Abstract
Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are new tools for producing gene knockout (KO) animals. The current study reports produced genetically modified pigs, in which two endogenous genes were knocked out. Porcine fibroblast cell lines were derived from homozygous α1,3-galactosyltransferase (GalT) KO pigs. These cells were subjected to an additional KO for the cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) gene. A pair of ZFN-encoding mRNAs targeting exon 8 of the CMAH gene was used to generate the heterozygous CMAH KO cells, from which cloned pigs were produced by somatic cell nuclear transfer (SCNT). One of the cloned pigs obtained was re-cloned after additional KO of the remaining CMAH allele using the same ZFN-encoding mRNAs to generate GalT/CMAH-double homozygous KO pigs. On the other hand, the use of TALEN-encoding mRNAs targeting exon 7 of the CMAH gene resulted in efficient generation of homozygous CMAH KO cells. These cells were used for SCNT to produce cloned pigs homozygous for a double GalT/CMAH KO. These results demonstrate that the combination of TALEN-encoding mRNA, in vitro selection of the nuclear donor cells and SCNT provides a robust method for generating KO pigs.
Keywords: Cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH), Double knockout pig, H-D antigen, Transcription activator-like effector nucleases (TALENs), Zinc-finger nucleases (ZFNs)
Gene knockout (KO) mediated by homologous recombination (HR) has been utilized as a standard method for the analysis of gene functions and generation of gene-deficient animals [1,2,3]. However, the generation of KO pigs is a much more laborious process than the generation of KO mice and rats. The major restraint is the unavailability of embryonic stem (ES) cells [4,5,6].
However, the development of somatic cell nuclear transfer (SCNT) facilitated the production of KO pigs [7]. This technology is based on the use of HR-mediated KO primary culture cells, followed by nuclear transfer of the KO cells, without using ES cells. In fact, the first α1,3-galactosyltransferase (GalT) KO pig was developed using this method [4, 5]. However, production of gene KO pigs using the combination of HR and SCNT has a low success rate and is technically challenging. Therefore, only a few groups around the world, including our laboratory, have succeeded in creating GalT KO pigs [4, 5, 8, 9]. In addition to organ transplantation, KO pigs could be used to develop models of inherited genetic disorders. However, the number of practical models created so far has been limited due to the largest bottleneck, that is, the inefficiency of HR [10, 11].
The development of genome editing technology significantly advanced the KO technology. The efficiency of creating KO cells by genome editing technologies, such as zinc-finger nucleases (ZFNs) [12,13,14], transcription activator-like effector nucleases (TALENs) [15,16,17] and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated caspase 9 (Cas9) [18,19,20], is significantly higher compared with the conventional HR method. The use of SCNT for production of genetically modified KO pigs has proven to be a highly reproducible technology [21,22,23]. Therefore, a realistic of production of gene-KO cloned pigs is possible if the efficiency of creating cells with gene KO can be sufficiently increased. In fact, many groups reported the development of gene-KO cloned pigs using genome editing technology in recent years [14, 17, 24].
In the present study, we attempted to create genetically modified cloned pigs by combining two types of genome editing technologies, ZFNs and TALENs, and SCNT. We chose the cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) gene as a target gene. We previously reported the generation of GalT KO pigs, in which the α-galactosyl (α-Gal) epitope (Galα1-3Galβ1-4GlcNAc-R) mediating xenograft rejection was removed [9, 25]. It has also been suggested that removal of the other xeno-epitope, the Hanganutziu-Deicher (H-D) antigen, is also necessary [26, 27]. CMAH is responsible for the synthesis of the H-D antigen [28, 29]. Therefore, development of pigs with a double KO of GalT and CMAH could significantly contribute to the xenograft research.
Materials and Methods
Animal care and chemicals
All of the animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Meiji University (IACUC-12-0007, -12-0008). All chemicals were purchased from Sigma-Aldrich Corporation (St. Louis, MO, USA) unless otherwise indicated.
Design of ZFNs and TALENs and mRNA preparation
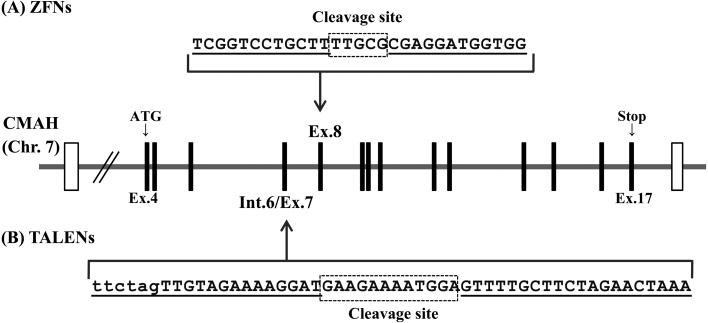
Custom ZFN and TALEN plasmids for pig CMAH were obtained from ToolGen (Seoul, South Korea). The design and validation of these ZFNs and TALENs were performed at ToolGen. The constructed ZFNs and TALENs were designed to target exon 8 and exon 7 of the porcine CMAH gene, respectively. The exon numbers correspond to those of the mouse gene. Each of the ZFN and TALEN domains recognize 12 and 20 DNA bases, respectively (Fig. 1).
Fig. 1.
Schematic diagram of ZFNs and TALENs targeting the porcine CMAH locus. (A) Recognition sites of the ZFNs (pig-CMAH-ZFN) and (B) TALENs (pig-CMAH-TALEN) are indicated by underlining. The coding and intron sequences are indicated by uppercase and lowercase letters, respectively. The cleavage sites indicated by the dotted box are 5 bps and 12 bps, respectively. Translated and untranslated regions in the porcine CMAH gene are indicated by vertical bars and white boxes, respectively.
For the production of mRNAs encoding ZFNs and TALENs, each of the plasmids was digested with the restriction enzymes XhoI and PvuII, respectively. The linearized plasmids were then purified with phenol/chloroform to generate a high-quality DNA template for use in in vitro transcription. Capped ZFN and TALEN mRNAs were produced from the linearized DNA templates via in vitro transcription using MessageMAX T7 ARCA-Capped Message Transcription Kit (Cambio, Cambridge, United Kingdom). A poly (A) tail was then added to each mRNA using the Poly (A) Polymerase Tailing Kit (Cambio). The poly (A)-tailed ZFN and TALEN-encoding mRNAs were then purified with a MEGAclear Kit (Life Technologies, Carlsbad, CA, USA) and resuspended in RNase-free water at a concentration of 400 ng/μl.
Establishment of CMAH KO cells
Skin fibroblast cells were isolated from female GalT KO pigs as previously described [25]. Male fetal fibroblast cells carrying homozygous GalT KO [25, 30] were seeded onto type I collagen-coated dishes or plates (Asahi Glass, Tokyo, Japan) and cultured in MEMα (Life Technologies) supplemented with 15% fetal bovine serum (FBS) (Bovogen Biologicals Pty, Victoria, Australia) and 1% antibiotic-antimycotic solution (Life Technologies) in a humidified atmosphere containing 5% CO2 at 37 C. The fibroblasts were cultured to 70–90% confluency, washed twice with D-PBS(-) (Life Technologies) and treated with 0.05% trypsin-EDTA (Life Technologies) to collect the isolated cells. The cells (4 × 105) were then resuspended in a mix containing 40 μl of R buffer (supplied as part of the Neon Transfection System, Life Technologies) and 2 μl of ZFN- or TALEN-encoding mRNA solution (400 ng/μl). The cells were then subjected to electroporation under the following conditions: pulse voltage, 1,100 V; pulse width, 30 ms; and pulse number, 1 (program #6). Following electroporation, the cells were cultured at 32 C for 2 (ZFNs) or 3 days (TALENs) (transient cold shock) and then without antibiotics for 24 h, after which the antibiotics were added to the media [31]. After the transient cold shock treatment, the cells were cultured at 37 C, and limiting dilution was performed to obtain single-cell-derived clones in five 96-well plates. At 14 days after limiting dilution, the cells with a confluency of more than 50% in each well were selected and divided for further culture and mutation analysis. The cells with a low confluency after limiting dilution were not used in further experiments.
Analysis of ZFN- and TALEN-induced mutations in nuclear donor cells and newborn piglets
The target regions of CMAH ZFNs and TALENs were amplified by direct polymerase chain reaction (PCR) from the clone cells using MightyAmp DNA polymerase (Takara Bio, Shiga, Japan). The primer sequences for ZFN were 5′-TAGAATCCTGTAGTCTCTGC-3′ and 5′-AGAGGCTATGCAAATGCAAGC-3′. The primer sequences for TALEN were 5′-TGCCACAGGATGAAATCCAGAC-3′ and 5′-TCAGGTTCAGTGCCTGGTCTG-3′. Nested PCR was then performed using PrimeSTAR HS DNA Polymerase (Takara Bio), and the appropriate primers were 5′-ACATCCTGAGTGAGTCCGCAAG-3′and 5′-TACCTCAGAATGAGCAGTG-3′ for ZFN and 5′-TGAACATCCAGCTCTCCCATG-3′ and 5′-AGCTGAGATCCACATCAAGC-3′ for TALEN. Subsequently, the PCR fragment including the target region was examined using the BigDye Terminator Cycle Sequencing Kit and an ABI PRISM 3100 Genetic Analyzer (Life Technologies). The sequencing primer was 5′-TCTTGAGTCCTGTGTCATTG-3′ for ZFN and 5′-ATCTGCGATCTCATGAGTTC-3′ for TALEN.
For analysis of the mutations in the newborn piglets, genomic DNA was extracted from the tail biopsies using a DNeasy Tissue and Blood Kit (QIAGEN, Hilden, Germany), and DNA sequencing was then performed as described above.
SCNT and embryo transfer
SCNT was performed as described previously with slight modifications [25]. Briefly, in vitro-matured oocytes containing the first polar body were enucleated via gentle aspiration of the polar body and the adjacent cytoplasm using a beveled pipette in 10 mM HEPES-buffered Tyrode lactose medium containing 0.3% (w/v) polyvinylpyrrolidone, 0.1 mg/ml demecolcine, 5 mg/ml cytochalasin B (CB) and 10% FBS. CMAH KO fibroblasts were used as nuclear donors following cell cycle synchronization via serum starvation for 2 days. A single donor cell was inserted into the perivitelline space of an enucleated oocyte. The donor cell-oocyte complexes were placed in a solution of 280 mM mannitol (Nacalai Tesque, Kyoto, Japan) (pH 7.2) containing 0.15 mM MgSO4, 0.01% (w/v) PVA and 0.5 mM HEPES and were held between 2 electrode needles. Membrane fusion was induced with a somatic hybridizer (LF201; NEPA GENE, Chiba, Japan) by applying a single direct-current (DC) pulse (267 V/mm, 20 μs) and a pre- and post-pulse alternating current (AC) field of 2 V at 1 MHz for 5 sec. The reconstructed embryos were cultured in porcine zygote medium-5 (PZM-5, Research Institute for the Functional Peptides, Yamagata, Japan) supplemented with 4 mg/ml of bovine serum albumin (BSA) for 1–1.5 h, followed by electrical activation. The reconstructed embryos were then washed twice in an activation solution containing 280 mM mannitol, 0.05 mM CaCl2, 0.1 mM MgSO4 and 0.01% (w/v) PVA and were aligned between 2 wire electrodes (1.0 mm apart) of a fusion chamber slide filled with the activation solution. A single DC pulse of 150 V/mm was applied for 100 msec using an electrical pulsing machine (Multiporator; Eppendorf, Hamburg, Germany). After activation, the reconstructed embryos were cultured in PZM-5 for 3 h in the presence of 5 μg/ml CB and 500 nM scriptaid and then with 500 nM scriptaid for another 12–15 h. After the treatments, the SCNT embryos were cultured in PZM-5 for 1–2 days until transfer or for 5–6 days for in vitro development analysis. Embryo culture was performed in a humidified atmosphere containing 5% CO2, 5% O2 and 90% N2 at 38.5 C. After reaching the morula stage, embryos were cultured in PZM-5 supplemented with 10% FBS.
Crossbred (Large White/Landrace × Duroc) prepubertal gilts weighing between 100 and 105 kg were used as recipients for the SCNT embryos. The gilts were given a single intramuscular injection of 1,000 IU of equine chorionic gonadotropin (eCG, ASKA Pharmaceutical, Tokyo, Japan) to induce estrus. Ovulation was induced by an intramuscular injection of 1,500 IU of human chorionic gonadotropin (hCG, Kyoritsu Seiyaku, Tokyo, Japan) 66 h after the injection of eCG. The SCNT embryos cultured for 1–2 days were surgically transferred into the oviducts of recipient gilts 53 h after the hCG injection under general anesthesia.
Immunohistological analysis
A homozygous GalT/CMAH double KO pig (M129-2), a homozygous GalT KO pig (M71-5) and an age-matched wild-type (WT) pig were sacrificed 2 days after birth. Harvested samples were used for immunohistological analysis of the H-D antigen. The dissected organs were fixed in a 4% paraformaldehyde solution (Wako Pure Chemical Industries, Osaka, Japan), embedded in paraffin, sectioned and stained with hematoxylin using standard methods.
The fixed sections were also incubated with blocking solution (2% BSA/D-PBS) for 1 h and then treated with a chicken anti-H-D antibody (a generous gift from Prof N Wakamiya, Asahikawa Medical University) for 1 h. After removal of the excess antibody, the sections were incubated with biotin-conjugated goat anti-chicken IgY (Abcam, Cambridge, United Kingdom) for 30 min. For detection of the α-Gal antigen, the sections were incubated for 1 h with biotin-conjugated Griffonia simplicifolia I (GS-IB4) lectin (Life Technologies). Each section was also incubated with Streptavidin-Horseradish Peroxidase (HRP) (DAKO, Tokyo, Japan) for 15 min and 3,3′-Diaminobenzidine tetrahydrochloride (DAB) (DAKO) for 5 min. The slides were visualized using a Biorevo BZ-9000 microscope (Keyence, Osaka, Japan).
Flow cytometry analysis
Peripheral blood mononuclear cells (PBMCs) were harvested from whole blood of the GalT/CMAH double homozygous KO pig (M129-1) using an erythrocyte lysis solution (Pharm Lyse, Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Cells (1 × 106) were incubated with the chicken anti-H-D antibody for 20 min on ice, followed by incubation with a solution of R-phycoerythrin (PE)-labeled rabbit anti-chicken IgY H&L (Abcam) as a secondary antibody for 20 min on ice. The harvested PBMCs were also incubated with GS-IB4 lectin Alexa Fluor 488 (Life Technologies) for 20 min on ice. After incubation, the cell suspension was washed, and the cells were then resuspended in D-PBS(-) supplemented with 1% FBS (w/v). In all instances, the granulocyte population from the peripheral blood of the GalT/CMAH double homozygous KO pigs was gated, and the gated 1 × 104 events per sample were acquired and analyzed using a Cell Sorter (SH800, Sony, Tokyo, Japan).
Results
Generation of heterozygous CMAH KO cells
Heterozygous CMAH-KO cells were generated by introduction of ZFN-encoding mRNAs into the skin fibroblast cells of a female GalT-KO pig (DK3-9) [25]. Of the 96 single cell-derived cell colonies obtained after the limiting dilution, 6 clones (6/96, 6.3%) with heterozygous CMAH KO were established. Cell clone #75 was selected as the nuclear donor for SCNT based on cell morphology, proliferation ability and normality of the chromosome numbers. Sufficient numbers of heterozygous KO cells were prepared for SCNT after 3 weeks of culture.
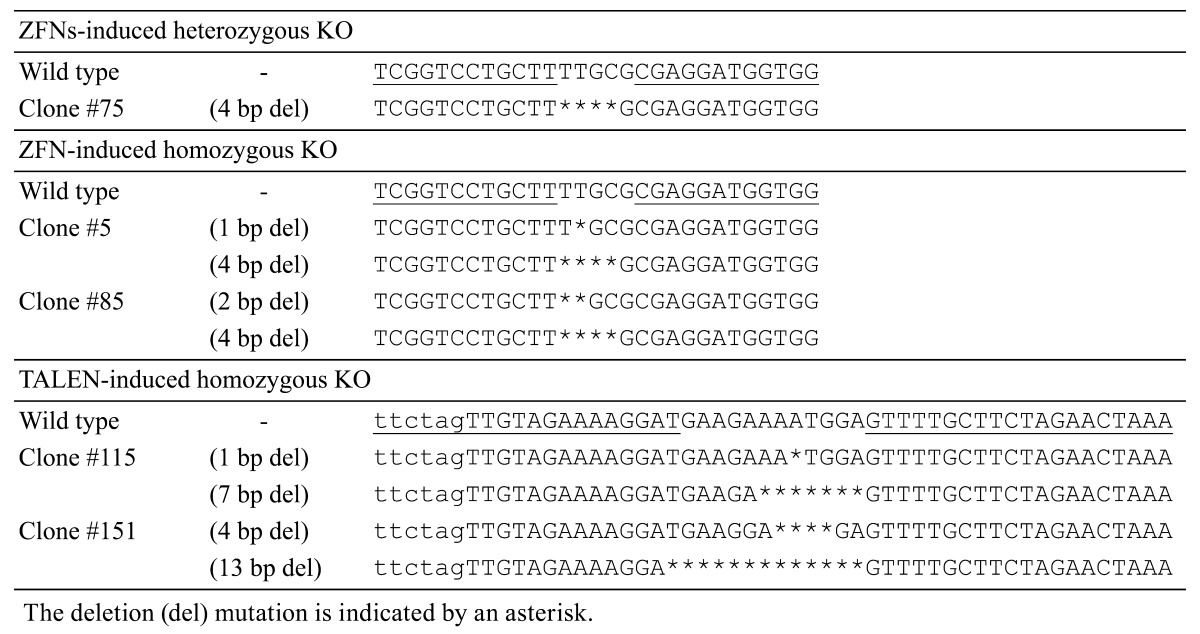
DNA sequence analyses showed that cell clone #75 carried a 4 base pair (bp) deletion (Table 1) causing a loss-of-function mutation of the CMAH gene owing to a frameshift mutation.
Table 1. ZFN and TALEN-induced mutation of nuclear donor cells.
Production of heterozygous CMAH KO pigs
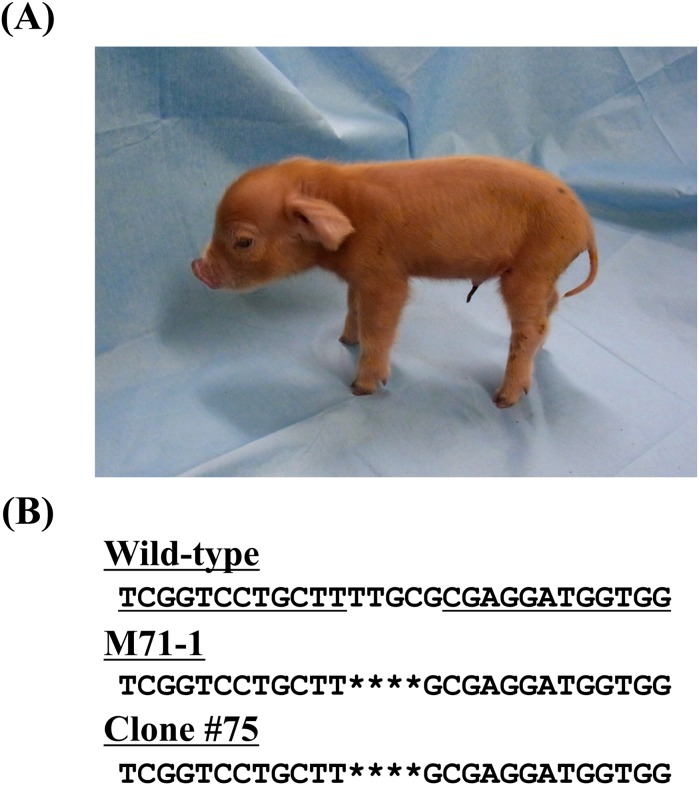
The developmental competence of the SCNT embryos reconstructed with the heterozygous CMAH KO cells was first examined in vitro. The cleavage and blastocyst formation rates of reconstructed SCNT embryos were 81.4% (48/59) and 78.0% (46/59), respectively. This blastocyst formation rate was comparable to values found in our previous studies [32, 33]. Then, the 65 embryos obtained by SCNT were subjected to transfer to an estrus-synchronized recipient gilt. On day 116 of gestation, 4 cloned pigs with heterozygous CMAH KO were produced (Fig. 2A).
Fig. 2.
Generation of heterozygous CMAH KO cloned pigs. (A) A ZFN-induced heterozygous CMAH KO cloned piglet (M71-1) at 2 days after birth. (B) DNA sequence analysis of CMAH in the cloned pig (M71-1) showing precisely the same mutation as that of the nuclear donor cell (clone #75). The deletion mutation is indicated by an asterisk.
Generation of homozygous CMAH KO cells
Skin fibroblast cells were established from one of the heterozygous CMAH KO cloned piglets (female, M71-1). Homozygous CMAH KO cells were then generated in precisely the same manner as before using the same ZFN-encoding mRNAs (Fig. 2B). Out of the 177 cell clones obtained after limiting dilution, 9 lines (5.1%) of homozygous CMAH KO cells were established (Table 2).
Table 2. Targeting efficiency of ZFNs and TALENs.
| Target cells | Sex | Genome editing tool | Cell cloned with mutations (%) | Heterozygous KO (%) | Homozygous KO (%) |
| DK3-9 | F | ZFN* | 6.3 (6/96) | 6.3 (6/96) | 0 (0/96) |
| M71-1 (heterozygous KO) | F | ZFN* | 5.1 (9/177) | 5.1 (9/177) | – |
| #158-5 (DK3-1 line) | M | TALEN | 41.0 (68/166) | 24.1 (40/166) | 3.0 (5/166) |
* Same ZFNs used.
To generate male lines of CMAH KO cells, TALEN-encoding mRNAs targeting exon 7 of the pig CMAH gene were introduced into male fetal fibroblast cells (#158-5; DK3-1 line). This resulted in 40 cell clones (24.1%) with heterozygous CMAH KO and 5 homozygous CMAH KO clones (3.0%) among 166 cell colonies (Table 2).
Two lines of female GalT/CMAH double homozygous KO cells (Table 1, #5 and #85) and two male GalT/CMAH double homozygous KO cells (Table 1, #115 and #151) were used as nuclear donors for SCNT. Sufficient numbers of KO cells for SCNT were obtained after culture for 3 weeks from both the female and male cell lines. DNA sequence analysis showed that these clones carried loss-of-function deletions ranging from 1 to 13 bps.
Production and analysis of GalT/CMAH double homozygous KO cloned pigs
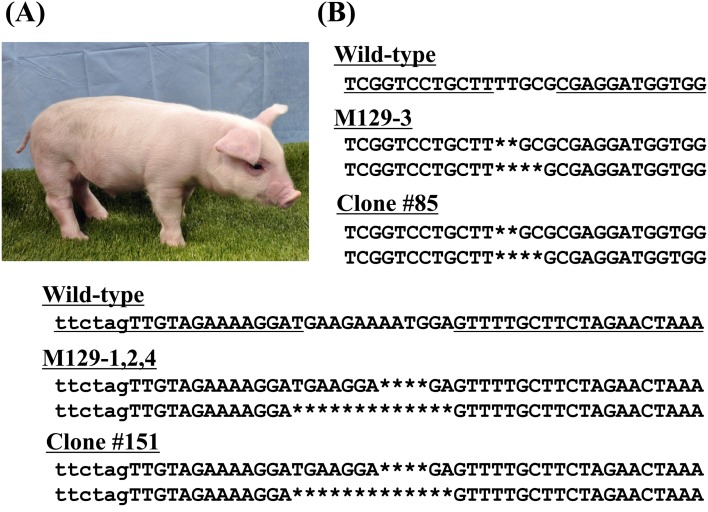
In total, 137 embryos obtained by SCNT of the above-mentioned GalT/CMAH double homozygous KO cells (#5, #85, #115, #151) were transferred into a recipient gilt (Table 3). On day 116 of gestation, one female and three male cloned piglets were farrowed (Fig. 3A). Analysis of genomic DNA isolated form tail tissues revealed that the female piglet (M129-3) was derived from cell clone #85 and that the males (M129-1, 2 and 4) were derived from cell clone #151 (Fig. 3B).
Table 3. Production of homozygous GalT/CMAH double homozygous KO cloned pigs.
| Genome editing tool | ZFNs | TALENs | ||
| Donor cell clone No. | #5 | #85 | #115 | #151 |
| SCNT embryos cultured | 50 | 49 | 47 | 47 |
| No. of normal embryos transferred (%)* | 32 (64.0) | 32 (65.3) | 37 (78.7) | 36 (76.6) |
| Recipient | M129 | |||
| Pregnancy | + | |||
| Total piglets obtained (%) | 4 (2.9, 4/137) | |||
| Genotype of piglets | 0 | 1 | 0 | 3 |
* On day 1–2.
Fig. 3.
Generation of homozygous CMAH KO cloned pigs. (A) A TALEN-induced homozygous CMAH KO male piglet, M129-1, at 20 days of age. (B) DNA sequence analysis of CMAH in the cloned pigs, M129-1, 2, 3 and 4. The sequences of M129-3 (female) and M129-1, 2 and 4 (male) showed the same mutation as the sequences of the nuclear donor cells, #85 and #151, respectively. The deletion mutation is indicated by an asterisk.
Phenotypic characterization of homozygous GalT/CMAH double KO pigs
Histological analysis of organs collected from a male piglet (M129-2) showed that the H-D antigen (Neu5Gc) and α-Gal antigen were absent in the homozygous GalT/CMAH double KO pig (Fig. 4A and 4B). In contrast, expression of the H-D antigen was clearly detected in the GalT-KO pig.
Fig. 4.
Immunohistochemical and flow cytometry analysis of the homozygous GalT/CMAH double KO cloned pigs. Tissue sections of lung, heart, pancreas and kidney from the homozygous GalT KO (M71-5), homozygous GalT/CMAH double KO (M129-2) and WT pigs were stained with anti-H-D antibody (A) and GS-IB4 lectin (B). Absence of H-D antigen was clearly shown in the tissue sections of the homozygous GalT/CMAH double KO pig. Scale bars in (A) and (B) = 100 μm. Flow cytometry analysis of the granulocytes also indicated the absence of the H-D and α-Gal antigens in the homozygous GalT/CMAH double KO pig (M129-1). The granulocytes were stained by anti-H-D antibody (C) and GS-IB4 lectin (D).
A blood sample was collected from one male cloned piglet (M129-1). Flow cytometry analysis of the peripheral blood showed that the granulocytes of the homozygous GalT/CMAH double KO pig (M129-1) also did not express Neu5Gc (Fig. 4C). The lack of GalT expression was confirmed in both of the GalT/CMAH double KO pigs (Fig. 4D).
Discussion
Recent development of genome editing technologies has opened a new avenue for generation of genetically modified (GM) pigs [34, 35]. Major applications of GM pigs involve research and development of disease models [36, 37], as well as regenerative medicine [32] and organ/tissue transplantation [38]. Use of pig-to-human xenograft technology is considered one of the most practical strategies for overcoming the worldwide shortage of organs. Based on current evidence, pig islet xenografts appear to have the greatest potential in clinical transplantation and have already been used in various human clinical trials [39]. Pigs represent an ideal animal source for xenografts for a variety of reasons, including those relating to anatomical, physiological and ethical considerations [40, 41]. However, severe immune rejection of a discordant xenograft is one of the major concerns in clinical application of pig-to-human tissue/organ transplantation. Such rejections are strongly related to the presence of species-specific glycoantigens. The major xenoantigen is the α-Gal epitope [27, 42], a carbohydrate expressed in most mammalian cells, including pigs, but absent in birds, monkeys, apes and humans. A number of attempts have been made to produce GalT KO pigs using HR [4, 5, 8, 9] and genome editing technologies [14, 17, 43].
Hauschild et al. first applied the ZFN system to knockout the GalT gene in pigs [14]. The ZFN system showed superior efficiency in producing GalT KO cells compared with the HR system, indicating the emergence of a new stage for the production of gene-targeted animals. Luts et al. applied the ZFN system to the production of GalT/CMAH double KO pigs [44]. On the other hand, Li et al. focused on the CRISPR/Cas9 system and succeeded in producing the pigs with a GalT/CMAH/iGb3 triple KO [20].
In the GalT KO pigs with the eliminated α-Gal epitope, however, many non-Gal antigens remain, including the H-D antigen [45, 46]. The H-D antigen, N-glycolylneuraminic acid (Neu5Gc), is widely distributed in mammalian species, including in pigs, monkeys and apes, but not in humans and birds [47]. The expression of Neu5Gc is controlled by cytidine monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) hydroxylase [28, 29], an enzyme that catalyzes the conversion of N-acetylneuraminic acid (Neu5Ac) to Neu5Gc. After extensive research [26, 48, 49], it is now recognized that the majority of human anti-non-Gal antibodies are specific for the H-D antigen [48] and that the H-D antigen may elicit a significant humoral response and play an indispensable role in a delayed form of xenograft rejection [50]. In addition, the silencing of pig CMAH by siRNA confirmed the importance of the H-D antigen as a major non-Gal antigen [51]. Therefore, the CMAH gene became the immediate target to KO in xenotransplantation research. In this study, the ZFN and TALEN systems together were shown to be effective in the production of CMAH KO pigs using fibroblasts from GalT KO pigs.
Direct microinjection of ZFN- and TALEN-encoding mRNA into fertilized eggs has been used for the creation of gene KO animals, mainly because of the simplicity of this process [12, 15]. However, drawbacks of the microinjection method include inefficiency and occurrence of mutation mosaicism [52] in the resultant embryos or fetuses. Therefore, the SCNT system was used for the generation of CMAH KO pigs in this study.
In some reports, SCNT was performed without the selection of targeted cells to avoid a lengthy in vitro period of culture for the transfected cells [20]. However, this procedure could result in various types of offspring, including homozygous and heterozygous KO animals, as well as WTs. In this study, in contrast, nuclear donor cells were examined for the presence of induced mutations by a DNA sequence analysis before production of the cloned animals by SCNT. This prescreening is useful to avoid the wasteful production of undesired animals. In addition, the high efficiency of the ZFN- and TALEN-system allowed for generation of CMAH KO cells without antibiotic selection, which is similar to our previous work on production of interleukin-2 receptor gamma gene KO pigs [24]. In fact, sufficient numbers of nuclear donor cells for SCNT could be obtained in a short time frame (approximately 3 weeks) in this study. Furthermore, the TALAN system yielded an apparent higher incidence of loss-of-function mutations compared with the ZFN system, although the difference in the cells used needs to be considered.
In this study, the homozygous CMAH-KO cells generated by the TALEN-encoding mRNA were used in the direct production of a live cloned pig without rejuvenation of the nuclear donor cells and subsequent re-cloning [53]. An additional advantage of TALEN-encoding mRNA is the transient TALEN expression, which may reduce the incidence of off-target mutations, a potential limitation of gene KO by genome editing [16, 54].
Since no off-target sites were analyzed in the present study, we cannot completely eliminate the possibility of off-target mutations affecting the development of the KO pig. However, given the growth of the KO cloned pig was similar to that of the wild type (non-knockout) clones, we believe that the CMAH knockout pig produced in the present study was not adversely affected by any off-target mutations.
Recently, remarkable targeting efficiency has been reported for the CRISPR/Cas9 system. This system may be useful for simultaneous KO of multiple genes. However, the possibility of a high incidence of off-target events in the CRISPR/Cas9 system needs to be clarified. In the meantime, the TALEN system is considered a feasible option in generating gene KO pigs without genomic integration.
In conclusion, this study demonstrated a strategy for generating KO pigs using ZFN- and TALEN-encoding mRNA. The combination of TALEN-encoding mRNA, in vitro selection of the nuclear donor cells and SCNT provides a robust method for generating KO pigs in a short time frame. To our knowledge, this is the first study to demonstrate the generation of CMAH KO pigs from gene KO cells prepared in vitro using TALEN-encoding mRNAs. Although further studies are required, these findings represent a very useful technique that has a great potential with respect to development of a pig xenograft model in the future.
Acknowledgments
We thank Dr Milton S Feather for his editing of the manuscript and H Kadoi for his help with maintenance of the pigs. This work was supported by Health and Labor Sciences Research Grants, Japan, the Meiji University International Institute for Bio-Resource Research (MUIIBR), JSPS Grant-in-Aid for Young Scientists (B) Grant Number 26870630 and the JST/ERATO Nakauchi Stem Cell and Organ Regeneration Project.
References
- 1.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 2005; 6: 507–512. [DOI] [PubMed] [Google Scholar]
- 2.Guan C, Ye C, Yang X, Gao J. A review of current large-scale mouse knockout efforts. Genesis 2010; 48: 73–85. [DOI] [PubMed] [Google Scholar]
- 3.Misra RP, Duncan SA. Gene targeting in the mouse: advances in introduction of transgenes into the genome by homologous recombination. Endocrine 2002; 19: 229–238. [DOI] [PubMed] [Google Scholar]
- 4.Dai Y, Vaught TD, Boone J, Chen SH, Phelps CJ, Ball S, Monahan JA, Jobst PM, McCreath KJ, Lamborn AE, Cowell-Lucero JL, Wells KD, Colman A, Polejaeva IA, Ayares DL. Targeted disruption of the alpha1,3-galactosyltransferase gene in cloned pigs. Nat Biotechnol 2002; 20: 251–255. [DOI] [PubMed] [Google Scholar]
- 5.Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, Murphy CN, Carter DB, Hawley RJ, Prather RS. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 2002; 295: 1089–1092. [DOI] [PubMed] [Google Scholar]
- 6.Koller BH, Hagemann LJ, Doetschman T, Hagaman JR, Huang S, Williams PJ, First NL, Maeda N, Smithies O. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci USA 1989; 86: 8927–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature 1997; 385: 810–813. [DOI] [PubMed] [Google Scholar]
- 8.Nottle MB, Beebe LF, Harrison SJ, McIlfatrick SM, Ashman RJ, O’Connell PJ, Salvaris EJ, Fisicaro N, Pommey S, Cowan PJ, d’Apice AJ. Production of homozygous alpha-1,3-galactosyltransferase knockout pigs by breeding and somatic cell nuclear transfer. Xenotransplantation 2007; 14: 339–344. [DOI] [PubMed] [Google Scholar]
- 9.Fujimura T, Takahagi Y, Shigehisa T, Nagashima H, Miyagawa S, Shirakura R, Murakami H. Production of alpha 1,3-galactosyltransferase gene-deficient pigs by somatic cell nuclear transfer: a novel selection method for gal alpha 1,3-Gal antigen-deficient cells. Mol Reprod Dev 2008; 75: 1372–1378. [DOI] [PubMed] [Google Scholar]
- 10.Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, Rogan MP, Pezzulo AA, Karp PH, Itani OA, Kabel AC, Wohlford-Lenane CL, Davis GJ, Hanfland RA, Smith TL, Samuel M, Wax D, Murphy CN, Rieke A, Whitworth K, Uc A, Starner TD, Brogden KA, Shilyansky J, McCray PB, Jr, Zabner J, Prather RS, Welsh MJ. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 2008; 321: 1837–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klymiuk N, Blutke A, Graf A, Krause S, Burkhardt K, Wuensch A, Krebs S, Kessler B, Zakhartchenko V, Kurome M, Kemter E, Nagashima H, Schoser B, Herbach N, Blum H, Wanke R, Aartsma-Rus A, Thirion C, Lochmüller H, Walter MC, Wolf E. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum Mol Genet 2013; 22: 4368–4382. [DOI] [PubMed] [Google Scholar]
- 12.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 2009; 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe M, Umeyama K, Matsunari H, Takayanagi S, Haruyama E, Nakano K, Fujiwara T, Ikezawa Y, Nakauchi H, Nagashima H. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun 2010; 402: 14–18. [DOI] [PubMed] [Google Scholar]
- 14.Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA 2011; 108: 12013–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 2011; 29: 695–696. [DOI] [PubMed] [Google Scholar]
- 16.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 2012; 109: 17382–17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y, Zou Q, Yan Q, Zeng Y, Lai L. Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS ONE 2013; 8: e84250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Estrada JL, Burlak C, Montgomery J, Butler JR, Santos RM, Wang ZY, Paris LL, Blankenship RL, Downey SM, Tector M, Tector AJ. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation 2015; 22: 20–31. [DOI] [PubMed] [Google Scholar]
- 21.Kurome M, Hisatomi H, Matsumoto S, Tomii R, Ueno S, Hiruma K, Saito H, Nakamura K, Okumura K, Matsumoto M, Kaji Y, Endo F, Nagashima H. Production efficiency and telomere length of the cloned pigs following serial somatic cell nuclear transfer. J Reprod Dev 2008; 54: 254–258. [DOI] [PubMed] [Google Scholar]
- 22.Kurome M, Geistlinger L, Kessler B, Zakhartchenko V, Klymiuk N, Wuensch A, Richter A, Baehr A, Kraehe K, Burkhardt K, Flisikowski K, Flisikowska T, Merkl C, Landmann M, Durkovic M, Tschukes A, Kraner S, Schindelhauer D, Petri T, Kind A, Nagashima H, Schnieke A, Zimmer R, Wolf E. Factors influencing the efficiency of generating genetically engineered pigs by nuclear transfer: multi-factorial analysis of a large data set. BMC Biotechnol 2013; 13: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurome M, Kessler B, Wuensch A, Nagashima H, Wolf E. Nuclear transfer and transgenesis in the pig. Methods Mol Biol 2015; 1222: 37–59. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe M, Nakano K, Matsunari H, Matsuda T, Maehara M, Kanai T, Kobayashi M, Matsumura Y, Sakai R, Kuramoto M, Hayashida G, Asano Y, Takayanagi S, Arai Y, Umeyama K, Nagaya M, Hanazono Y, Nagashima H. Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS ONE 2013; 8: e76478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsunari H, Watanabe M, Umeyama K, Nakano K, Ikezawa Y, Kurome M, Kessler B, Wolf E, Miyagawa S, Nakauchi H, Nagashima H. Cloning of homozygous α1,3-galactosyltransferase gene knock-out pigs by somatic cell nuclear transfer. In: Miyagawa S (ed.), Xenotransplantation. Rijeka, Croatia: InTech; 2012: 37–54.
- 26.Miwa Y, Kobayashi T, Nagasaka T, Liu D, Yu M, Yokoyama I, Suzuki A, Nakao A. Are N-glycolylneuraminic acid (Hanganutziu-Deicher) antigens important in pig-to-human xenotransplantation? Xenotransplantation 2004; 11: 247–253. [DOI] [PubMed] [Google Scholar]
- 27.Miyagawa S, Ueno T, Nagashima H, Takama Y, Fukuzawa M. Carbohydrate antigens. Curr Opin Organ Transplant 2012; 17: 174–179. [DOI] [PubMed] [Google Scholar]
- 28.Song KH, Kang YJ, Jin UH, Park YI, Kim SM, Seong HH, Hwang S, Yang BS, Im GS, Min KS, Kim JH, Chang YC, Kim NH, Lee YC, Kim CH. Cloning and functional characterization of pig CMP-N-acetylneuraminic acid hydroxylase for the synthesis of N-glycolylneuraminic acid as the xenoantigenic determinant in pig-human xenotransplantation. Biochem J 2010; 427: 179–188. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda K, Yamamoto A, Nanjo A, Inuinaka C, Takama Y, Ueno T, Fukuzawa M, Nakano K, Matsunari H, Nagashima H, Miyagawa S. A cloning of cytidine monophospho-N-acetylneuraminic acid hydroxylase from porcine endothelial cells. Transplant Proc 2012; 44: 1136–1138. [DOI] [PubMed] [Google Scholar]
- 30.Nakano K, Matsunari H, Matsuda T, Kanai T, Hayashida G, Matsumura Y, Kobayashi M, Kuramoto M, Asano Y, Sakai R, Uchikura A, Arai Y, Watanabe M, Umeyama K, Nagaya M, Nagashima H. Application of blastocyst complementation to development of genetically modified pig for xenotransplantation. In: Program of the 12th Congress of International Xenotransplantation Association; 2013, Osaka, Japan. Abstract 459.
- 31.Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 2010; 7: 459–460. [DOI] [PubMed] [Google Scholar]
- 32.Matsunari H, Nagashima H, Watanabe M, Umeyama K, Nakano K, Nagaya M, Kobayashi T, Yamaguchi T, Sumazaki R, Herzenberg LA, Nakauchi H. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc Natl Acad Sci USA 2013; 110: 4557–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe M, Kobayashi M, Nagaya M, Matsunari H, Nakano K, Maehara M, Hayashida G, Takayanagi S, Sakai R, Umeyama K, Watanabe N, Onodera M, Nagashima H. Production of transgenic cloned pigs expressing the far-red fluorescent protein monomeric Plum. J Reprod Dev 2015; 61: 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Lin L, Bolund L, Jensen TG, Sørensen CB. Genetically modified pigs for biomedical research. J Inherit Metab Dis 2012; 35: 695–713. [DOI] [PubMed] [Google Scholar]
- 35.Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E. Genetically engineered pig models for human diseases. Annu Rev Anim Biosci2013; 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umeyama K, Watanabe M, Saito H, Kurome M, Tohi S, Matsunari H, Miki K, Nagashima H. Dominant-negative mutant hepatocyte nuclear factor 1alpha induces diabetes in transgenic-cloned pigs. Transgenic Res 2009; 18: 697–706. [DOI] [PubMed] [Google Scholar]
- 37.Aigner B, Renner S, Kessler B, Klymiuk N, Kurome M, Wünsch A, Wolf E. Transgenic pigs as models for translational biomedical research. J Mol Med (Berl) 2010; 88: 653–664. [DOI] [PubMed] [Google Scholar]
- 38.Cooper DKC, Satyananda V, Ekser B, van der Windt DJ, Hara H, Ezzelarab MB, Schuurman H-J. Progress in pig-to-non-human primate transplantation models (1998–2013): a comprehensive review of the literature. Xenotransplantation 2014; 21: 397–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elliott RB, Escobar L, Tan PL, Muzina M, Zwain S, Buchanan C. Live encapsulated porcine islets from a type 1 diabetic patient 9.5 yr after xenotransplantation. Xenotransplantation 2007; 14: 157–161. [DOI] [PubMed] [Google Scholar]
- 40.Lunney JK. Advances in swine biomedical model genomics. Int J Biol Sci 2007; 3: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsunari H, Nagashima H. Application of genetically modified and cloned pigs in translational research. J Reprod Dev 2009; 55: 225–230. [DOI] [PubMed] [Google Scholar]
- 42.Galili U. α1,3Galactosyltransferase knockout pigs produce the natural anti-Gal antibody and simulate the evolutionary appearance of this antibody in primates. Xenotransplantation 2013; 20: 267–276. [DOI] [PubMed] [Google Scholar]
- 43.Li P, Estrada JL, Burlak C, Tector AJ. Biallelic knockout of the α-1,3 galactosyltransferase gene in porcine liver-derived cells using zinc finger nucleases. J Surg Res 2013; 181: e39–e45. [DOI] [PubMed] [Google Scholar]
- 44.Lutz AJ, Li P, Estrada JL, Sidner RA, Chihara RK, Downey SM, Burlak C, Wang ZY, Reyes LM, Ivary B, Yin F, Blankenship RL, Paris LL, Tector AJ. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose α-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation 2013; 20: 27–35. [DOI] [PubMed] [Google Scholar]
- 45.Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem 1998; 273: 15866–15871. [DOI] [PubMed] [Google Scholar]
- 46.Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci USA 1998; 95: 11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varki A. Multiple changes in sialic acid biology during human evolution. Glycoconj J 2009; 26: 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu A, Hurst R. Anti-N-glycolylneuraminic acid antibodies identified in healthy human serum. Xenotransplantation 2002; 9: 376–381. [DOI] [PubMed] [Google Scholar]
- 49.Magnusson S, Månsson JE, Strokan V, Jussila R, Kobayashi T, Rydberg L, Romano E, Breimer ME. Release of pig leukocytes during pig kidney perfusion and characterization of pig lymphocyte carbohydrate xenoantigens. Xenotransplantation 2003; 10: 432–445. [DOI] [PubMed] [Google Scholar]
- 50.Basnet NB, Ide K, Tahara H, Tanaka Y, Ohdan H. Deficiency of N-glycolylneuraminic acid and Galα1-3Galβ1-4GlcNAc epitopes in xenogeneic cells attenuates cytotoxicity of human natural antibodies. Xenotransplantation 2010; 17: 440–448. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto A, Ikeda K, Wang D, Nakatsu S, Takama Y, Ueno T, Nagashima H, Kondo A, Fukuzawa M, Miyagawa S. Trial using pig cells with the H-D antigen knocked down. Surg Today 2013; 43: 782–786. [DOI] [PubMed] [Google Scholar]
- 52.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE 2010; 5: e8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujimura T, Murakami H, Kurome M, Takahagi Y, Shigehisa T, Nagashima H. Effects of recloning on the efficiency of production of alpha 1,3-galactosyltransferase knockout pigs. J Reprod Dev 2008; 54: 58–62. [DOI] [PubMed] [Google Scholar]
- 54.Whyte JJ, Prather RS. Cell Biology Symposium: Zinc finger nucleases to create custom-designed modifications in the swine (Sus scrofa) genome. J Anim Sci 2012; 90: 1111–1117. [DOI] [PubMed] [Google Scholar]