Abstract
Diploid germ cells are thought to have pluripotency potential. We recently described a method to derive pluripotent stem cells (PSCs) from cultured spermatogonial stem cells (SSCs) by depleting Trp53 and Dmrt1, both of which are known suppressors of teratomas. In this study, we used this technique to analyze the effect of this protocol in deriving PSCs from the male germline at different developmental stages. We collected primordial germ cells (PGCs), gonocytes and spermatogonia, and the cells were transduced with lentiviruses expressing short hairpin RNA against Dmrt1 and/or Trp53. We found that PGCs are highly susceptible to reprogramming induction and that only Trp53 depletion was sufficient to induce pluripotency. In contrast, gonocytes and spermatogonia were resistant to reprogramming by double knockdown of Dmrt1 and Trp53. PSCs derived from PGCs contributed to chimeras produced by blastocyst injection, but some of the embryos showed placenta-only phenotypes suggestive of epigenetic abnormalities of PGC-derived PSCs. These results show that PGCs and gonocytes/spermatogonia have distinct reprogramming potential and also suggest that fresh and cultured SSCs do not necessarily have the same properties.
Keywords: Dmrt1, Pluripotency, Primordial germ cells, Spermatogonia, Trp53
Diploid germ cells have a unique ability to convert into pluripotent stem cells (PSCs) [1]. The ability to experimentally induce pluripotency in germ cells was demonstrated originally by Leroy Stevens, who showed that genital ridges of specific strains of mice (129 or A) between 8.5 and 12.5 days post coitum (dpc) can form teratomas upon transplantation into testes of different animals [2]. Since embryos with a reduced number of primordial germ cells (PGCs) do not produce teratomas, PGCs in embryos may have unique potential to develop into PSCs. Although the reprogramming mechanisms remained unclear, it was subsequently found in 1992 that PGCs could transform into pluripotent embryonic germ cells (EGCs) when cultured with fibroblast growth factor 2 (FGF2), stem cell factor (SCF) and leukemia inhibitory factor (LIF) [3, 4]. EGCs are not only similar to embryonic stem cells (ESCs) in their morphology and phenotype but also contribute to germline chimeras when microinjected into blastocysts [5, 6]. Interestingly, the ability to develop into EGCs was limited to PGCs from 8.5 to 12.5 dpc embryos. Postnatal spermatogonia did not give rise to EGCs even when cultured under the same conditions [5]. Moreover, the frequency of conversion into EGCs decreases with age [5]. This suggests that germ cells lose pluripotency as they mature into gonocytes.
In 2003, a long-term culture system was described for spermatogonial stem cells (SSCs) [7], which are the only stem cells in the male germline that produce sperm throughout their lifetime [8, 9]. SSCs undergo self-renewal divisions in vitro in the presence of cytokines, which include glial cell line-derived neurotrophic factor (GDNF) [10]. Cultured cells, designated as germline stem cells (GSCs), proliferate as grape-like clusters of spermatogonia but can reinitiate spermatogenesis upon introduction into seminiferous tubules of infertile mice [7]. In the course of establishing GSCs, we noticed that ESC-like cells appear in culture. These cells, designated as multipotent GSCs (mGSCs), proliferate without GNDF and can differentiate in vitro into various types of somatic cells and contribute to the male germline by microinjection into blastocysts [11].
Although these results showed that the ability to become PSCs persists in postnatal male germline cells, studies on mGSCs are difficult because of the relatively low frequency of derivation of these cells. They appeared in 1 out of 30 testes used to establish GSCs [11]. Although we noted that Trp53 deficiency could increase the frequency, it was still difficult to derive mGSCs in an efficient manner from wild-type mice. To overcome this problem, we screened germ cell tumor candidate genes for their activity to induce pluripotency in GSCs and found that double depletion of Dmrt1 and Trp53 induces mGSCs [12]. Both genes are implicated in germ cell tumor development [13]. However, Dmrt1 is a critical gene involved in sexual differentiation, and Dmrt1 overexpression is thought to cause spermatocytic seminomas [14, 15]. In GSCs, Dmrt1 depletion causes massive apoptosis of GSCs, but co-depletion with Trp53 could rescue apoptosis and induce pluripotency [12]. Further analysis revealed that downregulation of Dmrt1 and Trp53 increased the expression of Sox2, which probably collaborated with Pou5f1 to activate the pluripotency program [12]. Thus, development of this new protocol allows us to study the regulation of pluripotency in GSCs.
In the present study, we used this technique and examined the impact of Dmrt1 and Trp53 genes in reprogramming male germline cells at different stages. Although both Dmrt1 and Trp53 are expressed from the early stages of the male germline cells, the difference between embryonic and postnatal germline cells in the reprogramming protocol suggested that distinct machineries operate to regulate pluripotency. To address this, gonads of different stages were collected, and their abilities to reprogram into PSCs were examined by transfecting knockdown (KD) vectors for Dmrt1 and Trp53.
Materials and Methods
Animal and cell collection
PGCs and gonocytes were collected from pregnant ICR mice (Japan SLC, Shizuoka, Japan) that were maintained in a controlled environment with 12:12-h light:dark cycles (lights on from 0800 to 2000 h). The day on which a copulation plug was found was designated as 0.5 dpc. The sex of the embryos was determined using a polymerase chain reaction (PCR)-based genotyping method and the Ube1 gene [16], and only male embryos were used in the current study. Cells were collected from germ cell-containing regions by dissection of the posterior thirds of 8.5 dpc embryos, the mesentery and urogenital sinuses of 10.5 dpc embryos and the genital ridges of 12.5, 15.5 and 18.5 dpc embryos. Tissues were digested in 0.25% trypsin/1 mM EDTA for 10 min. For collection of pup testis cells, we used 8- to 10-day-old ICR mice (Japan SLC). Testis cells were dissociated using a two-step enzymatic digestion protocol with collagenase type IV and trypsin, as described previously [17]. Dissociated testis cells were then incubated with biotin-conjugated anti-CD9 antibody (MZ3; BioLegend, San Diego, CA, USA) for 10 min at 4 C. Magnetic cell sorting (MACS) was then performed using streptavidin-conjugated Dynabeads according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA).
Cell culture
GSCs with an ICR background were cultured in StemPro-34 medium (Invitrogen), as described previously [7]. FGF2 and GDNF for GSC culture were purchased from PeproTech (London, UK). All PSCs were cultured in DMEM/15% fetal bovine serum (FBS) on mouse embryonic fibroblasts (MEFs). ESGRO was routinely added to PSC cultures (1,000 U/ml; Millipore, Billerica, MA, USA). We also used PD0325901 (2 μM; Selleck Chemicals, Houston, TX, USA) and CHIR99021 (3 μM; BioVision, Inc., Milpitas, CA, USA) before blastocyst injection experiments. Embryoid body formation was performed as described previously [12].
Lentivirus transfection
For lentivirus-mediated gene KD, we used pSicoR Trp53 (Addgene, Cambridge, MA, USA) and pLKO.1 Dmrt1 (Open Biosystems, Huntsville, AL, USA; TRCN0000084388, TRCN0000084389, TRCN0000084390, TRCN0000084391, TRCN0000084392), as described previously [12]. We used pSicoR and pLKO-luciferase as controls. For immunostaining experiments, Gfp cDNA was excised from pSicoR plasmids and used as a control. We also produced a tetracycline-inducible short hairpin RNA (Tet-shRNA) against Dmrt1 and Trp53 by cloning the same shRNA sequence (Dmrt1, TRCN0000084388; Trp53, pSicoR Trp53) into CS-H1tetO-ETR, which contains red fluorescent protein (RFP) as a marker (gift from Dr H Miyoshi, RIKEN BRC, Tsukuba, Japan). For transfection of Yamanaka factors, Pou5f1, Sox2, Klf4 and Myc cDNAs were cloned into CSII-EF-IRES2-Puro vectors (gift from Dr H Miyoshi). Constitutively active Akt1 (gift from Dr T Nakano, Osaka University, Osaka, Japan) was also cloned into CSII-EF-IRES2-Puro vector. Lentivirus particles were produced by transfection into 293T cells with pCMV-VSV-G-RSV-Rev and pCAG-HIVgp. For titration of lentiviruses, a Lenti-X p24 Rapid Titer Kit was used according to the manufacturer’s instructions (Clontech, Mountain View, CA, USA). All transfection experiments were performed by plating cells on MEFs, and plates were centrifuged at 3,000 × g for 1 h at 32 C, as described previously [18]. The multiplicities of infection (MOIs) were adjusted to 10 for Trp53 KD, 4 for Dmrt1 KD and 1 for overexpression of Yamanaka factors. For experiments using CS-H1tetO-ETR, MOIs were adjusted to 10 for both Trp53 and Dmrt1. Doxycycline (1 μg/ml; Sigma, St. Louis, MO, USA) was added on the day following transfection.
For small interfering RNA (siRNA) transfection, PGCs were seeded on MEFs at a density of 1.5 × 105 cells/3.8 cm2. The culture medium was changed the following day, and siRNA transfection was conducted according to the manufacturer’s instructions. Stealth siRNAs for Dmrt1 and Trp53 were purchased from Invitrogen (Dmrt1, Dmrt1-MSS284782; Trp53, Trp53-MSS212108) and were used at 25 nM.
Immunostaining
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde (PFA) for 15 min at 4 C. After washing three times with phosphate-buffered saline (PBS), cells were permeabilized with 0.1% Triton X-100 for 10 min and incubated in blocking solution (10% goat serum or 10% donkey serum, 0.1% Tween 20 and 0.1% bovine serum albumin in PBS) for 1 h at room temperature. The cells were then stained overnight at 4 C with the indicated primary antibodies. After washing in PBS/0.1% Tween 20 three times, samples were incubated at room temperature for 1 h before staining with the secondary antibody. The nuclei were counterstained with Hoechst 33342 (2 μg/ml; Sigma). Images were acquired on a confocal laser scanning microscope (FV1000-D; Olympus, Tokyo, Japan) or a fluorescence microscope equipped with a CCD camera (DP-70; Olympus). The antibodies used are listed in Supplementary Table 1 (online only).
Flow cytometry
Cells were suspended in PBS/1% FBS. All antibodies used are listed in Supplementary Table 1. For ANXA5 staining, Annexin V Apoptosis Detection Kit FITC (eBioscience, San Diego, CA, USA) was used. Cells were analyzed on a FACSCalibur (BD Biosciences, San Jose, CA, USA).
Reverse transcription (RT)-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen). First-strand cDNA was produced using a Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) for RT-PCR. For real-time PCR, a StepOnePlusTM Real-Time PCR System (Applied Biosystems, Warrington, UK) and FastStart Universal SYBR Green Master Mix (Roche Applied Science) were used according to the manufacturers’ protocols. Transcript levels were normalized relative to those of Hprt. The real-time PCR conditions were 95 C for 10 min, 40 cycles at 95 C for 15 sec, and then 60 C for 1 min. Each PCR was performed at least in triplicate, and gene expression levels were determined with at least three biological repeats. For RT-PCR, the PCR conditions were 95 C for 5 min followed by 30 cycles at 94 C for 30 sec, 60 C for 30 sec and 72 C for 30 sec. The primers used for PCR are listed in Supplementary Table 2 (online only).
Western blotting
Samples were separated by SDS-PAGE and transferred onto Hybond-P membranes (GE Healthcare, Piscataway, NJ, USA). The membrane was incubated with primary antibodies. Antibodies used in the experiments are listed in Supplementary Table 1. Band intensity was quantified using the ImageJ 1.43r software (US National Institutes of Health, Bethesda, MD, USA).
Combined bisulfite restriction analysis (COBRA) and bisulfite sequencing
Genomic DNA was treated with sodium bisulfite, which deaminates unmethylated cytosines to uracils, and used as a template for amplification of differentially methylated regions (DMRs) of the indicated genes by PCR. The PCR products were subsequently digested with the indicated restriction enzymes, which had recognition sequences containing CpG in the original unconverted DNA. The intensity of the digested bands was assessed using the ImageJ 1.43r software (US National Institutes of Health). Bisulfite sequencing was performed as described previously [19]. The PCR primers used are listed in Supplementary Table 2.
Karyotype analysis
Cultured cells were incubated in colcemid solution (50 ng/ml; KaryoMAX, Invitrogen) for 1 h. After recovery by trypsin digestion, cells were treated with 75 mM KCl for 7 min. Metaphase spreads were prepared after fixing the cells with methanol/acetic acid (3:1). The slides were counterstained with Hoechst 33352 (2 μg/ml; Sigma). At least 20 cells were counted for each cell type.
Transplantation
For producing teratomas, 3 × 106 cells were injected subcutaneously into KSN nude mice (Japan SLC). Teratomas were recovered 1 month after transplantation for histological analysis by paraffin sectioning. The Institutional Animal Care and Use Committee of Kyoto University approved all animal experimentation protocols.
Chimera formation
For chimera production, PSCs were maintained in culture medium supplemented with LIF + CHIR99021 + PD0325901. These cells were injected into the blastocoels of 3.5 dpc blastocysts that resulted from in vitro fertilization using oocytes from C57BL/6 × DBA/2 (BDF1) females and spermatozoa from C57BL/6 males. A piezo-driven micromanipulator was used for microinjection [20]. The blastocysts were returned to the uteri of 2.5 dpc pseudopregnant ICR foster mothers on the day of microinjection. Pups were produced by natural parturition or caesarean section at 19.5 dpc.
Statistical analyses
All results are presented as the means ± SEM. Significant differences between means for single comparisons were identified using the Student’s t-test. Multiple comparison analyses were performed using ANOVA followed by Tukey’s HSD (honestly significant difference) test.
Results
Induction of PSCs from PGCs by Trp53 depletion
To examine the effect of Dmrt1 and Trp53 in different stages of germ cells, PGCs or gonocytes were collected from different stages of embryos (Supplementary Fig. 1A: online only). These tissues were dissociated by trypsin. For postnatal germ cells, we used testes from 8 to 10 days postpartum (dpp) pups, which are enriched for mitotically active SSCs. Unlike embryonic gonads, postnatal testes contain more differentiating germ cells. Therefore, SSCs were enriched by MACS using an antibody against CD9, which is expressed on SSCs [21] (Supplementary Fig. 1A and B: online only).
Recovered cells were then transduced with lentivirus vectors that express shRNA against Dmrt1 and/or Trp53. GSCs were infected with the same lentiviruses as the positive control. We also transduced CD9-selected SSCs with Yamanaka factors (Pou5f1, Sox2, Klf4 and Myc) because we could consistently obtain ESC-like colonies by transfection of Yamanaka factors in pup testis cell culture in our previous study [22]. To mimic protocol for derivation of PSCs from GSCs (GSC-PSC) [12], all transfected cells were cultured in medium containing FGF2 and GDNF, both of which are self-renewal factors for SSCs [10]. Unlike the typical EGC induction method, neither SCF nor LIF was added to the cultures. As expected from our previous studies, Dmrt1 KD induced extensive apoptosis of the germ cells of all stages within 4 days after lentivirus infection (Supplementary Fig. 1C: online only). However, we noted that depletion of Trp53 had different effects on PGCs and gonocytes/spermatogonia. While many PGCs underwent apoptosis after Trp53 depletion, the same treatment did not show any effect on gonocytes and spermatogonia. Apoptosis of PGCs was confirmed by flow cytometric analysis using an EPCAM antibody and ANXA5 staining (Supplementary Fig. 1C). However, apoptosis caused by Dmrt1 undergoes a switch in terms of Trp53 dependency; while Trp53 inhibition did not show any effect on Dmrt1-depleted PGC cultures, apoptosis caused by Dmrt1 depletion could be suppressed by simultaneous depletion of Trp53 in CD9-selected SSCs. Although real-time PCR analysis confirmed the reduction in target gene expression, no apparent correlative changes between Dmrt1 and Trp53 were observed in any cell types (Supplementary Fig. 1D: online only). These results suggested that Dmrt1 and Trp53 are regulated independently.
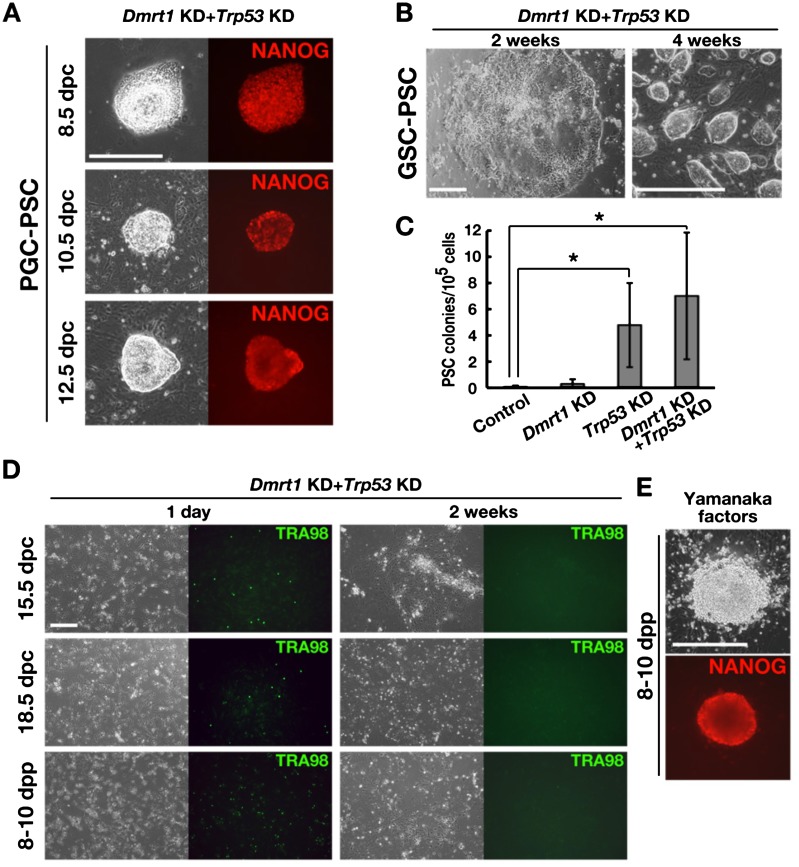
After the initial extensive apoptosis, cells were incubated for more than 2 weeks to monitor the development of PSC colonies. PSC colonies were derived efficiently from PGCs (PGC-PSCs) (Table 1). However, the pattern of PGC-PSC formation was distinct from dedifferentiation of GSCs into mGSCs. While PGC-PSC colonies were similar to ESCs from the beginning of colony formation, GSC-PSCs were typically found as epiblast-like cell sheets and developed an ESC-like morphology (Fig. 1A and B). Moreover, unlike EGCs that typically develop within 4–5 days after culture initiation [3], PGC-PSCs developed within as little as 2 weeks, suggesting that reprogramming occurs more slowly. In contrast to the EGC formation that occurs at higher frequency from younger embryos [5], we noted a significant increase in PGC-PSC formation from 12.5 dpc embryos compared with PGC-PSC formation from 8.5 or 10.5 dpc embryos (Table 1). This was not due to the higher frequency of total PGC numbers in cell suspensions prepared from 12.5 dpc embryos, because the same trend was still observed when the number of infected cells was normalized by POU5F1 immunostaining (Table 1).
Table 1. PSC induction efficiency by double KD of Dmrt1 and Trp53.
| Cell type | Number of experiments |
Number of infected cells (× 104) |
Number of PSC colonies /105 cells |
Number of PSC colonies /105 germ cellsa) |
| 8.5 dpc | 4 | 19.3 ± 0.8 | 0.3 ± 0.2 | 28.1 ± 16.3 |
| 10.5 dpc | 4 | 19.0 | 0.2 ± 0.2 | 24.9 ± 17.0 |
| 12.5 dpc | 6 | 32.1 ± 5.5 | 9.7 ± 4.7 | 123.9 ± 59.8 |
| 15.5 dpc | 5 | 50.0 | 0 | 0 |
| 18.5 dpc | 4 | 50.0 | 0 | 0 |
| 8–10 dpp | 10 | 50.0 | 0 | 0 |
| GSC | 10 | 50.0 | 0.4 ± 0.3 | 0.4 ± 0.3 |
Values are shown as the mean ± SEM. a) These values for fresh germ cells were normalized by the number of POU5F1-positive (8.5 to 12.5 dpc) or TRA98-positive (15.5 dpc to 8–10 dpp) cells, as determined by immunostaining (Supplementary Fig. 1B: online only).
Fig. 1.
Development of PSC colonies by double KD of Dmrt1 and Trp53. (A) NANOG expression in PSC colonies that developed from PGCs of different stages. (B) Development of PSC colonies from GSCs by double KD of Dmrt1 and Trp53. (C) Frequency of PSC colony development after KD of the indicated genes in PGCs from 12.5 dpc embryos (n = 10). (D) Lack of PSC colony development from 15.5 and 18.5 dpc embryos and CD9-selected SSCs from 8–10 dpp pups. TRA98 immunostaining was used to detect surviving germ cells. (E) NANOG expression in a PSC colony developed by transfections of Yamanaka factors into CD9-selected SSCs from 8–10 dpp pups. Bar = 200 μm (A, B, D, E).
Although apoptosis of PGCs was similarly induced by depletion of Dmrt1 or Trp53, PGC-PSC colonies were found when only Trp53 was depleted (Fig. 1C). On the other hand, Dmrt1 depletion produced few colonies. PSC development was not significantly increased by double depletion of Dmrt1 and Trp53. The average numbers of colonies generated were 0.1, 0.3, 4.8 and 7.0 for the control, Dmrt1 KD, Trp53 KD and Dmrt1 KD + Trp53 KD (n = 10), respectively. These results suggested that Dmrt1 is essential for PGC survival but is not involved in suppression of pluripotency in vitro.
For gonocytes and postnatal spermatogonia, double depletion of Trp53 and Dmrt1 caused extensive apoptosis, and few cells survived by 2 weeks after transfection (Fig. 1D). No PSC colonies were observed in these experiments. However, consistent with our previous study [12], double depletion of Dmrt1 and Trp53 in GSCs could successfully produce PSCs (GSC-PSCs), which formed epiblast-like colonies before turning into typical ESC-like PSC colonies (Fig. 1B). The frequency of GSC-PSC colony formation was significantly lower than that of PGC-PSC colonies (Table 1), and it occurred only when both Dmrt1 and Trp53 were depleted. Although double depletion of Dmrt1 and Trp53 could not induce reprogramming in CD9-selected SSCs, transfection of Yamanaka factors (Pou5f1, Sox2, Klf4 and Myc) directly induced ESC-like cells without forming epiblast-like colonies (Fig. 1E). These ESC-like colonies probably originated from germ cells, because significantly fewer colonies were generated in our previous study using germ cell-deficient WBB6F1-W/Wv mice [22]. However, Yamanaka factor transfection did not yield ESC-like colonies from GSCs.
Because AKT activation induces EGC formation in PGCs [23], we transfected pup testis cells and GSCs with constitutively activated AKT. However, neither of these cells produced PSC colonies. We also cultured pup testis cells and GSCs with LIF and two small molecule inhibitors of MAP2K1 signaling and GSK3 (CHIR99021 + PD0325901, 2i), which causes increased EGC formation [24, 25]. These factors also did not induce PSC formation. These results suggest that spermatogonia/GSCs are significantly more resistant than PGCs in reprogramming and that they do not necessarily share the same mechanism that suppresses potential pluripotency.
Sox2 is upregulated in PGCs after Trp53 depletion
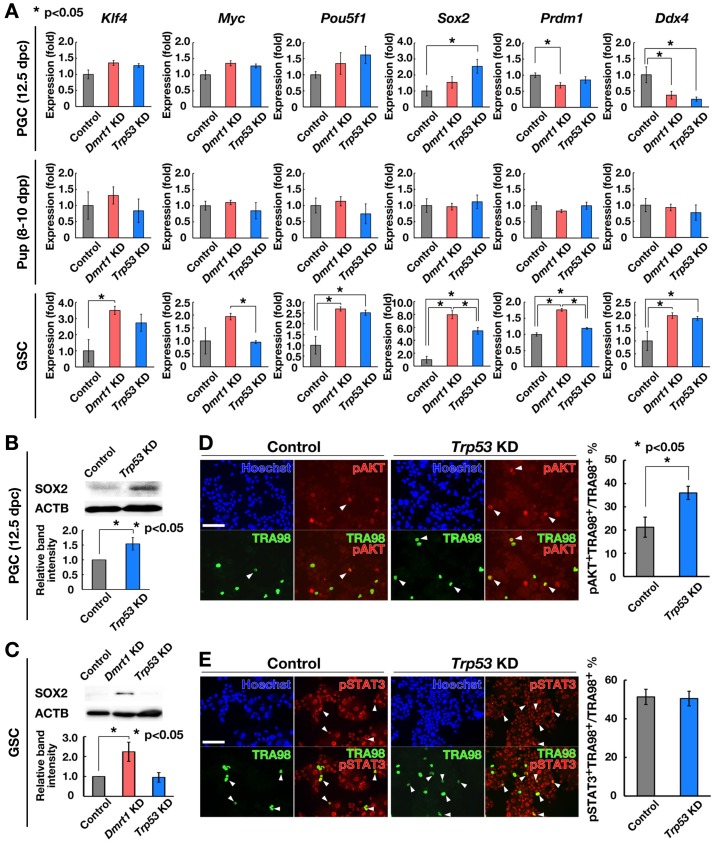
Enhanced PGC-PSC colony formation by Trp53 KD suggested that Trp53 is important for regulating pluripotency in PGCs. However, because PGC-PSC derivation occurred regardless of apoptosis, suppression of apoptosis did not appear to be required for pluripotency induction. To explore the mechanism of PGC-PSC formation caused by Trp53 depletion, we analyzed the expression of PSC and germ cell markers. We collected PGCs from 12.5 dpc embryos by digestion of male gonads in 0.25% trypsin/1 mM EDTA (Supplementary Fig. 1A). Immunocytochemistry showed that PGCs comprised ~7.8 ± 0.5% (n = 4) of the total cell number (Supplementary Fig. 1B). Using this cell population, Trp53 was depleted by shRNA, and the effect was compared with that of Dmrt1 depletion. Real-time PCR analysis of the cultured cells showed that Dmrt1 depletion decreased the expression of Prdm1 and Ddx4, which probably reflected apoptosis of PGCs or conversion of PGCs into PSCs (Fig. 2A). In addition, Sox2 was upregulated by Trp53 depletion in PGCs. Although Dmrt1 depletion slightly increased Sox2 expression, the difference was not statistically significant. In contrast, no significant changes were observed in pup testis cell cultures as a result of depletion of Dmrt1 or Trp53, which may explain the lack of PSC colonies after depletion of these molecules. However, when we analyzed GSCs, Dmrt1 depletion caused upregulation of many pluripotent and germ cell markers, including Sox2. Trp53 depletion also upregulated Pou5f1 and Sox2 expression as well as germ cell markers. These results suggested that PGCs and GSCs have different mechanisms for regulating Sox2 expression.
Fig. 2.
Phenotypic changes induced by Trp53 KD. (A) Real-time PCR analysis of PSC and germ cell markers after KD of the indicated genes. Cells were recovered 2 days after KD (n = 5). Results of five experiments. (B) Western blot analysis of SOX2 in PGCs from 12.5 dpc embryos after Trp53 KD (n = 4). Cells were collected 2 days after KD. (C) Western blot analysis of SOX2 in GSCs after Dmrt1 or Trp53 KD (n = 4). Cells were collected 2 days after KD. (D, E) Double immunostaining of PGCs from 12.5 dpc embryos with antibodies against pAKT (D; n = 4) or pSTAT3 (E; n = 4) 2 days after Trp53 KD. TRA98 was also used to detect PGCs. At least 100 cells were counted for each sample. Results of four experiments. Bar = 100 μm (D, E). Stain: Hoechst 33342 (D, E).
Because Sox2 is commonly upregulated in PGCs and GSCs at the mRNA level, Western blot analysis was performed to confirm the increase at the protein level. We found that SOX2 is expressed in gonadal cells from 12.5 dpc embryos, which could be increased by Trp53 KD within 2 days after transfection (Fig. 2B). On the other hand, GSCs did not show SOX2 before transfection, but they clearly expressed SOX2 upon Dmrt1 KD (Fig. 2C). A small increase in SOX2 was observed after Trp53 KD. Because Sox2 transfection can induce pluripotency in PGCs [26], these results suggested that upregulation of Sox2 expression induced pluripotency after Trp53 depletion in PGCs.
We then examined the relationship between TRP53 and AKT. AKT activation is critical for acquisition of pluripotency in PGCs [23, 27]. Moreover, AKT was shown to inactivate Trp53 during EGC formation [23]. Therefore, we examined the phosphorylation status of AKT after Trp53 KD. Dot-like signals of activated phosphorylated (Ser473-pAKT) near the cell membrane were counted as pAKT-positive (pAKT+) cells [27]. In contrast to EGC formation, AKT phosphorylation significantly increased after Trp53 depletion (Fig. 2D). We also analyzed the phosphorylation status of STAT3 (Tyr705-pSTAT3), another critical molecule for EGC derivation [25]. However, we did not observe a significant difference in phosphorylation levels of STAT3 (Fig. 2E). These results suggested that PGC-PSC formation is distinct from EGC formation and that TRP53 acts upstream of AKT to induce pluripotency.
Phenotypic analysis of PSCs
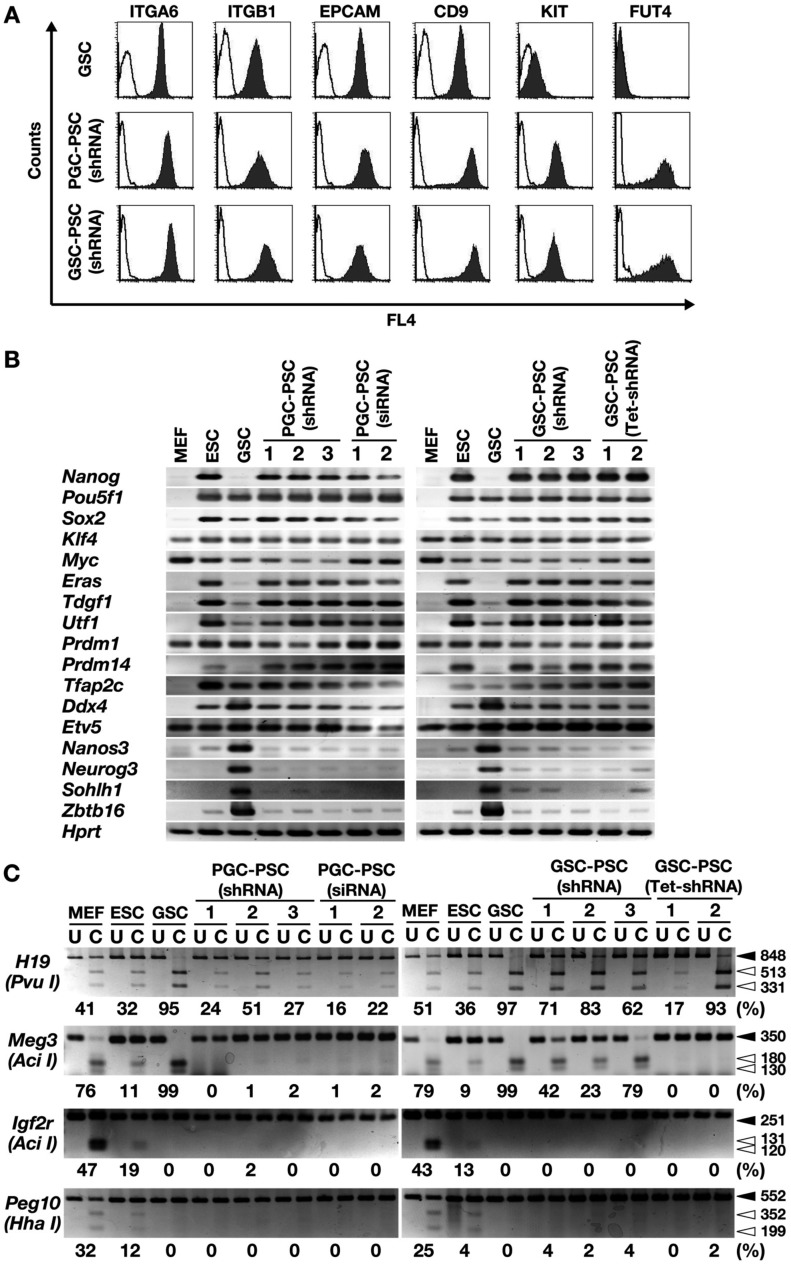
We compared the phenotypes of GSC-PSCs and PGC-PSCs derived from 12.5 dpc embryos. Flow cytometric analysis of germ cell surface markers did not show apparent differences, and all cell types showed strong expression of ITGA6, ITGB1, EPCAM and CD9 (Fig. 3A). GSCs were distinct from these PSCs in that the former showed weak expression of KIT and completely lacked expression of FUT4. RT-PCR was also performed to examine the expression of germ cell and PSC markers (Fig. 3B). Overall, both PGC-PSCs and GSC-PSCs showed similar gene expression patterns and were indistinguishable. Weak spermatogonia marker expression was detected in both cell types. Although ESCs and GSCs showed many similar genes, the former was distinct from the latter in that they expressed Nanog and Eras [11]. Similar to ESCs, both PGC-PSCs and GSC-PSCs strongly expressed both Nanog and Eras. These phenotypic analyses suggested that PGC-PSCs and GSC-PSCs are more closely related to ESCs than to GSCs.
Fig. 3.
Phenotypic characterization of PSCs derived from PGCs and GSCs. (A) Flow cytometric analysis of cell surface markers. Open histograms indicate controls. (B) RT-PCR analysis. (C) COBRA of imprinted genes. Open arrowheads, methylated DNA; closed arrowheads, unmethylated DNA. Enzymes used to cleave each locus are indicated in parentheses. Levels of percentage methylation, estimated based on the intensity of each band, are indicated below the gels. U, uncleaved; C, cleaved.
We next analyzed the DNA methylation patterns in imprinted genes based on COBRA (Fig. 3C). This was because GSC-PSC formation is often accompanied by dynamic changes in DNA methylation patterns in imprinted genes [11, 28]. In GSC-PSCs, all three clones showed demethylation in H19 and Meg3 DMRs, but no methylation was observed in DMRs of Peg10 and Igf2r. On the other hand, DMRs in PGC-PSCs showed slightly different patterns between H19 and Meg3 DMRs: all three clones of PGC-PSCs showed no DNA methylation in Meg3 DMRs, while DMRs in H19 were significantly less methylated than those in GSC-PSCs. No apparent methylation was observed in Peg10 or Igf2r DMRs of PGC-PSCs. These results suggested that PGC-PSCs and GSC-PSCs have distinct epigenetic properties.
Chimera formation by blastocyst injection
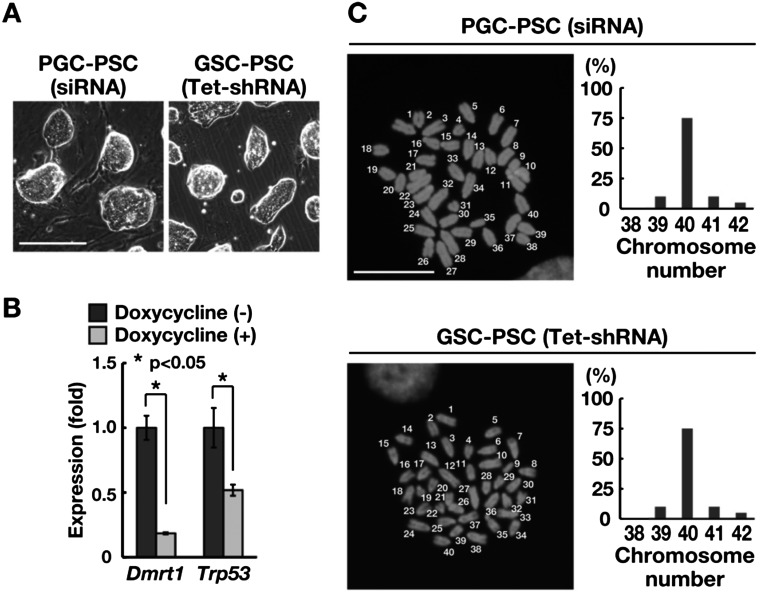
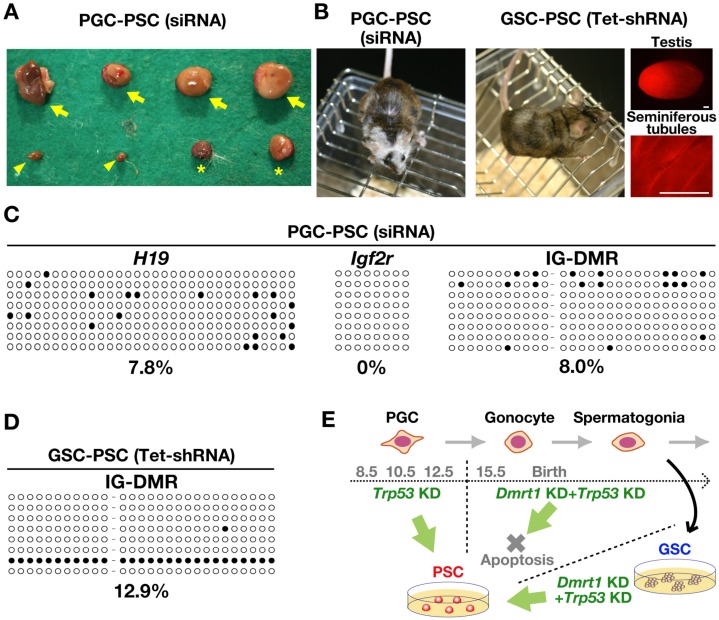
To determine whether PGC-PSCs and GSC-PSCs are capable of differentiation into somatic tissues, we used siRNA for reprogramming to avoid the possible negative effect of constitutive KD of Dmrt1 and Trp53 on differentiation. Suppression of Dmrt1 and Trp53 by siRNA resulted in successfull derivation of PGC-PSCs (Supplementary Table 3: online only). However, we were unable to obtain GSC-PSCs using the same siRNA treatment, which may reflect the lower reprogramming efficiency of GSCs. Therefore, a doxycycline-regulated KD vector (Tet-shRNA) was used to suppress Dmrt1 and Trp53 to derive GSC-PSCs for differentiation experiments. These PSC lines showed similar appearances based on morphology (Fig. 4A), and real-time PCR analysis confirmed depletion of the target genes by doxycycline (Fig. 4B). The phenotypes of these cells were also comparable to those produced by constitutive KD vectors and expressed ESC markers (Fig. 3B and C). Karyotype analysis using Hoechst 33342 showed that > 65% of the cells were euploid (Fig. 4C).
Fig. 4.
Differentiation potential of PSCs. (A) Appearance of PSCs derived by siRNA or tet-inducible shRNA. (B) Real-time PCR analysis of the target gene in GSCs by tet-inducible shRNA following doxycycline treatment (n = 3). Cells were recovered 7 days after transfection. (C) Distribution of metaphase spreads with different chromosome numbers. At least 20 cells were counted. The numbers in the images indicate that 40 chromosomes are present in these cells. Bar = 100 μm (A), 10 μm (C). Stain: Hoechst 33342 (C).
Using these cell types, we first produced embryoid bodies in vitro and examined the type of differentiating cells (Supplementary Fig. 2A: online only). Immunohistological staining showed positive staining of ACTA2 (mesoderm marker), AFP (endoderm marker) and TUBB3 (ectoderm marker), suggesting that both PGC-PSCs and GSC-PSCs could differentiate into cells of the three germ layers. We also confirmed their differentiation potential by injecting PGC-PSCs and GSC-PSCs subcutaneously into nude mice (Supplementary Fig. 2B: online only). Histological analysis showed typical teratoma formation from both PGC-PSCs and GSC-PSCs 4 weeks after injection.
We then injected both PGC-PSCs and GSC-PSCs into blastocysts to examine their differentiation potential in vivo. In the first set of experiments using PGC-PSCs, a total of 132 blastocysts were microinjected. After embryo transfer, thirty-eight embryos were implanted, and nine offspring were born by caesarean section. However, there were abnormal embryos that died during the midgestation period, and placenta-only conceptuses were also identified (Fig. 5A). Nevertheless, seven of the nine born offspring became normal adults, and one of the females showed coat color chimerism (Fig. 5B). In the second set of experiments using GSC-PSCs, a total of 112 blastocysts were injected. A total of 27 embryos were implanted, and 17 offspring were born. Unlike PGC-PSCs, neither abnormal embryos nor placenta-only conceptuses were observed in these experiments. One of the male offspring showed coat color chimerism (Fig. 5B). The testes of this male showed RFP fluorescence under UV light. However, we were unable to confirm donor cell-derived RFP expression in spermatogenic cells by histological analysis. No donor cell-derived offspring were obtained from PGC-PSCs (42 offspring) or GSC-PSCs (72 offspring) by natural mating of the chimeric animals, both of which lived more than a year without apparent abnormalities. These results suggested that neither cell type contributed to germ cells.
Fig. 5.
Production of chimeric animals. (A) Abnormal embryo development after PGC-PSC injection. Arrows and asterisks indicate placenta-only conceptuses with or without a degenerating fetus, respectively. Arrowheads indicate deciduae, which were found at the implantation sites. (B) Chimeric offspring born after microinjection of the indicated cell types into blastocysts. The white coat color represents the contribution of the donor cells (ICR background). RFP fluorescence also shows the contribution of GSC-PSCs to the testes of chimeric offspring. (C) Bisulfite sequencing analysis of H19, Igf2r and IG-DMR in PGC-PSCs. (D) Bisulfite sequencing analysis of the IG-DMR in GSC-PSCs. (E) Summary of the experiment. Bar = 250 μm (B).
Although these results showed that double depletion of Dmrt1 and Trp53 induces pluripotency in PGCs and GSCs, the increased number of placenta-only embryos in the PGC-PSC experiments suggested that PGC-PSCs have more epigenetic abnormalities. Such placenta-only phenotypes have often been observed in cloned animals [29, 30]. Since more than half of the cloned animals showed hypermethylation in IG-DMR and Dlk1 is consistently upregulated in cloned animals [29, 30], we examined the DNA methylation status of H19, Igf2r and IG-DMRs in PGC-PSCs by bisulfite sequencing (Fig. 5C and D). Both H19 and Igf2r DMRs were demethylated in PGC-PSCs. Contrary to our expectations, the IG-DMR region was hypomethylated, which was comparable to the results for GSC-PSC. These results suggested that abnormalities in IG-DMR methylation are not responsible for the placenta-only phenotype of PGC-PSCs.
Discussion
Although germ cells are thought to have potential for pluripotency, the mechanism underlying pluripotency suppression has long remained unknown. In this study, we examined the feasibility of reprogramming germline cells of different stages by manipulating Dmrt1 and Trp53 levels, which can induce GSC-PSCs [12]. Pluripotency is thought be specific to PGCs [31] because no animal models have clearly demonstrated initiation of teratoma formation from postnatal germ cells in vivo. In fact, all teratomas are thought to be of embryonic origin in humans [32]. However, our previous in vitro studies have shown that postnatal spermatogonia can convert into teratoma-forming PSCs. Because of the extensive epigenetic reprogramming during germline development, it is not surprising that PGCs, which proliferate more actively than postnatal germ cells, are epigenetically unstable and can more easily convert into EGCs or teratomas. In contrast, GSCs show stable epigenetic properties [33], and it is likely that spermatogonia have additional mechanisms to suppress pluripotency compared with PGCs. Our recent development of a pluripotency induction protocol in GSCs suggested that germ cells in prenatal stages acquire pluripotency more easily than GSCs by depleting Dmrt1 and Trp53. The present study explored this hypothesis.
PGC transfection experiments revealed at least three differences from GSC reprogramming. First, PGCs directly become ESC-like colonies, while GSCs become PSCs via epiblast-like colonies. In addition, our gene expression analyses showed differences, because Prdm1 and Ddx4 were decreased immediately in PGCs, while they were upregulated in GSCs. Second, PGCs are reprogrammed by depleting only Trp53, while Dmrt1 depletion causes apoptosis. Third, PGCs, but not gonocytes, are more readily reprogrammed than GSCs. Interestingly, reprogramming occurred most efficiently when we used PGCs from 12.5 dpc embryos than embryos of earlier developmental stages. This result was counterintuitive, because PGCs in younger embryos are more easily converted into EGCs by cytokine treatment [5]. The exact mechanisms underlying these phenomena remain unclear, but one factor that needs to be considered is that the cells were cultured without SCF and LIF in the current experiments. GSC culture conditions may have induced premature expression of postnatal germ cell genes, which can influence teratoma formation [34]. It is also possible that PGCs/gonocytes were more susceptible to damage caused by lentivirus-mediated gene transfection.
While PGCs are more easily reprogrammed than GSCs, Trp53 was the only common factor in regulating the pluripotency of both cell types. Although both Dmrt1 and Trp53 are implicated in teratoma formation [13, 35], our KD experiments did not show correlative changes in their expression levels, suggesting that they function independently. Importantly, Trp53 is distinct from Dmrt1 in that Trp53 KO mice have teratomas in non-129 backgrounds [35]. In contrast, Dmrt1 deficiency induces germ cell loss in non-129 backgrounds, and teratomas are found only in the 129 background. This pattern of teratoma formation resembles Dnd1, whose deficiency in the 129 background causes teratomas [36, 37]. Although Pten deficiency or AKT activation also induces teratomas or EGCs in non-129 backgrounds [23, 38], neither has effects on spermatogonia reprogramming [39,40,41,42]. Therefore, Trp53 is the only factor that acts in both embryonic and postnatal germ cells to suppress pluripotency. However, the role of Trp53 appears to be different between these stages because Trp53 depletion causes apoptosis only in PGCs. This was in contrast to Dmrt1, whose depletion caused apoptosis in both stages. Suppression of apoptosis by Bax depletion in GSCs could not induce GSC-PSCs [12]. This was in contrast to PGCs, because Trp53 depletion could not suppress apoptosis of PGCs caused by Dmrt1 depletion, but Bax deficiency could promote teratoma formation [37]. These results suggest that the role of apoptosis in pluripotency induction is different in these cell types.
Further analysis suggested that Sox2 is a prime target for Trp53 function in PGCs. SOX2 has a unique expression pattern during germ cell development. It is expressed in PGCs but gradually disappears during the gonocyte stage, and no protein expression is observed in spermatogonia, including GSCs [28, 43, 44]. The mechanism by which SOX2 expression is regulated remains unknown. Analysis in this study showed that Sox2 was upregulated in GSCs when either Dmrt1 or Trp53 was depleted. In contrast, Trp53 depletion alone could upregulate Sox2 in PGCs. Therefore, one critical function of Dmrt1 in GSCs was to prevent upregulation of Sox2, which may act as an additional barrier to pluripotency in these cells. Besides Trp53, Sox2 is probably regulated by multiple factors in PGCs because recent experiments showed that overexpression of any of the Yamanaka factors could induce pluripotency in PGCs [26]. The same group also showed that PGCs express Pou5f1, Sox2 and Myc, but not Klf4, which suggested that a relatively low level of Klf4 expression prevents pluripotency induction in PGCs [26]. However, given these facts and the robust increase of Sox2 in PGCs after Trp53 KD, we speculate that the increase in Sox2, with no increase in Klf4, is responsible for pluripotent cell conversion after Trp53 KD in PGCs. Considering that GSCs, which express POU5F1, MYC and KLF4, but not SOX2, are more resistant to reprogramming than PGCs, the difference in Sox2 regulation may explain the critical change in pluripotency regulation and inefficiency of reprogramming during germ cell development.
Another notable finding in this study was the failure to reprogram freshly isolated testis cells by double KD by Dmrt1 and Trp53. This result was unexpected because we believed that reprogramming of postnatal spermatogonia would occur in a similar manner as in GSCs, which were derived from SSCs. This result suggests that GSCs and freshly isolated SSCs do not have the same properties [45]. However, it should be noted that our results do not necessarily suggest that GSCs are more easily reprogrammed than fresh spermatogonia. In our previous study, we found that transfection of Yamanaka factors did not transform GSCs but instead induced transformation of fresh testis cells and produced ESC-like colonies [22]. Although transfection of Pou5f1 or Sox2 could reprogram GSCs, the efficiency was significantly lower, and it did not occur without suppression of Trp53 [12]. Therefore, the optimal reprogramming protocols for GSCs and freshly isolated testis cells are clearly different. It remains unclear why these two cell populations behave in distinct manners, but it is possible that GSCs are derived from specific subpopulations of SSCs, which can be propagated in vitro [46]. Alternatively, this phenomenon may reflect the ageing of SSCs due to in vitro culture. These hypotheses need to be explored in future experiments.
Pluripotency of the established cells was examined by chimera production experiments. Although chimeric offspring were born from both cell types, we observed many placenta-only conceptuses and aborted fetuses when we microinjected PGC-PSCs. Lack of germline transmission could be due to the small number of chimeras produced, but the abnormal placenta-only phenotype is probably not due to the transfection procedure because we never observed such embryos when we used siRNA to obtain GSC-PSCs in our previous studies [11, 12, 28]. Skeletal abnormalities were reported in EGC chimeras [5, 47]. However, to the best of our knowledge, the placenta-only phenotype has not been observed in other PSC lines, including ESCs or induced PSCs, suggesting that the PGC-PSCs had distinct epigenetic properties or underwent incomplete reprogramming. Because this placenta-only phenotype is characteristic of cloned animals [29, 30], we examined abnormalities in DNA methylation patterns in IG-DMR but failed to identify any apparent defects. More extensive analyses of the epigenetic profile of PGC-PSCs will be useful to understand the mechanism of dynamic epigenetic reprogramming that occurs during PGC development.
Using a newly developed technique for reprogramming GSCs, our study revealed distinct reprogramming potentials of different stages of germ cells (Fig. 5E). Although derivation of PSCs from germline cells has been described for more than 20 years, few studies have compared pluripotency regulation between PGCs and spermatogonia. Our study also provides additional evidence for functional differences between spermatogonia and GSCs. Comparing our results with those of studies on EGC formation or teratoma development will provide a novel venue to dissect the regulation of the pluripotency program in germline cells. Thus analyzing the pluripotency regulation in the germline will increase our understanding of germ cell development and may provide important information for prevention of germ cell tumors.
Supplementary
Acknowledgments
This research was supported by the Uehara Foundation for Medical and Pharmaceutical Research, Japan Science and Technology Agency (PRESTO), Takeda Science Foundation and Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. We thank Ms Y Ogata for technical assistance.
References
- 1.Sabour D, Schöler HR. Reprogramming and the mammalian germline: the Weismann barrier revisited. Curr Opin Cell Biol 2012; 24: 716–723. [DOI] [PubMed] [Google Scholar]
- 2.Illmensee K, Stevens LC. Teratomas and chimeras. Sci Am 1979; 240: 120–132. [DOI] [PubMed] [Google Scholar]
- 3.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 1992; 70: 841–847. [DOI] [PubMed] [Google Scholar]
- 4.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature 1992; 359: 550–551. [DOI] [PubMed] [Google Scholar]
- 5.Labosky PA, Barlow DP, Hogan BL. Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development 1994; 120: 3197–3204. [DOI] [PubMed] [Google Scholar]
- 6.Stewart CL, Gadi I, Bhatt H. Stem cells from primordial germ cells can reenter the germ line. Dev Biol 1994; 161: 626–628. [DOI] [PubMed] [Google Scholar]
- 7.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003; 69: 612–616. [DOI] [PubMed] [Google Scholar]
- 8.Meistrich ML, van Beek MEAB. Spermatogonial stem cells. In: Desjardins C, Ewing LL (eds.), Cell and Molecular Biology of the Testis. New York: Oxford University Press; 1993: 266–295.
- 9.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21: 776–798. [PubMed] [Google Scholar]
- 10.Kanatsu-Shinohara M, Shinohara T. Spermatogonial stem cell self-renewal and development. Annu Rev Cell Dev Biol 2013; 29: 163–187. [DOI] [PubMed] [Google Scholar]
- 11.Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, Toyoshima M, Niwa O, Oshimura M, Heike T, Nakahata T, Ishino F, Ogura A, Shinohara T. Generation of pluripotent stem cells from neonatal mouse testis. Cell 2004; 119: 1001–1012. [DOI] [PubMed] [Google Scholar]
- 12.Takashima S, Hirose M, Ogonuki N, Ebisuya M, Inoue K, Kanatsu-Shinohara M, Tanaka T, Nishida E, Ogura A, Shinohara T. Regulation of pluripotency in male germline stem cells by Dmrt1. Genes Dev 2013; 27: 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey M, McArthur MJ, Montgomery CA, Jr, Bradley A, Donehower LA. Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J 1993; 7: 938–943. [DOI] [PubMed] [Google Scholar]
- 14.Looijenga LH, Hersmus R, Gillis AJ, Pfundt R, Stoop HJ, van Gurp RJ, Veltman J, Beverloo HB, van Drunen E, van Kessel AG, Pera RR, Schneider DT, Summersgill B, Shipley J, McIntyre A, van der Spek P, Schoenmakers E, Oosterhuis JW. Genomic and expression profiling of human spermatocytic seminomas: primary spermatocyte as tumorigenic precursor and DMRT1 as candidate chromosome 9 gene. Cancer Res 2006; 66: 290–302. [DOI] [PubMed] [Google Scholar]
- 15.Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 2011; 476: 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuma S, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Hosokawa M, Nakatsuji N, Ogura A, Shinohara T. Spermatogenesis from epiblast and primordial germ cells following transplantation into postnatal mouse testis. Development 2005; 132: 117–122. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa T, Aréchaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol 1997; 41: 111–122. [PubMed] [Google Scholar]
- 18.Kanatsu-Shinohara M, Muneto T, Lee J, Takenaka M, Chuma S, Nakatsuji N, Horiuchi T, Shinohara T. Long-term culture of male germline stem cells from hamster testes. Biol Reprod 2008; 78: 611–617. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Kanatsu-Shinohara M, Ogonuki N, Miki H, Inoue K, Morimoto T, Morimoto H, Ogura A, Shinohara T. Heritable imprinting defect caused by epigenetic abnormalities in mouse spermatogonial stem cells. Biol Reprod 2009; 80: 518–527. [DOI] [PubMed] [Google Scholar]
- 20.Kawase Y, Iwata T, Watanabe M, Kamada N, Ueda O, Suzuki H. Application of the piezo-micromanipulator for injection of embryonic stem cells into mouse blastocysts. Contemp Top Lab Anim Sci 2001; 40: 31–34. [PubMed] [Google Scholar]
- 21.Kanatsu-Shinohara M, Toyokuni S, Shinohara T. CD9 is a surface marker on mouse and rat male germline stem cells. Biol Reprod 2004; 70: 70–75. [DOI] [PubMed] [Google Scholar]
- 22.Morimoto H, Lee J, Tanaka T, Ishii K, Toyokuni S, Kanatsu-Shinohara M, Shinohara T. In vitro transformation of mouse testis cells by oncogene transfection. Biol Reprod 2012; 86: 148–: 1–11.. [DOI] [PubMed] [Google Scholar]
- 23.Kimura T, Tomooka M, Yamano N, Murayama K, Matoba S, Umehara H, Kanai Y, Nakano T. AKT signaling promotes derivation of embryonic germ cells from primordial germ cells. Development 2008; 135: 869–879. [DOI] [PubMed] [Google Scholar]
- 24.Leitch HG, Blair K, Mansfield W, Ayetey H, Humphreys P, Nichols J, Surani MA, Smith A. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development 2010; 137: 2279–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitch HG, Nichols J, Humphreys P, Mulas C, Martello G, Lee C, Jones K, Surani MA, Smith A. Rebuilding pluripotency from primordial germ cells. Stem Cell Reports 2013; 1: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagamatsu G, Kosaka T, Saito S, Honda H, Takubo K, Kinoshita T, Akiyama H, Sudo T, Horimoto K, Oya M, Suda T. Induction of pluripotent stem cells from primordial germ cells by single reprogramming factors. Stem Cells 2013; 31: 479–487. [DOI] [PubMed] [Google Scholar]
- 27.Matsui Y, Takehara A, Tokitake Y, Ikeda M, Obara Y, Morita-Fujimura Y, Kimura T, Nakano T. The majority of early primordial germ cells acquire pluripotency by AKT activation. Development 2014; 141: 4457–4467. [DOI] [PubMed] [Google Scholar]
- 28.Kanatsu-Shinohara M, Lee J, Inoue K, Ogonuki N, Miki H, Toyokuni S, Ikawa M, Nakamura T, Ogura A, Shinohara T. Pluripotency of a single spermatogonial stem cell in mice. Biol Reprod 2008; 78: 681–687. [DOI] [PubMed] [Google Scholar]
- 29.Hirasawa R, Matoba S, Inoue K, Ogura A. Somatic donor cell type correlates with embryonic, but not extra-embryonic, gene expression in postimplantation cloned embryos. PLoS ONE 2013; 8: e76422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okae H, Matoba S, Nagashima T, Mizutani E, Inoue K, Ogonuki N, Chiba H, Funayama R, Tanaka S, Yaegashi N, Nakayama K, Sasaki H, Ogura A, Arima T. RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum Mol Genet 2014; 23: 992–1001. [DOI] [PubMed] [Google Scholar]
- 31.Spiller CM, Bowles J, Koopman P. Nodal/Cripto signaling in fetal male germ cell development: implications for testicular germ cell tumors. Int J Dev Biol 2013; 57: 211–219. [DOI] [PubMed] [Google Scholar]
- 32.Looijenga LH, Verkerk AJ, Dekker MC, van Gurp RJ, Gillis AJ, Oosterhuis JW. Genomic imprinting in testicular germ cell tumours. APMIS 1998; 106: 187–195, discussion :196–197. [DOI] [PubMed] [Google Scholar]
- 33.Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, Miki H, Takehashi M, Toyokuni S, Shinkai Y, Oshimura M, Ishino F, Ogura A, Shinohara T. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development 2005; 132: 4155–4163. [DOI] [PubMed] [Google Scholar]
- 34.Heaney JD, Anderson EL, Michelson MV, Zechel JL, Conrad PA, Page DC, Nadeau JH. Germ cell pluripotency, premature differentiation and susceptibility to testicular teratomas in mice. Development 2012; 139: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krentz AD, Murphy MW, Kim S, Cook MS, Capel B, Zhu R, Matin A, Sarver AL, Parker KL, Griswold MD, Looijenga LH, Bardwell VJ, Zarkower D. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc Natl Acad Sci USA 2009; 106: 22323–22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu R, Bhattacharya C, Matin A. The role of dead-end in germ-cell tumor development. Ann N Y Acad Sci 2007; 1120: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook MS, Coveney D, Batchvarov I, Nadeau JH, Capel B. BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1(Ter/Ter) mice. Dev Biol 2009; 328: 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura T, Suzuki A, Fujita Y, Yomogida K, Lomeli H, Asada N, Ikeuchi M, Nagy A, Mak TW, Nakano T. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development 2003; 130: 1691–1700. [DOI] [PubMed] [Google Scholar]
- 39.Lee J, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Kimura T, Nakano T, Ogura A, Shinohara T. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development 2007; 134: 1853–1859. [DOI] [PubMed] [Google Scholar]
- 40.Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282: 25842–25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest 2011; 121: 3456–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Mao X, Boyce T, Zhu GZ. Dispensable role of PTEN in mouse spermatogenesis. Cell Biol Int 2011; 35: 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol 2006; 6: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Western PS, van den Bergen JA, Miles DC, Sinclair AH. Male fetal germ cell differentiation involves complex repression of the regulatory network controlling pluripotency. FASEB J 2010; 24: 3026–3035. [DOI] [PubMed] [Google Scholar]
- 45.Morimoto H, Kanatsu-Shinohara M, Takashima S, Chuma S, Nakatsuji N, Takehashi M, Shinohara T. Phenotypic plasticity of mouse spermatogonial stem cells. PLoS ONE 2009; 4: e7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takashima S, Kanatsu-Shinohara M, Tanaka T, Morimoto H, Inoue K, Ogonuki N, Jijiwa M, Takahashi M, Ogura A, Shinohara T. Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports 2015; 4: 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tada T, Tada M, Hilton K, Barton SC, Sado T, Takagi N, Surani MA. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol 1998; 207: 551–561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.