Abstract
Circulating microRNAs (miRNAs) are emerging as promising non-invasive biomarkers for human cancer. Head and neck squamous cell carcinoma (HNSCC) is a prevalent malignancy worldwide, but its overall survival has remained unchanged in the past 3 decades. Biomarkers for evaluating efficacy of cancer therapy are urgently needed. To explore circulating miRNAs as cancer therapy biomarkers, we initially identified that 8 miRNAs were distinctly dysregulated in cancerous tissues compared with adjacent non-cancerous counterparts from 16 patients, using microarray and real-time PCR. Based on this discovery, the comparison study was performed between pre- and 6 months post-operative paired plasma samples on 9 patients. MiR-99a, which was down-regulated in cancerous tissues, was significantly increased in plasma after operation. Meanwhile, oncomiR miR-21 and miR-223 that were up-regulated in cancerous tissues, were significantly reduced in post-operative plasma samples. We firstly report the significant changes of miR-99a in plasma of HNSCC patients after surgery. Furthermore, plasma miR-223 was inversely increased in a patient whose cancer relapsed within 6 months after operation. We conclude that these circulating miRNAs may serve as biomarkers to evaluate the efficacy of therapy and the prognosis of HNSCC.
Keywords: biomarker, circulating miRNA, head and neck squamous cell carcinoma, miR-99a, miR-223
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- miRNA
microRNA
Introduction
Head and neck cancer is a group of epithelial malignancies that originates most often from the oral cavity, oropharynx, hypopharynx, and the larynx. The predominant type of head and neck cancer is squamous cell carcinoma.1 Head and neck squamous carcinomas (HNSCC) represent one of the 6 most common cancer types worldwide. Although advanced surgical techniques and effective adjunctive therapeutic strategies have been developed, the overall survival of HNSCC has remained unchanged in the past 3 decades. Thus, it is necessary to search for better markers for cancer therapeutic evaluations.
MicroRNAs (miRNAs) are a class of 18–25 nucleotide noncoding RNAs that post-transcriptionally regulate numerous gene expressions.2 These RNA molecules are involved in many essential biological activities and miRNA dysregulation correlates with various human cancers.3 Tumor-derived circulating miRNAs exist in the tumor microenvironment, and serve as mediators of cell-cell communication, immune regulation and tumor invasion.4 Recently, circulating miRNAs have been demonstrated to be sufficiently stable in circulation fluid such as plasma and serum,5,6 and therefore they have potential to serve as biomarkers for cancers. Increasing attention is gradually being paid to the search for specific circulating miRNAs as non-invasive biomarkers that enable clinicians to monitor the cancer therapeutic efficiency.7
In the present study, to explore circulating miRNAs for cancer therapy biomarkers, we initially performed a profile study by miRNA microarray on 4 paired HNSCC tumors and adjacent normal tissues, and we confirmed expression levels of the miRNA candidates in operative samples from 16 HNSCC patients (Table 1) by quantitative real-time RT-PCR (qRT-PCR). Based on this discovery, we investigated the expression levels of these dysregulated miRNAs in paired plasma samples from 9 patients before and 6 months after tumor resections to clarify a potentially reliable therapeutic biomarker for HNSCC.
Results
miRNA profilihg in HNSCC tumor tissues versus adjacent non-tumor counterparts
The microarray platform determined 63 differentially (P < 0.05 by Student's paired t-test) expressed miRNAs in 4 HNSCC tumor tissues compared to the adjacent non-tumor counterparts. We selected the top 10 downregulated miRNAs (less than −1.5 in log2 scale) and top 7 upregulated miRNAs (more than 1.5 in log2 scale), signals of which could be detected in all 4 paired samples (Table 2) for further validations.
Table 2.
Candidate miRNAs selected from microarray assay analysis
| 1) Downregulated miRNAs in HNSCC tissues compared with the adjacent non-tumor tissues | |||
|---|---|---|---|
| systematic name | miRBase accession No | fold change (log2) | P-value |
| hsa-miR-99a | MIMAT0000097 | −2.608 | 0.034 |
| hsa-miR-1275 | MIMAT0005929 | −1.800 | 0.047 |
| hsa-miR-125b | MIMAT0000423 | −1.674 | 0.021 |
| hsa-miR-638 | MIMAT0003308 | −1.616 | 0.005 |
| hsa-let-7c | MIMAT0000064 | −1.604 | 0.035 |
| hsa-miR-1224 | MIMAT0005458 | −1.598 | 0.019 |
| hsa-miR-4299 | MIMAT0016851 | −1.572 | 0.015 |
| hsa-miR-572 | MIMAT0003237 | −1.544 | 0.006 |
| hsa-miR-100 | MIMAT0000098 | −1.538 | 0.044 |
| hsa-miR-2861 |
MIMAT0013802 |
−1.500 |
0.010 |
| 2) Upregulated miRNAs in HNSCC tissues compared with the adjacent non-tumor tissues | |||
|
systematic name |
miRBase accession No |
fold change (log2) |
P-value |
| hsa-miR-21 | MIMAT0000076 | 3.334 | 0.007 |
| hsa-miR-155 | MIMAT0000646 | 3.037 | 0.008 |
| hsa-miR-146b | MIMAT0002809 | 2.880 | 0.008 |
| hsa-miR-223 | MIMAT0000280 | 2.870 | 0.036 |
| hsa-miR-34b* | MIMAT0000685 | 1.940 | 0.003 |
| hsa-miR-4286 | MIMAT0016916 | 1.910 | 0.043 |
| hsa-miR-106b | MIMAT0000680 | 1.719 | 0.023 |
Evaluation of miRNA expression in HNSCC cancerous tissues by real-time qRT-PCR analysis
To confirm the results obtained by microarray analysis, we validated the miRNA expressions in HNSCC paired sample tissues (n = 16) by qRT-PCR. Our data demonstrated that 4 miRNAs (miR-21, miR-146b, miR-155, miR-223) were significantly up-regulated and 4 miRNAs (let-7c, miR-99a, miR-100, miR-125b) were down-regulated (Fig. 1) in HNSCC cancerous tissues compared to their adjacent non-tumor counterparts.
Figure 1.
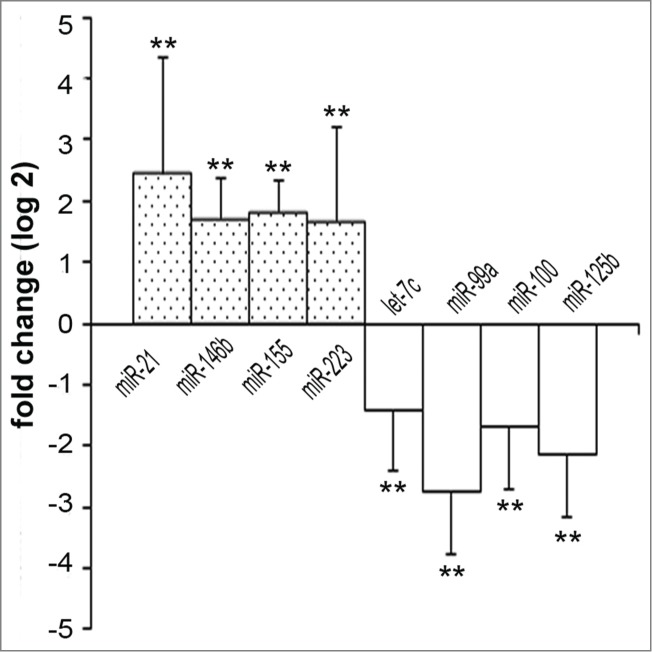
Differentially expressed miRNAs in HNSCC tissues. Dysregulated miRNAs in HNSCC tissues compared with the adjacent non-tumor tissues (n = 16). Fold changes of miRNAs in tumor tissues compared to non-tumor tissues are represented in log2 scale. Statistical analysis was determined by Student's paired t-test using a delta Ct value adjusted with the RNU6B value. **: P < 0.01.
Pre-operative and post-operative plasma miRNAs changes in HNSCC patients
We measured the expressions of 8 dysregulated miRNAs on 9 paired plasma samples and found that circulating miR-21 (P = 0.000, Fig. 2A) and miR-223 (P = 0.029, Fig. 2B) were significantly downregulated in plasma samples 6 months after surgery compared to the pre-operative levels. On the other hand, the expression of plasma miR-99a (P = 0.001) was significantly enhanced 6 months after the operation (Fig. 2C). Remarkably, the expression of miR-223 was upregulated in a patient whose cancer recurred within 6 months after operation (Fig. 2B, dotted line). The levels of other miRNAs did not significantly changed (data not shown).
Figure 2.
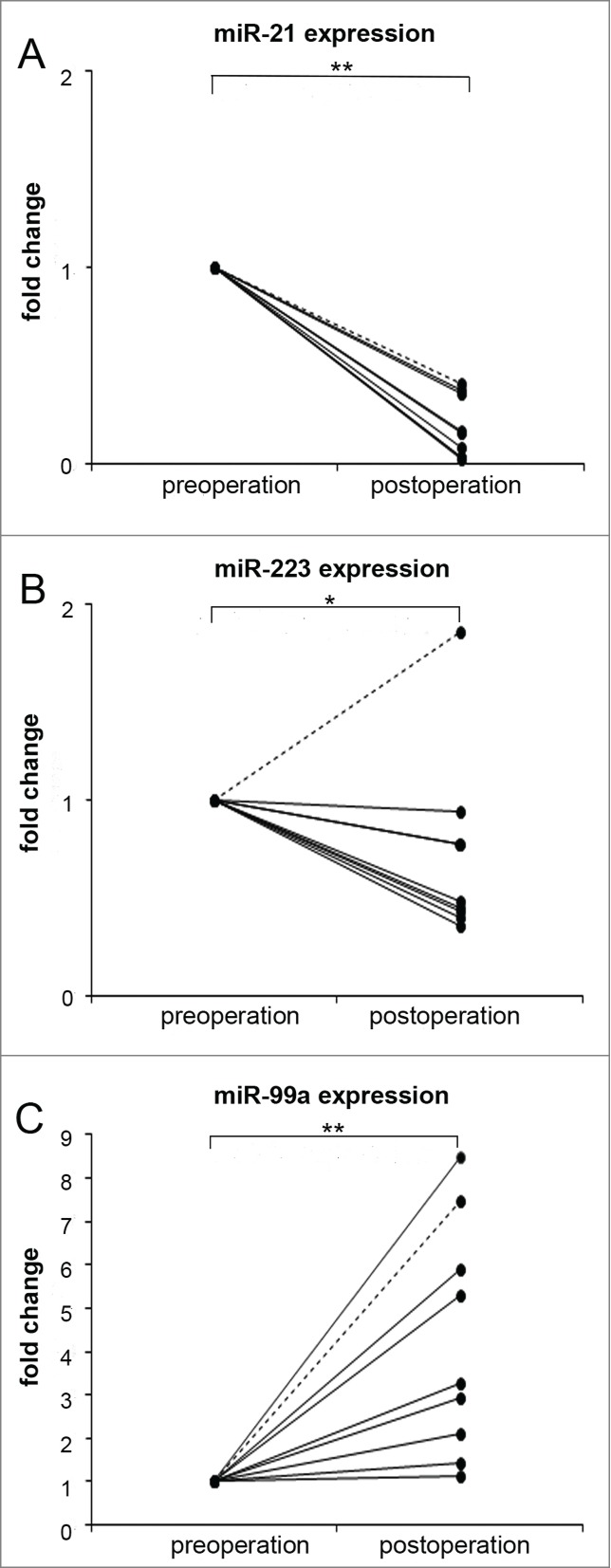
Dysregulated plasma miRNAs in HNSCC patients. Plasma expression levels of miR-21 (A), miR-223 (B) and miR-99a (C) in patients followed for more than 6 months after surgery (n = 9). The pre-operation value of plasma miRNA of each subject is designated as 1, and the post-operation fold changes of miRNA are obtained. Statistical analysis was determined by Student's paired t-test using delta Ct value adjusted with the average value of 4 reference miRNAs. Disease free survival patient: solid line, cancer recurrence patient: dotted line. *: P < 0.05, **: P < 0.01.
Discussion
In the present study, we demonstrated that 63 miRNAs were dysregulated between HNSCC tumors and non-tumor counterparts by miRNA microarray. We then examined the top 17 dysregulated miRNAs independently by RT-qPCR and confirmed that 4 miRNAs (miR-21, miR-146b, miR-155, miR-223) were up-regulated and 4 miRNAs (let-7c, miR-99a, miR-100, miR-125b) were down-regulated in the HNSCC cancerous tissues. Recently, tumor-derived miRNAs were illustrated to be released into the circulation.6 In other words, circulating miRNAs could reflect the miRNA disturbance in tumor tissue and may hold much potential as novel non-invasive biomarkers for cancer processes. Therefore, we expected the changes of miRNA levels in plasma after the surgical removal of the primary tumor. Consequently, we found that the dysregulated miR-21, miR-223 and miR-99a in cancerous tissues were recovered in plasma samples 6 months after the operation. MiR-21 is known as an oncomiR, which is over-expressed miRNA in many cancers, and functions to regulate cell proliferation and apoptosis during neoplastic progression in a variety of cancers.8,9 The decrement of miR-21 and miR-223 after surgery was concordant with previous report in HNSCC.10,11 However, we found that the expression of miR-223 failed to recover in a cancer recurrence patient after surgery. To our knowledge, this is the first report to demonstrate the recovery of miR-99a in plasma samples of HNSCC patients after surgery and the potential changes of miR-223 in the patient with cancer recurrence.
The roles of miR-223 appear to be cancer type specific in carcinogenesis. Decrement of miR-223 is often observed in leukemia disease,12 while increased expression of miR-223 is mainly seen in solid cancer types including HNSCC.13,14 In the present study, we consistently found that miR-223 expression was enhanced in HNSCC cancerous tissues. In addition, a reduction of plasma miR-223 was observed in HNSCC patients who were free from recurrent tumor at 6 months after tumor dissection. However, the expression of plasma miR-223 remained upregulated in a patient with cancer recurrence. A recent report has shown that miR-223 functions as an oncogene and is highly expressed in advanced stage gastric cancer, promoting gastric cancer invasion and metastasis by targeting FBXW7/hCdc4.13 Moreover, Laios et al.15 revealed that miR-223 was up-regulated in recurrent ovarian cancers compared to the primary cancer and its target list was enriched for genes involved in FGF cell signaling, which is crucial for collective invasion of carcinoma cells. Taken together, the dysregulation of plasma miR-223 may serve as a cancer recurrence biomarker in HNSCC.
Decreased miR-99a is consistently detected in HNSCC and various other cancer types.16,17 Tumor suppressor miRNAs are often downregulated in cancer cells. Enhanced expression of miR-99a could lead to a reduction in cell proliferation, cell migration and increased apoptosis, while the down-regulated expression of miR-99a attenuates the inhibitory effect of cell migration and invasion in HNSCC cell lines.18 In our study, miR-99a was the most downregulated miRNA in terms of fold change among HNSCC cancerous tissues compared to their normal counterparts. Moreover, we demonstrated that circulating miR-99a was restored in plasma after tumor resection. A number of functional studies have suggested that miR-99a could suppress oncogenesis post-transcriptionally by targeting MTMR3, IGF1R, mTOR, and SMARCA5.17-20 Taken together, this dysregulation of circulating miR-99a could evaluate the therapeutic efficacy after tumor resection in HNSCC.
Li et al. developed starBase v2.0 (http://starbase.sysu.edu.cn/) to systematically identify the RNA-RNA and protein-RNA interaction networks from 108 CLIP-Seq (PAR-CLIP, HITS-CLIP, iCLIP, CLASH) data sets generated by 37 independent studies on many kinds of cancers.21 By using this database, we checked RNA and miRNA interaction network in HNSCC for above mentioned target genes and miRNAs. The database showed that miR-223 was significantly upregulated and FBXW7 was down-regulated in cancerous tissues of patients with HNSCC compared with normal control, while there was no significant correlation between miR-223 and FBXW7. Among the target genes (MTMR3, IGF1R, mTOR, and SMARCA518-20) of miR-99a, SMARCA5 was up-regulated but there was no significant correlation with miR-99a, and the expression levels of MTMR3 and mTOR were not significantly different between cancerous tissues and normal tissues in HNSCC patients. On the other hand, the database indicated that IGF1R mRNA was significantly increased in HNSCC cancerous tissues and given a negative correlation with miR-99a. Recently, Yen et al. have shown that IGF1R is a target of miR-99a in oral squamous cell carcinoma cells by using lentiviral infection of miR-99a.22 In addition, we confirmed a up-regulation of IGF1R protein expression level in HNSCC cancerous tissues compared to the non-tumor counterparts (Fig. S1, the relative expression levels of IGF1R/GAPDH (n = 12), T: 0.44 ± 0.24, N: 0.20 ± 0.11, P = 0.022 by Student's paired t-test). This indicated the possibility of IGF1R as a putative target for miR-99a in HNSCC.
This study is the first to report that circulating miR-99a can serve as a biomarker for HNSCC therapy. In addition, the alteration of plasma miR-223 may also reflect cancer recurrence. Our results shed light on the basis of circulating miRNA study in HNSCC. Future studies with larger numbers of participants are needed.
Materials and Methods
Patient recruitment and sample collection
Paired tissue samples were collected from 16 patients with HNSCC who were undergoing surgery at the Department of Otorhinolaryngology-Head and Neck Surgery, Mie University Hospital, between 2012 and 2013. Tissue specimens were immediately steeped in RNAlater® (Ambion Inc., Austin, TX, USA) at 4°C for 24 h and subsequently stored at −80°C until analysis. Subsequently, we collected 9 paired plasma samples among 16 patients before and 6 months after tumor resections for perioperative comparison. Clinical characteristics including patient age, sex, and TNM staging are listed in Table 1. One patient was pathologically confirmed as cancer recurrence within 6 months after cancer resection. This study was approved by the Mie University Graduate School of Medicine Ethical Committee (No. 2445). Written informed consent was obtained from each patient before the study.
Table 1.
Clinical features of HNSCC patients
| Group | Patients for operation samples | Patients for pre- and post-operation plasma samples |
|---|---|---|
| Sex | ||
| Male | 15 | 8 |
| Female | 1 | 1 |
| Age | ||
| Range | 48 – 80 | 48 – 80 |
| Mean ± SD | 69.3 ± 8.7 | 69.2 ± 9.2 |
| Sites | ||
| Maxillary sinus | 1 | 0 |
| Oral cavity | 4# | 2# |
| Mesopharynx | 4 | 4 |
| Hypopharynx | 5 | 3 |
| Larynx | 2 | 0 |
| TNM stage | ||
| I | 0 | 0 |
| II | 3 | 2 |
| III | 2 | 0 |
| IV | 11 | 7 |
| Recurrence# | 1 | 1 |
One patient with cancer recurrence.
RNA extraction from tissues
Tissues were disrupted by TissueLyser with 5 mm Stainless Steel Beads (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. RNA was isolated using a mirVana™ miRNA Isolation kit (Ambion, Life Technologies, USA). The integrity and quantity of total RNA were confirmed by gel electrophoresis and a NanoDrop 2000 spectrophotometer (NanoDrop, Wilmington, DE USA).
MiRNA microarray assay
miRNA expression profiling were performed on 4 paired HNSCC tissue samples by Hokkaido System Science (Sapporo, Japan) using a Sureprint G3 Human v16 miRNA, 8 × 60K (miRBase release 16.0) microarray platform (Agilent Technologies) which includes more than 1000 mature miRNAs. Differential expression analysis was performed using GeneSpring GX (Agilent Technologies).
RNA extraction from plasma
RNA was extracted from 200 μl of plasma using a miRNeasy Serum/Plasma Kit (QIAGEN) in the automated QIAcube (QIAGEN), according to the manufacturer's instructions. Synthetic C. elegans miR-39 miRNA mimic (QIAGEN) and carrier RNA (0.94 µg, MS2 bacteriophage total RNA, Roche Applied Sciences, Indianapolis, USA) were spiked-in before RNA extraction. Isolated RNA was eluted in 15 μl RNase-free water.
Reverse transcription and qRT-PCR
For the HNSCC tissues, cDNA was synthesized from 500 ng RNA containing miRNA using a miScript Reverse Transcription Kit (QIAGEN) according to the manufacturer's protocol. qRT-PCR was performed in duplicate using a miScript SYBR Green PCR kit (QIAGEN) for the samples with miScript Universal Primer and the miRNA-specific forward primers. Expression of miRNA was normalized by snRNA RNU6B. With the plasma RNA samples, in order to get higher specificity, we performed universal reverse-transcription reaction using locked nucleic acid (LNA) PCR primers and Exiqon's miRCURY LNA™ miRNA PCR System. Three microliters of RNA eluate was reverse transcribed in 10 μl reactions using the miRCURY LNA™ Universal RT cDNA synthesis kit (Exiqon, Vedbaek, Denmark). Four microliters of 40-fold diluted cDNA was assayed in 10 μl PCR reactions according to the protocol for miRCURY LNA™ Universal RT miRNA PCR. All amplifications were carried out in an ABI Step One Plus Real-time PCR System (Applied Biosystems, Singapore, Singapore). Amplification curves were analyzed using Applied Biosystems SDS software 2.2.2.
As no absolute internal controls exist due to the cell-free conditions, we selected so-called “invariant” miRNAs among several miRNAs as endogenous controls. We evaluated the stability of candidate internal control miRNAs, including spike-in cel-miR-39, RNU6B and 5 reference miRNAs (miR-93, miR-103a, miR-191, miR-423–5p, miR-425) according to refFinder of EST database (http://www.leonxie.com/). Finally, the average value of 4 reference miRNAs (miR-93, miR-103a, miR-191, miR-423) was recommended as an endogenous reference.
Statistical analysis
All statistical analyses were performed using the SPSS 19 statistical software. Two-group comparisons of delta Ct value between tumor tissues and the adjacent non-tumor tissues as well as pre-operation and post-operation plasma were performed using the Student's paired t-test. A P value of less than 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was partly supported by Grants-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 25293149 and No. 23659327) and Mie University Hospital Seed Grant Program 2012, 2013, 2014.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Polanska H, Raudenska M, Gumulec J, Sztalmachova M, Adam V, Kizek R, Masarik M. Clinical significance of head and neck squamous cell cancer biomarkers. Oral Oncol 2014; 50:168-77; PMID:24382422; http://dx.doi.org/ 10.1016/j.oraloncology.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev 2013; 23:3-11; PMID:23465882; http://dx.doi.org/ 10.1016/j.gde.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101:2087-92; PMID:20624164; http://dx.doi.org/ 10.1111/j.1349-7006.2010.01650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011; 8:467-77; PMID:21647195; http://dx.doi.org/ 10.1038/nrclinonc.2011.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al.. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105:10513-8; PMID:18663219; http://dx.doi.org/ 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 2012; 32:326-48; PMID:22383180; http://dx.doi.org/ 10.1002/med.20215 [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, Li X, Tang H. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res 2009; 19:828-37; PMID:19546886; http://dx.doi.org/ 10.1038/cr.2009.72 [DOI] [PubMed] [Google Scholar]
- 9.Reis S, Pontes-Junior J, Antunes A, Dall'Oglio M, Dip N, Passerotti C, Rossini G, Morais D, Nesrallah A, Piantino C, et al.. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol 2012; 12:14; PMID:22642976; http://dx.doi.org/ 10.1186/1471-2490-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol 2012; 33:1933-42; PMID:22811001; http://dx.doi.org/ 10.1007/s13277-012-0454-8 [DOI] [PubMed] [Google Scholar]
- 11.Maclellan SA, Lawson J, Baik J, Guillaud M, Poh CF, Garnis C. Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med 2012; 1:268-74; PMID:23342275; http://dx.doi.org/ 10.1002/cam4.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 2009; 113:5237-45; PMID:19144983; http://dx.doi.org/ 10.1182/blood-2008-11-189407 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Guo Y, Liang X, Sun M, Wang G, De W, Wu W. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol 2012; 138:763-74; PMID:22270966; http://dx.doi.org/ 10.1007/s00432-012-1154-x [DOI] [PubMed] [Google Scholar]
- 14.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B, Waldron J, et al.. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res 2010; 16:1129-39; PMID:20145181; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-2166 [DOI] [PubMed] [Google Scholar]
- 15.Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al.. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer 2008; 7:35; PMID:18442408; http://dx.doi.org/ 10.1186/1476-4598-7-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I, Wang C, Liu X, Zhou X. Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral Oncol 2012; 48:686-91; PMID:22425712; http://dx.doi.org/ 10.1016/j.oraloncology.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, Jensen RV, Moskaluk CA, Dutta A. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res 2011; 71:1313-24; PMID:21212412; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo YZ, Tai YH, Lo HI, Chen YL, Cheng HC, Fang WY, Lin SH, Yang CL, Tsai ST, Wu LW. MiR-99a exerts anti-metastasis through inhibiting myotubularin-related protein 3 expression in oral cancer. Oral Dis 2014; 20:e65-75; PMID:23731011; http://dx.doi.org/ 10.1111/odi.12133 [DOI] [PubMed] [Google Scholar]
- 19.Doghman M, El Wakil A, Cardinaud B, Thomas E, Wang J, Zhao W, Peralta-Del Valle MH, Figueiredo BC, Zambetti GP, Lalli E. Regulation of insulin-like growth factor-mammalian target of rapamycin signaling by microRNA in childhood adrenocortical tumors. Cancer Res 2010; 70:4666-75; PMID:20484036; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM, et al.. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol 2010; 24:447-63; PMID:20081105; http://dx.doi.org/ 10.1210/me.2009-0295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42:D92-7; PMID:24297251; http://dx.doi.org/ 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen YC, Shiah SG, Chu HC, Hsu YM, Hsiao JR, Chang JY, Hung WC, Liao CT, Cheng AJ, Lu YC, et al.. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol Cancer 2014; 13:6; PMID:24410957; http://dx.doi.org/ 10.1186/1476-4598-13-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


