, McIvor RS, Wagner JE, et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood 2009; 114:1319-30; PMID:19414858; http://dx.doi.org/ 10.1182/blood-2009-03-210005 [DOI] [PubMed] [Google Scholar]
- 20. Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, Largaespada DA, Scheetz TE, Jenkins NA, Copeland NG. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 2009; 69:8150-6; PMID:19808965; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene 2000; 244:47-54; PMID:10689186; http://dx.doi.org/ 10.1016/S0378-1119(00)00008-1 [DOI] [PubMed] [Google Scholar]
- 22. Semprini S, Troup TJ, Kotelevtseva N, King K, Davis JR, Mullins LJ, Chapman KE, Dunbar DR, Mullins JJ. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques. Nucleic Acids Res 2007; 35:1402-10; PMID:17284462; http://dx.doi.org/ 10.1093/nar/gkl1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lou DQ, Molina T, Bennoun M, Porteu A, Briand P, Joulin V, Vasseur-Cognet M, Cavard C. Conditional hepatocarcinogenesis in mice expressing SV 40 early sequences. Cancer Lett 2005; 229:107-14; PMID:16157222; http://dx.doi.org/ 10.1016/j.canlet.2004.12.032 [DOI] [PubMed] [Google Scholar]
- 24. Ito A, Asamoto M, Hokaiwado N, Shirai T. Regulation of cell proliferation by induction of p21/WAF1 in rat bladder carcinoma cells using the Cre–loxP system. Cancer Lett 2003; 193:183-8; PMID:12706876; http://dx.doi.org/ 10.1016/S0304-3835(03)00007-7 [DOI] [PubMed] [Google Scholar]
- 25. Sin YY, Ballantyne LL, Mukherjee K, St Amand T, Kyriakopoulou L, Schulze A, Funk CD. Inducible arginase 1 deficiency in mice leads to hyperargininemia and altered amino acid metabolism. PLoS One 2013; 8:e80001; PMID:24224027; http://dx.doi.org/ 10.1371/journal.pone.0080001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fischer JM, Schepers AG, Clevers H, Shibata D, Liskay RM. Occult progression by Apc-deficient intestinal crypts as a target for chemoprevention. Carcinogenesis 2014; 35:237-46; PMID:23996931; http://dx.doi.org/ 10.1093/carcin/bgt296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 2005; 436:221-6; PMID:16015321; http://dx.doi.org/ 10.1038/nature03691 [DOI] [PubMed] [Google Scholar]
- 28. Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 2005; 436:272-6; PMID:16015333; http://dx.doi.org/ 10.1038/nature03681 [DOI] [PubMed] [Google Scholar]
- 29. Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, Silverstein KA, Sarver A, Starr TK, Akagi K, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol 2009; 27:264-74; PMID:19234449; http://dx.doi.org/ 10.1038/nbt.1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009; 323:1747-50; PMID:19251594; http://dx.doi.org/ 10.1126/science.1163040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bell JB, Aronovich EL, Schreifels JM, Beadnell TC, Hackett PB. Duration of expression and activity of Sleeping Beauty transposase in mouse liver following hydrodynamic DNA delivery. Mol Ther 2010; 18:1796-802; PMID:20628359; http://dx.doi.org/ 10.1038/mt.2010.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet 2000; 25:35-41; PMID:10802653; http://dx.doi.org/ 10.1038/75568 [DOI] [PubMed] [Google Scholar]
- 33. Horie K, Kuroiwa A, Ikawa M, Okabe M, Kondoh G, Matsuda Y, Takeda J. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc Natl Acad Sci U S A 2001; 98:9191-6; PMID:11481482; http://dx.doi.org/ 10.1073/pnas.161071798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Transposon mutagenesis of the mouse germline. Genetics 2003; 165:243-56; PMID:14504232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis 2001; 30:82-8; PMID:11416868; http://dx.doi.org/ 10.1002/gene.1037 [DOI] [PubMed] [Google Scholar]
- 36. Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol 2003; 23:9189-207; PMID:14645530; http://dx.doi.org/ 10.1128/MCB.23.24.9189-9207.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci U S A 2001; 98:6759-64; PMID:11381141; http://dx.doi.org/ 10.1073/pnas.121569298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dupuy AJ, Clark K, Carlson CM, Fritz S, Davidson AE, Markley KM, Finley K, Fletcher CF, Ekker SC, Hackett PB, et al. Mammalian germ-line transgenesis by transposition. Proc Natl Acad Sci U S A 2002; 99:4495-9; PMID:11904379; http://dx.doi.org/ 10.1073/pnas.062630599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, Nelsestuen G, Key NS, McIvor RS, Hackett PB, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood 2005; 105:2691-8; PMID:15576475; http://dx.doi.org/ 10.1182/blood-2004-09-3496 [DOI] [PubMed] [Google Scholar]
- 40. Belur LR, Frandsen JL, Dupuy AJ, Ingbar DH, Largaespada DA, Hackett PB, Scott McIvor R. Gene insertion and long-term expression in lung mediated by the Sleeping Beauty transposon system. Mol Ther 2003; 8:501-7; PMID:12946324; http://dx.doi.org/ 10.1016/S1525-0016(03)00211-9 [DOI] [PubMed] [Google Scholar]
- 41. Dupuy AJ. Transposon-based screens for cancer gene discovery in mouse models. Semin Cancer Biol 2010; 20:261-8; PMID:20478384; http://dx.doi.org/ 10.1016/j.semcancer.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, Yusa K, Kokubu C, Kinoshita T, Akagi K, Jenkins NA, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods 2005; 2:763-9; PMID:16179923; http://dx.doi.org/ 10.1038/nmeth795 [DOI] [PubMed] [Google Scholar]
- 43. Karsi A, Moav B, Hackett P, Liu Z. Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar Biotechnol (NY) 2001; 3:241-5; PMID:14961361; http://dx.doi.org/ 10.1007/s101260000072 [DOI] [PubMed] [Google Scholar]
- 44. Wilson MH, Coates CJ, George AL Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther 2007; 15:139-45; PMID:17164785; http://dx.doi.org/ 10.1038/sj.mt.6300028 [DOI] [PubMed] [Google Scholar]
- 45. Mikkelsen JG, Yant SR, Meuse L, Huang Z, Xu H, Kay MA. Helper-independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther 2003; 8:654-65; PMID:14529839; http://dx.doi.org/ 10.1016/S1525-0016(03)00216-8 [DOI] [PubMed] [Google Scholar]
- 46. Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol 2000; 302:93-102; PMID:10964563; http://dx.doi.org/ 10.1006/jmbi.2000.4047 [DOI] [PubMed] [Google Scholar]
- 47. Dalsgaard T, Moldt B, Sharma N, Wolf G, Schmitz A, Pedersen FS, Mikkelsen JG. Shielding of sleeping beauty DNA transposon-delivered transgene cassettes by heterologous insulators in early embryonal cells. Mol Ther 2009; 17:121-30; PMID:18985029; http://dx.doi.org/ 10.1038/mt.2008.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garrison BS, Yant SR, Mikkelsen JG, Kay MA. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol 2007; 27:8824-33; PMID:17938204; http://dx.doi.org/ 10.1128/MCB.00498-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moldt B, Yant SR, Andersen PR, Kay MA, Mikkelsen JG. Cis-acting gene regulatory activities in the terminal regions of sleeping beauty DNA transposon-based vectors. Hum Gene Ther 2007; 18:1193-204; PMID:17988194; http://dx.doi.org/ 10.1089/hum.2007.099 [DOI] [PubMed] [Google Scholar]
- 50. Walisko O, Schorn A, Rolfs F, Devaraj A, Miskey C, Izsvak Z, Ivics Z. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther 2008; 16:359-69; PMID:18071335; http://dx.doi.org/ 10.1038/sj.mt.6300366 [DOI] [PubMed] [Google Scholar]
- 51. Liu G, Aronovich EL, Cui Z, Whitley CB, Hackett PB. Excision of Sleeping Beauty transposons: parameters and applications to gene therapy. J Gene Med 2004; 6:574-83; PMID:15133768; http://dx.doi.org/ 10.1002/jgm.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell 2004; 13:279-90; PMID:14759372; http://dx.doi.org/ 10.1016/S1097-2765(03)00524-0 [DOI] [PubMed] [Google Scholar]
- 53. Yant SR, Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol 2003; 23:8505-18; PMID:14612396; http://dx.doi.org/ 10.1128/MCB.23.23.8505-8518.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elick TA, Bauser CA, Fraser MJ. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica 1996; 98:33-41; PMID:8765680; http://dx.doi.org/ 10.1007/BF00120216 [DOI] [PubMed] [Google Scholar]
- 55. Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 2004; 36:283-7; PMID:14981521; http://dx.doi.org/ 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- 56. Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell 2002; 108:135-44; PMID:11832204; http://dx.doi.org/ 10.1016/S0092-8674(02)00621-9 [DOI] [PubMed] [Google Scholar]
- 57. Quintana RM, Dupuy AJ, Bravo A, Casanova ML, Alameda JP, Page A, Sánchez-Viera M, Ramírez A, Navarro M. A transposon-based analysis of gene mutations related to skin cancer development. J Invest Dermatol 2013; 133:239-48; PMID:22832494; http://dx.doi.org/ 10.1038/jid.2012.245 [DOI] [PubMed] [Google Scholar]
- 58. O'Donnell KA, Keng VW, York B, Reineke EL, Seo D, Fan D, Silverstein KA, Schrum CT, Xie WR, Mularoni L, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci U S A 2012; 109:E1377-86; PMID:22556267; http://dx.doi.org/ 10.1073/pnas.1115433109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, Australian Pancreatic Cancer Genome I, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A 2012; 109:5934-41; PMID:22421440; http://dx.doi.org/ 10.1073/pnas.1202490109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rahrmann EP, Watson AL, Keng VW, Choi K, Moriarity BS, Beckmann DA, Wolf NK, Sarver A, Collins MH, Moertel CL, et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet 2013; 45:756-66; PMID:23685747; http://dx.doi.org/ 10.1038/ng.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 2012; 482:529-33; PMID:22343890; http://dx.doi.org/ 10.1038/nature10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc Natl Acad Sci U S A 2005; 102:17059-64; PMID:16286660; http://dx.doi.org/ 10.1073/pnas.0502974102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jung S, Ro SW, Jung G, Ju HL, Yu ES, Son WC. Sleeping Beauty transposon system harboring HRAS, c-Myc and shp53 induces sarcomatoid carcinomas in mouse skin. Oncol Rep 2013; 29:1293-8; PMID:23380875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kitada K, Keng VW, Takeda J, Horie K. Generating mutant rats using the Sleeping Beauty transposon system. Methods 2009; 49:236-42; PMID:19398007; http://dx.doi.org/ 10.1016/j.ymeth.2009.04.010 [DOI] [PubMed] [Google Scholar]
- 65. Park JS, Kim BH, Park SG, Jung SY, Lee H, Son WC. Induction of rat liver tumor using the Sleeping Beauty transposon and electroporation. Biochem Biophys Res Commun 2013; 434:589-93; PMID:23583385; http://dx.doi.org/ 10.1016/j.bbrc.2013.03.119 [DOI] [PubMed] [Google Scholar]
- 66. Garrels W, Mates L, Holler S, Dalda A, Taylor U, Petersen B, Niemann H, Izsvak Z, Ivics Z, Kues WA. Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS One 2011; 6:e23573; PMID:21897845; http://dx.doi.org/ 10.1371/journal.pone.0023573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 2013; 45:72-5; PMID:23242368; http://dx.doi.org/ 10.1038/ng.2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nam RK, Zhang W, Siminovitch K, Shlien A, Kattan MW, Klotz LH, Trachtenberg J, Lu Y, Zhang J, Yu C, et al. New variants at 10q26 and 15q21 are associated with aggressive prostate cancer in a genome-wide association study from a prostate biopsy screening cohort. Cancer Biol Ther 2011; 12:997-1004; PMID:22130093; http://dx.doi.org/ 10.4161/cbt.12.11.18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kashyap MK, Marimuthu A, Kishore CJ, Peri S, Keerthikumar S, Prasad TS, Mahmood R, Rao S, Ranganathan P, Sanjeeviah RC, et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther 2009; 8:36-46; PMID:18981721; http://dx.doi.org/ 10.4161/cbt.8.1.7090 [DOI] [PubMed] [Google Scholar]
- 70. Aquino G, Pannone G, Santoro A, Liguori G, Franco R, Serpico R, Florio G, De Rosa A, Mattoni M, Cozza V, et al. pEGFR-Tyr 845 expression as prognostic factors in oral squamous cell carcinoma: a tissue-microarray study with clinic-pathological correlations. Cancer Biol Ther 2012; 13:967-77; PMID:22825335; http://dx.doi.org/ 10.4161/cbt.20991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013; 501:506-11; PMID:24037378; http://dx.doi.org/ 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Keng VW, Tschida BR, Bell JB, Largaespada DA. Modeling hepatitis B virus X-induced hepatocellular carcinoma in mice with the Sleeping Beauty transposon system. Hepatology 2011; 53:781-90; PMID:21374658; http://dx.doi.org/ 10.1002/hep.24091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Emerenciano M, Kowarz E, Karl K, de Almeida Lopes B, Scholz B, Bracharz S, Meyer C, Pombo-de-Oliveira MS, Marschalek R. Functional analysis of the two reciprocal fusion genes MLL-NEBL and NEBL-MLL reveal their oncogenic potential. Cancer Lett 2013; 332:30-4; PMID:23340173; http://dx.doi.org/ 10.1016/j.canlet.2012.12.023 [DOI] [PubMed] [Google Scholar]
- 74. Riordan JD, Keng VW, Tschida BR, Scheetz TE, Bell JB, Podetz-Pedersen KM, Moser CD, Copeland NG, Jenkins NA, Roberts LR, et al. Identification of rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet 2013; 9:e1003441; PMID:23593033; http://dx.doi.org/ 10.1371/journal.pgen.1003441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rahrmann EP, Collier LS, Knutson TP, Doyal ME, Kuslak SL, Green LE, Malinowski RL, Roethe L, Akagi K, Waknitz M, et al. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res 2009; 69:4388-97; PMID:19401450; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dey J, Dubuc AM, Pedro KD, Thirstrup D, Mecham B, Northcott PA, Wu X, Shih D, Tapscott SJ, LeBlanc M, et al. MyoD is a tumor suppressor gene in medulloblastoma. Cancer Res 2013; 73:6828-37; PMID:24092238; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0730-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mumert M, Dubuc A, Wu X, Northcott PA, Chin SS, Pedone CA, Taylor MD, Fults DW. Functional genomics identifies drivers of medulloblastoma dissemination. Cancer Res 2012; 72:4944-53; PMID:22875024; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang X, Guo H, Kang J, Choi S, Zhou TC, Tammana S, Lees CJ, Li ZZ, Milone M, Levine BL, et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther 2008; 16:580-9; PMID:18227839; http://dx.doi.org/ 10.1038/sj.mt.6300404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, Hackett PB, Kohn DB, Shpall EJ, Champlin RE, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res 2008; 68:2961-71; PMID:18413766; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kebriaei P, Huls H, Jena B, Munsell M, Jackson R, Lee DA, Hackett PB, Rondon G, Shpall E, Champlin RE, et al. Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Hum Gene Ther 2012; 23:444-50; PMID:22107246; http://dx.doi.org/ 10.1089/hum.2011.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Torikai H, Reik A, Liu PQ, Zhou Y, Zhang L, Maiti S, Huls H, Miller JC, Kebriaei P, Rabinovitch B, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood 2012; 119:5697-705; PMID:22535661; http://dx.doi.org/ 10.1182/blood-2012-01-405365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther 2009; 16:1042-9; PMID:19494842; http://dx.doi.org/ 10.1038/gt.2009.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Huang G, Yu L, Cooper LJ, Hollomon M, Huls H, Kleinerman ES. Genetically modified T cells targeting interleukin-11 receptor alpha-chain kill human osteosarcoma cells and induce the regression of established osteosarcoma lung metastases. Cancer Res 2012; 72:271-81; PMID:22075555; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Belur LR, Podetz-Pedersen KM, Sorenson BS, Hsu AH, Parker JB, Carlson CS, Saltzman DA, Ramakrishnan S, McIvor RS. Inhibition of angiogenesis and suppression of colorectal cancer metastatic to the liver using the Sleeping Beauty Transposon System. Mol Cancer 2011; 10:14; PMID:21310067; http://dx.doi.org/ 10.1186/1476-4598-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ohlfest JR, Demorest ZL, Motooka Y, Vengco I, Oh S, Chen E, Scappaticci FA, Saplis RJ, Ekker SC, Low WC, et al. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther 2005; 12:778-88; PMID:16150649; http://dx.doi.org/ 10.1016/j.ymthe.2005.07.689 [DOI] [PubMed] [Google Scholar]
- 86. Song J, Kim C, Ochoa ER. Sleeping Beauty-mediated suicide gene therapy of hepatocellular carcinoma. Biosci Biotechnol Biochem 2009; 73:165-8; PMID:19129627; http://dx.doi.org/ 10.1271/bbb.80581 [DOI] [PubMed] [Google Scholar]
- 87. Wu A, Oh S, Ericson K, Demorest ZL, Vengco I, Gharagozlou S, Chen W, Low WC, Ohlfest JR. Transposon-based interferon gamma gene transfer overcomes limitations of episomal plasmid for immunogene therapy of glioblastoma. Cancer Gene Ther 2007; 14:550-60; PMID:17415381; http://dx.doi.org/ 10.1038/sj.cgt.7701045 [DOI] [PubMed] [Google Scholar]
- 88. Clinical Trials Use Sleeping Beauty Gene Transfer to Create CAR T cells [Internet]. Houston, TX, USA: MD Anderson Cancer Center; 2013 Dec 8. 1 p. Available from: http://www.mdanderson.org/newsroom/news-releases/2013/car-t-cells.html [Google Scholar]


