Abstract
Krüppel-like protein Gli-similar 1 (GLIS1) is known as a direct reprogramming factor for the generation of induced pluripotent stem cells. The objective of this study was to investigate the role of GLIS1 in the preimplantation development of bovine embryos. GLIS1 transcripts in in vitro-matured oocytes and 1-cell to 4-cell stage embryos were detected, but they were either absent or at trace levels at the 8-cell to blastocyst stages. We attempted GLIS1 downregulation of bovine early embryos by RNA interference and evaluated developmental competency and gene transcripts, which are involved in zygotic gene activation (ZGA) in GLIS1-downregulated embryos. Injection of specific siRNA resulted in a distinct decrease in GLIS1 transcript in bovine embryos at the 4-cell stage. Although the bovine embryos injected with GLIS1-siRNA could develop to the 16-cell stage, these embryos had difficulty in developing beyond the 32-cell stage. Gene transcripts of PDHA1 and HSPA8, which are transcribed after ZGA, showed lower level in GLIS1 downregulated embryos. It is possible that GLIS1-downregulated embryos fail to initiate ZGA. Our results indicated that GLIS1 is an important factor for the preimplantation development of bovine embryos.
Keywords: Bovine embryo, Early development, Gene expression, GLIS1, RNA interference
Although the procedures for in vitro production (IVP) of domestic animal embryos such as in vitro fertilization (IVF) and somatic cell nuclear transfer (SCNT) have improved, the efficiencies of embryo development and offspring production after embryo transfer are not high. In particular, neonatal and postnatal aberrations have been observed at varying incidence levels after the use of bovine IVF and SCNT procedures [1,2,3,4]. Although the cause of such abnormalities is unknown, the phenomenon may be correlated with abnormal epigenetic status [5,6,7].
During early development, the epigenetic status of embryos, such as DNA methylation levels, changes considerably [8]. This process of epigenetic reprogramming in early embryos removes gamete-specific epigenetic patterns inherited from the parents [8,9,10]. Similarly, SCNT requires the epigenetic information of the donor nucleus to be reprogrammed to an embryonic state [11]. It is a persuasive argument that aberrant epigenetic reprogramming of IVP embryos, such as those produced by IVF and SCNT is responsible for the developmental failure of these embryos. Oocytes can remarkably induce transcription in sperm after fertilization and in somatic nuclei after SCNT. Therefore, oocytes can efficiently reprogram transplanted somatic nuclei to an embryonic state [12, 13]. However, the reprogramming factor(s) in oocytes have not yet been determined, and limited information concerning the mechanism of nuclear reprogramming in oocytes is available.
The Krüppel-like protein Gli-similar 1 (GLIS1) constitutes a subfamily of Krüppel-like zinc finger transcription factors that are closely related to the Gli family [14,15,16]. Recently, GLIS1 has been shown to be a direct reprogramming factor for somatic cell nuclei, in that, GLIS1 markedly enhances the generation of induced pluripotent stem cells (iPSCs) from both mouse and human fibroblasts when it is expressed along with Pou5f1 (OCT3/4), SOX2 and KLF4 [17]. GLIS1 is able to replace oncogenic MYC, resulting in decreased tumorigenicity as well as improved safety and efficiency of iPSCs production [17]. Notably, Glis1 transcript is enriched in unfertilized oocytes and 1-cell stage embryos in mice [17]. Furthermore, Glis1 was reported to be both temporally and spatially regulated, suggesting that it may play a role in the regulation of embryonic development programs at specific stages [15, 18]. However, the role of GLIS1 in preimplantation development of bovine embryos is unclear.
The objectives of this study were to investigate the expression status of the GLIS1 gene in bovine embryos at preimplantation stages and to investigate the role of GLIS1 during the early development of bovine embryos using RNA interference targeted to GLIS1.
Materials and Methods
Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated.
Oocyte collection and in vitro maturation
Cow ovaries were collected at a local slaughterhouse and maintained at room temperature during transport to the laboratory. Cumulus-oocyte complexes (COCs) were aspirated from follicles of 2–8 mm. Ten bovine COCs were matured in a 100 µl drop of IVMD-101 medium (Research Institute for the Functional Peptides, Yamagata, Japan) [19] at 39 C in a humidified atmosphere containing 5% CO2 in air for 22 h.
In vitro fertilization and in vitro culture
After in vitro maturation, COCs were washed with IVF-100 medium (Research Institute for the Functional Peptides, Yamagata, Japan) [19]. Cryopreserved semen was thawed, and sperm were washed twice by centrifugation (at 1800 rpm for 5 min) in IVF-100 medium. The sperm were resuspended in the IVF-100, and 50 µl of this suspension was added to 50 µl drops of IVF-100. The final concentration of sperm was adjusted to 5.0 × 106/ml. COCs (15–20 COCs/drop) were placed into each sperm suspension drop. COCs and sperm were incubated for 6 h at 39 C in a humidified atmosphere containing 5% CO2 in air.
Following microinjection of siRNA, embryos were cultured in modified TALP (mTALP) medium [20], with 0.1% BSA at 39 C in 5% CO2, 5% O2, and 90% N2. On day 2 (IVF = day 0), embryos were transferred to mTALP supplemented with 3% newborn calf serum (Invitrogen, Carlsbad, CA, USA) and subsequently cultured at 39 C in 5% CO2, 5% O2, and 90% N2 until day 7. Rates of embryo development were assessed on day 2 (2-cell ≤), day 3 (8-cell ≤), day 4 (16-cell ≤), day 5 (32-cell ≤), day 6 (morula ≤), and day 7 (blastocyst).
Design of siRNA and microinjection into embryos
The target sites of the GLIS1 transcript were selected from bovine sequences (GenBank accession number: XM_002686397.1). The specific siRNA (GLIS1-siRNA) was designed using siRNA design software, BLOCK-iT RNAi Designer (http://rnaidesigner.invitrogen.com/rnaiexpress/). Both sense and antisense RNA sequences for siRNA were commercially synthesized (Table 1). After insemination, cumulus cells and excess sperm were removed from presumptive zygotes by pipetting. These embryos were subsequently transferred to a 20 µl drop of mTALP containing 1 mg/ml BSA (fraction V) for microinjection. Approximately 10 pl of 50 µM specific siRNA duplexes were injected into the cytoplasm of each embryo using a Transjector 5246 (Eppendorf, Hamburg, Germany). Approximately 10 pl of 20 µM nonsilencing siRNA (AllStars Negative Control siRNA, Qiagen, Tokyo, Japan) was injected as control siRNA by the same method. The embryos were washed three times immediately after microinjection and cultured as described above.
Table 1. Primers and siRNA sequences.
| Name | Nucleotide sequences (5’–3’) | Annealing temperature (C) | Fragment size (bp) | GenBank accession no. |
| GLIS1 | F- ACTGCTCCCAGGCATGTATC | 60 | 216 | XM_002686397.1 |
| R- CCCTTCAGTGGACTGACCAT | ||||
| PGK1 | F- CTGCTGTTCCAAGCATCAAA | 60 | 202 | NM_001034299 |
| R- GCACAAGCCTTCTCCACTTC | ||||
| PDHA1 | F- TGGTCAGGAAGCTTGTTGTG | 60 | 177 | NM_001101046 |
| R- TGCATTGATCCTCCTTTTCC | ||||
| HSPA8 | F- CTGCTTGTGAGCGTGCTAAG | 60 | 226 | NM_174345 |
| R- GAGCCACCAACCAAGACAAT | ||||
| XIST | F- AATAATGCGACAGGCAAAGG | 58 | 168 | AF104906 |
| R- TCCCGCTCATTTTCCATTAG | ||||
| Histone H2A | F- AGGACGACTAGCCATGGACGTGTG | 60 | 208 | NM_174809 |
| R- CCACCACCAGCAATTGTAGCCTTG | ||||
| GLIS1-siRNA | S- GCAUGUAUCCUGGCUCCAUTT | N/A | N/A | N/A |
| AS- AUGGAGCCAGGAUACAUGCTT | N/A | N/A | N/A |
F, forward; R, reverse; S, sense strand; AS, antisense strand.
Determination of the relative abundance of gene transcripts in bovine embryos
In vitro matured (IVM) oocytes and embryos developed to the 1-cell [12 h after in vitro culture (IVC)], 2-cell (18–24 h after IVC), 4-cell (24–48 h after IVC), 8-cell (24–48 h after IVC), 16-cell (48–72 h after IVC), morula (96–120 h after IVC), and blastocyst (144–168 h after IVC) stages were treated with 0.1% protease in 1% PVP-PBS for 5 min, and washed seven times in 1% PVP-PBS. Pools of 20 IVM oocytes, 10 (1-cell to 16-cell stages) or five (morula and blastocyst stages) embryos were added to 5 µl of lysis buffer [0.8% Igepal (ICN Biomedicals, Aurora, OH, USA), 5 mM DTT (Invitrogen) and 1 U[l of RNasin (Promega, Madison, WI, USA)], snap-frozen in liquid nitrogen and stored at –80 C. RNA samples were heated to 80 C for 5 min and treated for reverse transcription (RT) using a QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. Real-time PCRs were performed using a StepOneTM system (Applied Biosystems, Tokyo, Japan), and the products were detected with SYBR Green included in the QuantiTect SYBR Green PCR Master Mix (Qiagen). A 2 µl aliquot of the RT product was used for each quantification. The amplification program was as follows: preincubation at 95 C for 15 min to activate HotStarTaq DNA Polymerase (Qiagen), followed by 45 cycles of denaturation at 94 C for 15 sec, annealing of primers at different temperatures (Table 1) for 30 sec, and elongation at 72 C for 30 sec. After the end of the last cycle, a melting curve was generated by starting fluorescence acquisition at 60 C and recording measurements at 0.3 C increments up to 95 C.
A standard curve was generated for each amplicon by amplifying serial dilutions of a known quantity. PCR products for each gene were purified using a QIAquick PCR Purification Kit (Qiagen), quantified by measuring absorbance at 260 nm using NanoDrop (ND-1000; Thermo Fisher Scientific, Kanagawa, Japan), and diluted. Serial 10-fold dilutions for creating the standard curve were amplified in every real-time PCR run. The standards and cDNA samples were then co-amplified in the same reaction prepared from a master mix. Fluorescence was acquired at each cycle to determine the threshold cycle or the cycle during the log-linear phase of the reaction at which fluorescence increased above the background for each sample. Final quantification was performed using the StepOneTM quantification software. Expression of the target gene in each run was normalized to the internal standard, Histone H2A. Five or six RT samples were used for quantitative analysis, and each sample was run in duplicate for real-time PCR.
Statistical analysis
The percentage data for embryo development were subjected to arcsine transformation. The transformed values were analyzed by one-way analysis of variance, followed by multiple pairwise comparisons using the Tukey-Kramer method. GLIS1 mRNA expression levels in IVM oocytes and embryos at various stages were analyzed by the Kruskal-Wallis test, followed by multiple pairwise comparisons using the Scheffé method. Differences in all mRNA expression levels between control siRNA-injected embryos and GLIS1-siRNA injected embryos were analyzed by the F-test, followed by the Mann-Whitney’s U test or the Student’s t-test. A P value less than 0.05 denoted a statistically significant difference.
Results
Expression of GLIS1 mRNA in IVM oocytes and embryos at various stages
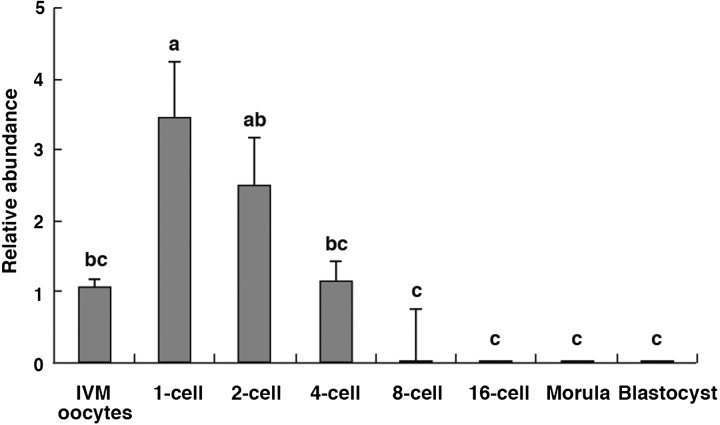
As shown in Fig. 1, GLIS1 mRNA levels in IVM oocytes and embryos at various stages are indicated. GLIS1 transcripts in the 1-cell stage embryos were significantly higher than that in IVM oocytes and 4-cell to blastocyst stage embryos (P < 0.05). GLIS1 transcript levels were decreased at the 8-cell stages, and it was difficult to detect in the embryos following the 8-cell stage (Fig. 1).
Fig. 1.
Relative abundance (mean ± SEM) of GLIS1 transcripts in bovine in vitro matured (IVM) oocytes and 1-cell to blastocyst stage embryos (n = 5 or 6). The relative abundance represents the normalized quantity compared with Histone H2A. a, b, c Different superscripts indicate a significant difference (P < 0.05).
Effect of siRNA injection on GLIS1 expression in bovine embryos
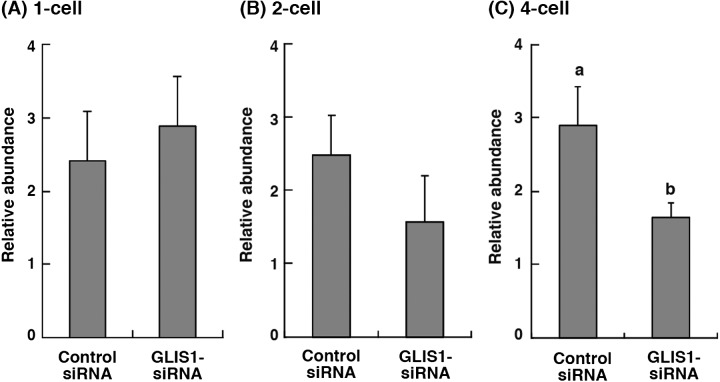
The expression levels of GLIS1 mRNA in 1-, 2- and 4-cell stage embryos obtained from control siRNA or GLIS1-siRNA injection were evaluated (Fig. 2). The relative abundance of GLIS1 in the 4-cell stage embryos injected with GLIS1-siRNA was significantly lower than that in control siRNA-injected embryos (P < 0.05, Fig. 2C). The GLIS1 transcript levels in both 1-, and 2-cell embryos did not differ between treatment groups (Fig. 2A and B).
Fig. 2.
Relative abundance (mean ± SEM) of GLIS1 transcripts in bovine (A) 1-cell, (B) 2-cell, and (C) 4-cell stage embryos obtained from control siRNA or GLIS1-siRNA injection (n = 5 or 6). The relative abundance represents the normalized quantity compared with Histone H2A. a, b Different superscripts indicate a significant difference (P < 0.05).
Effect of GLIS1 downregulation on the development of bovine embryos
In vitro developmental competence of GLIS1-siRNA-injected embryos was evaluated (Table 2). No differences in developmental rates for the 2-cell ≤ (day 2) to 16-cell ≤ (day 4) stages were observed between GLIS1-siRNA-injected and control (uninjected and control siRNA injected) embryos. However, the rate of GLIS1-siRNA-injected embryos that developed to 32-cell ≤ stage (day 5) was significantly lower than that of uninjected embryos (22.6 and 45.6%, respectively, P < 0.05). Both morula ≤ (day 6) and blastocyst (day 7) rates of GLIS1-siRNA-injected embryos (9.0 and 6.5%, respectively) were significantly (P < 0.05) lower than those of the uninjected (45.0 and 43.9%, respectively) and control siRNA-injected (36.9 and 34.5%, respectively) embryos.
Table 2. Effect of GLIS1 siRNA injection on in vitro development of bovine embryos*.
| Treatment | Number of embryos cultured | No. (%)† of embryos develop to |
|||||
| Day 2 |
Day 3 |
Day 4 |
Day 5 |
Day 6 |
Day 7 |
||
| 2-cell ≤ | 8-cell ≤ | 16-cell ≤ | 32-cell ≤ | Morula ≤ | Blastocyst | ||
| Uninjected | 171 | 141 (82.5) | 114 (66.7) | 83 (48.5) | 78 (45.6)a | 77 (45.0)a | 75 (43.9)a |
| Control siRNA | 168 | 142 (84.5) | 107 (63.7) | 62 (36.9) | 63 (37.5)ab | 62 (36.9)a | 58 (34.5)a |
| GLIS1-siRNA | 155 | 139 (89.7) | 94 (60.6) | 49 (31.6) | 35 (22.6)b | 14 (9.0)b | 10 (6.5)b |
* Experiments were replicated five times. † Percentages of the number of embryos cultured. a, b Values with different superscripts within each column differ significantly (P < 0.05).
Gene expressions in bovine embryos derived from GLIS1-siRNA injection
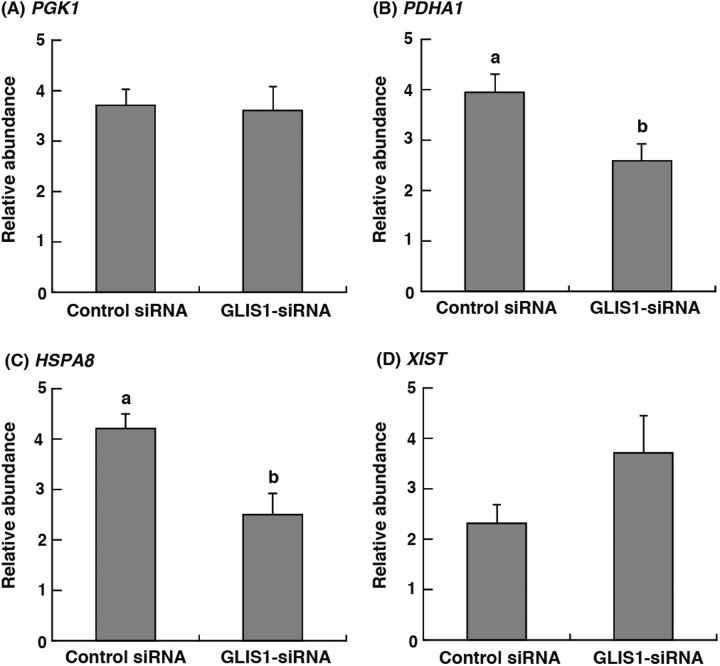
To clarify the effect of GLIS1 downregulation on gene transcripts, phosphoglycerate kinase 1 (PGK1), pyruvate dehydrogenase 1α (PDHA1), heat shock cognate protein 70 (HSPA8), and X inactive specific transcript (XIST) mRNA expression in 8- to 16-cell stage (48–72 h after IVC) embryos were examined. As shown in Fig. 3B and 3C, PDHA1 and HSPA8 transcript levels in GLIS1-siRNA-injected embryos were significantly lower than those in control siRNA-injected embryos (P < 0.05). On the other hand, the relative abundance of PGK1 and XIST did not differ between treatment groups (Fig. 3A and 3D).
Fig. 3.
Relative abundance (mean ± SEM) of (A) PGK1, (B) PDHA1, (C) HSPA8 and (D) XIST transcripts in bovine 8- to 16-cell stage embryos obtained from Control siRNA or GLIS1-siRNA injection (n = 6). The relative abundance represents the normalized quantity compared with Histone H2A. a, b Different superscripts indicate a significant difference (P < 0.05).
Discussion
GLIS1 has been reported as a reprogramming factor in generation of murine and human iPSCs, and it may have a role in the development of mouse embryos after implantation [18]. In the present study, we indicated the GLIS1 expression status and the necessity of GLIS1 transcription for the preimplantation development of bovine embryos.
Glis1 is expressed in a temporal and spatial manner during mouse development; Glis1 expression is most prominent in several defined structures of the mesodermal lineage of the fetus, including craniofacial regions or limb buds [18]. On the other hand, Glis1 mRNA is enriched in metaphase II oocytes and 1-cell stage murine embryos [17]. Glis1 expression levels in mouse embryos decreased at the 2-cell stage, and it was difficult to detect them from the 4-cell to blastocyst stages [17]. In our study, bovine GLIS1 transcripts were detected clearly in IVM oocytes and 1-cell to 4-cell stage embryos, and the GLIS1 transcript level in the 1-cell stage embryos was higher than that in the IVM oocytes. A rise in GLIS1 expression after fertilization has been observed in murine embryos [17]. Thus, it is possible that GLIS1 expression in mammalian embryos increases transiently just after fertilization. On the other hand, the GLIS1 expression in bovine embryos was either absent or at trace levels from the 8-cell to blastocyst stage. These findings indicated that in mammalian embryos, GLIS1 expression is limited to the early stage after fertilization and that GLIS1 transcription resumes after implantation. In the present study, downregulation of GLIS1 mRNA by siRNA injection was detected at the 4-cell stage, but there was no difference in both 1-cell and 2-cell stage embryos. It is well known that siRNA-based RNAi in mammalian cells varies considerably depending on the target sequences selected [21, 22] but that the time required for gene silencing after siRNA injection is unclear. It is possible that a certain amount of time is required to silence genes in early embryos.
In the present study, downregulation of GLIS1 expression in bovine embryos had no effects on development to the 16-cell stage, but embryo development to the 32-cell stage was inhibited. The markedly effects of GLIS1 downregulation also appeared in both development to the morula and blastocyst formation, and developmental competences to both stages were very low. In the mouse, Glis1 mutants were found to be viable and to have no obvious phenotype [23]. The cause of developmental arrest in bovine embryos with GLIS1 suppression was not clear; thus we focused on the developmental stage in which developmental arrest occurred in GLIS1 knockdown embryos. In bovine embryos, zygotic gene activation (ZGA) is well known as an important event that occurs around the 16-cell stage. After fertilization, the developmental program controlled by maternally inherited transcripts in oocytes is replaced by a program controlled by embryonic transcripts [24, 25]. In mammalian embryos, the transition from maternal to embryonic control is an essential event during early development and is termed ZGA [24]. ZGA occurs at the 2-cell stage in mouse embryos [26], at the 4- to 8-cell stage in human [27] and pig embryos [28] and at the 8- to 16-cell stage in bovine embryos [29, 30]. In our study, embryo development from the 16-cell to 32-cell stage was inhibited by GLIS1 downregulation, i.e., developmental arrest was observed at just after ZGA. Therefore, to test the hypothesis that bovine embryos obtained from GLIS1-siRNA injection are defective in terms of ZGA, we evaluated the transcript levels of four genes that start to be transcribed after ZGA in bovine embryos [31]. Consequently, the transcripts of two genes (PDHA1 and HSPA8) in GLIS1 downregulated-embryos were found to have lower levels than control embryos.
PGK1, PDHA1 and HSPA8 are essential housekeeping genes that regulate various physiological functions [31,32,33]. PGK1 and PDHA1 are involved in glucose metabolism, and play an important role in bovine and murine embryo development [34, 35]. HSPA8 encodes heat shock cognate protein 70 (HSC70) [33]. HSC70 has been reported to be involved in numerous of housekeeping and chaperoning functions, such as folding of nascent polypeptides, protein translocation across membranes and chaperone-mediated autophagy [36]. RNAi-based knockdown of HSC70 results in massive cell death in various cell types [37]. XIST is well known as a key factor for X chromosome inactivation [38]. Song et al. reported that in bovine IVF embryos, transcription levels of these four genes markedly increased after ZGA [31]. It has also been reported that PGK1, PDHA1 and HSC70-encoding genes began to express in nonhuman primate embryos after ZGA and that these transcriptions were inhibited by α-amanitin (RNA polymerase inhibitor) treatment [39]. It was suggested that aberrant expression of nuclear proteins causes abnormal nucleologenesis in bovine SCNT embryos [40], which leads to failed ZGA and developmental arrest [41]. In bovine SCNT embryos, after ZGA, the transcript levels of PGK1 and XIST increased, but the HSPA8 and PDHA1 levels did not increase [31]. In the present study, transcripts of HSPA8 and PDHA1 were downregulated in the GLIS1-siRNA-injected embryos, but PGK1 and XIST expressions did not differ from those of control embryos. Although it is possible that the XIST expression level in our results was affected by the sex ratio of the embryos used for gene expression analysis, at least three other genes in the GLIS1-siRNA-injected embryos showed expression patterns similar to SCNT embryos. Together with these results, it is possible that the developmental arrest manifested after GLIS1-siRNA injection is due to failed ZGA in embryos caused by GLIS1 downregulation.
ZGA is a nuclear reprogramming event that changes the transcriptionally inactive embryonic genome to an active one after fertilization [25]. Although little is known regarding the molecular mechanisms of ZGA, oocyte-derived mRNAs and proteins probably play an important role in this reprogramming [25]. As described in the Introduction, GLIS1 is a direct reprogramming factor for the generation of iPSCs from both mouse and human fibroblasts [17]. However, it is not clear whether GLIS1 is involved in nuclear reprogramming after fertilization. Further studies, such as those related to the relationship between GLIS1 and the epigenetic status of the nucleus after fertilization, are necessary. In addition, investigation of GLIS1 has been performed using a limited number of species. Further studies using various species are essential to elucidate of the function of GLIS1 in embryo development.
In conclusion, we found that GLIS1 is essential for early development of bovine embryos. The present study is the first to demonstrate the critical presence and importance of GLIS1 for bovine oocytes and embryos and may also provide the basis for understanding the role of mRNAs accumulated in oocytes for embryo development after fertilization.
Acknowledgments
This study was supported by JSPS KAKENHI Grant number 26292162.
References
- 1.Willadsen SM, Janzen RE, McAlister RJ, Shea BF, Hamilton G, MacDermand D. The viability of late morulae and blastocysts produced by nuclear transplantation in cattle. Theriogenology 1991; 35: 161–170. [Google Scholar]
- 2.Behboodi E, Anderson GB, BonDurant RH, Cargill SL, Kreuscher BR, Medrano JF, Murray JD. Birth of large calves that developed from in vitro-derived bovine embryos. Theriogenology 1995; 44: 227–232. [DOI] [PubMed] [Google Scholar]
- 3.Wilson JM, Williams JD, Bondioli KR, Looney CR, Westhudin ME, McCalla DF. Comparison of birth weight and growth characteristics of bovine calves produced by nuclear transfer (cloning), embryotransfer and natural mating. Anim Reprod Sci 1995; 38: 73–83. [Google Scholar]
- 4.Kruip AM, den Dass JHG. In vitro produced and cloned embryos: effects on pregnancy, parturition and offspring. Theriogenology 1997; 47: 43–52. [Google Scholar]
- 5.Sawai K, Kageyama S, Moriyasu S, Hirayama H, Minamihashi A, Onoe S. Analysis of mRNA transcripts for insulin-like growth factor receptors and binding proteins in bovine embryos derived from somatic cell nuclear transfer. Cloning Stem Cells 2005; 7: 189–198. [DOI] [PubMed] [Google Scholar]
- 6.Wrenzycki C, Herrmann D, Lucas-Hahn A, Korsawe K, Lemme E, Niemann H. Messenger RNA expression patterns in bovine embryos derived from in vitro procedures and their implications for development. Reprod Fertil Dev 2005; 17: 23–35. [DOI] [PubMed] [Google Scholar]
- 7.Sawai K, Takahashi M, Fujii T, Moriyasu S, Hirayama H, Minamihashi A, Hashizume T, Onoe S. DNA methylation status of bovine blastocyst embryos obtained from various procedures. J Reprod Dev 2011; 57: 236–241. [DOI] [PubMed] [Google Scholar]
- 8.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 1987; 99: 371–382. [DOI] [PubMed] [Google Scholar]
- 9.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development 1991; 113: 119–127. [DOI] [PubMed] [Google Scholar]
- 10.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000; 10: 475–478. [DOI] [PubMed] [Google Scholar]
- 11.Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 2003; 69: 902–914. [DOI] [PubMed] [Google Scholar]
- 12.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol 2011; 12: 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi G, Kim YS, Nakajima T, Jetten AM. Regulatory role for Kruppel-like zinc-finger protein Gli-similar 1 (Glis1) in PMA-treated and psoriatic epidermis. J Invest Dermatol 2006; 126: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang HS, ZeRuth G, Lichti-Kaiser K, Vasanth S, Yin Z, Kim YS, Jetten AM. Gli-similar (Glis) Kruppel-like zinc finger proteins: insights into their physiological functions and critical roles in neonatal diabetes and cystic renal disease. Histol Histopathol 2010; 25: 1481–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalesi E, Nakamura H, Lee KL, Putra AC, Fukazawa T, Kawahara Y, Makino Y, Poellinger L, Yuge L, Tanimoto K. The Kruppel-like zinc finger transcription factor, GLI-similar 1, is regulated by hypoxia-inducible factors via non-canonical mechanisms. Biochem Biophys Res Commun 2013; 441: 499–506. [PubMed] [Google Scholar]
- 17.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 2011; 474: 225–229. [DOI] [PubMed] [Google Scholar]
- 18.Kim YS, Lewandoski M, Perantoni AO, Kurebayashi S, Nakanishi G, Jetten AM. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J Biol Chem 2002; 277: 30901–30913. [DOI] [PubMed] [Google Scholar]
- 19.Abe H, Yamashita S, Itoh T, Satoh T, Hoshi H. Ultrastructure of bovine embryos developed from in vitro-matured and -fertilized oocytes: comparative morphological evaluation of embryos cultured either in serum-free medium or in serum-supplemented medium. Mol Reprod Dev 1999; 53: 325–335. [DOI] [PubMed] [Google Scholar]
- 20.Bavister BD, Leibfried ML, Lieberman G. Development of preimplantation embryos of the golden hamster in a defined culture medium. Biol Reprod 1983; 28: 235–247. [DOI] [PubMed] [Google Scholar]
- 21.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res 2002; 30: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 2004; 32: 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakashima M, Tanese N, Ito M, Auerbach W, Bai C, Furukawa T, Toyono T, Akamine A, Joyner AL. A novel gene, GliH1, with homology to the Gli zinc finger domain not required for mouse development. Mech Dev 2002; 119: 21–34. [DOI] [PubMed] [Google Scholar]
- 24.Schultz RM, Davis W, Jr, Stein P, Svoboda P. Reprogramming of gene expression during preimplantation development. J Exp Zool 1999; 285: 276–282. [DOI] [PubMed] [Google Scholar]
- 25.Minami N, Suzuki T, Tsukamoto S. Zygotic gene activation and maternal factors in mammals. J Reprod Dev 2007; 53: 707–715. [DOI] [PubMed] [Google Scholar]
- 26.Kidder GM, McLachlin JR. Timing of transcription and protein synthesis underlying morphogenesis in preimplantation mouse embryos. Dev Biol 1985; 112: 265–275. [DOI] [PubMed] [Google Scholar]
- 27.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988; 332: 459–461. [DOI] [PubMed] [Google Scholar]
- 28.Hyttel P, Laurincik J, Viuff D, Fair T, Zakhartchenko V, Rosenkranz C, Avery B, Rath D, Niemann H, Thomsen PD, Schellander K, Callesen H, Wolf E, Ochs RL, Greve T. Activation of ribosomal RNA genes in preimplantation cattle and swine embryos. Anim Reprod Sci 2000; 60-61: 49–60. [DOI] [PubMed] [Google Scholar]
- 29.Camous S, Kopecny V, Flechon JE. Autoradiographic detection of the earliest stage of [3H]-uridine incorporation into the cow embryo. Biol Cell 1986; 58: 195–200. [DOI] [PubMed] [Google Scholar]
- 30.Frei RE, Schultz GA, Church RB. Qualitative and quantitative changes in protein synthesis occur at the 8-16-cell stage of embryogenesis in the cow. J Reprod Fertil 1989; 86: 637–641. [DOI] [PubMed] [Google Scholar]
- 31.Song BS, Lee SH, Kim SU, Kim JS, Park JS, Kim CH, Chang KT, Han YM, Lee KK, Lee DS, Koo DB. Nucleologenesis and embryonic genome activation are defective in interspecies cloned embryos between bovine ooplasm and rhesus monkey somatic cells. BMC Dev Biol 2009; 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahl HH. Pyruvate dehydrogenase E1 alpha deficiency: males and females differ yet again. Am J Hum Genet 1995; 56: 553–557. [PMC free article] [PubMed] [Google Scholar]
- 33.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett 2007; 581: 3702–3710. [DOI] [PubMed] [Google Scholar]
- 34.Pliss L, Pentney RJ, Johnson MT, Patel MS. Biochemical and structural brain alterations in female mice with cerebral pyruvate dehydrogenase deficiency. J Neurochem 2004; 91: 1082–1091. [DOI] [PubMed] [Google Scholar]
- 35.Machado GM, Ferreira AR, Guardieiro MM, Bastos MR, Carvalho JO, Lucci CM, Diesel TO, Sartori R, Rumpf R, Franco MM, Dode MA. Morphology, sex ratio and gene expression of day 14 in vivo and in vitro bovine embryos. Reprod Fertil Dev 2013; 25: 600–608. [DOI] [PubMed] [Google Scholar]
- 36.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet 1988; 22: 631–677. [DOI] [PubMed] [Google Scholar]
- 37.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev 2005; 19: 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet 2001; 2: 59–67. [DOI] [PubMed] [Google Scholar]
- 39.Zheng P, Patel B, McMenamin M, Reddy SE, Paprocki AM, Schramm RD, Latham KE. The primate embryo gene expression resource: a novel resource to facilitate rapid analysis of gene expression patterns in non-human primate oocytes and preimplantation stage embryos. Biol Reprod 2004; 70: 1411–1418. [DOI] [PubMed] [Google Scholar]
- 40.Laurincik J, Zakhartchenko V, Stojkovic M, Brem G, Wolf E, Muller M, Ochs RL, Maddox-Hyttel P. Nucleolar protein allocation and ultrastructure in bovine embryos produced by nuclear transfer from granulosa cells. Mol Reprod Dev 2002; 61: 477–487. [DOI] [PubMed] [Google Scholar]
- 41.Svarcova O, Laurincik J, Avery B, Mlyncek M, Niemann H, Maddox-Hyttel P. Nucleolar development and allocation of key nucleolar proteins require de novo transcription in bovine embryos. Mol Reprod Dev 2007; 74: 1428–1435. [DOI] [PubMed] [Google Scholar]