Abstract
Bacterial lipoproteins possess diverse structure and functionality, ranging from bacterial physiology to pathogenic processes. As such many lipoproteins, originating from Brucella are exploited as potential vaccines to countermeasure brucellosis infection in the host. These membrane proteins are translocated from the cytoplasm to the cell membrane where they are anchored peripherally by a multifaceted targeting mechanism. Although much research has focused on the identification and classification of Brucella lipoproteins and their potential use as vaccine candidates for the treatment of Brucellosis, the underlying route for the translocation of these lipoproteins to the outer surface of the Brucella (and other pathogens) outer membrane (OM) remains mostly unknown. This is partly due to the complexity of the organism and evasive tactics used to escape the host immune system, the variation in biological structure and activity of lipoproteins, combined with the complex nature of the translocation machinery. The biosynthetic pathway of Brucella lipoproteins involves a distinct secretion system aiding translocation from the cytoplasm, where they are modified by lipidation, sorted by the lipoprotein localization machinery pathway and thereafter equipped for export to the OM. Surface localized lipoproteins in Brucella may employ a lipoprotein flippase or the β-barrel assembly complex for translocation. This review provides an overview of the characterized Brucella OM proteins that form part of the OM, including a handful of other characterized bacterial lipoproteins and their mechanisms of translocation. Lipoprotein localization pathways in gram negative bacteria will be used as a model to identify gaps in Brucella lipoprotein localization and infer a potential pathway. Of particular interest are the dual topology lipoproteins identified in Escherichia coli and Haemophilus influenza. The localization and topology of these lipoproteins from other gram negative bacteria are well characterized and may be useful to infer a solution to better understand the translocation process in Brucella.
Keywords: Brucella vaccine target, lipoprotein localization, Brucella lipoprotein, lipoprotein secretion, outer membrane protein, Lol pathway, pathogen-associated molecular patterns, toll-like receptors
Lipoprotein Localization
Bacteria are classified as gram negative or gram positive based on the structure of the cell wall. Gram negative bacteria have a cell envelope composed of the outer membrane (OM), a thin hydrophilic peptidoglycan layer lining the periplasmic space and the inner membrane (IM). Whereas, the gram positive bacteria cell envelope is composed of a thick hydrophilic peptidoglycan layer and IM. The IM of gram negative bacteria is a phospholipid bilayer while the OM is composed of an asymmetric lipid bilayer with lipopolysaccharides (LPS) orientated toward the outer surface and phospholipids orientated toward the periplasm (Mitchell, 1961; Bladen and Mergenhagen, 1964). Proteins are synthesized in the cytoplasm and may either span the IM with characteristic α-helical stretches causing their retention in the membrane, referred to as Imps or translocate across the IM by means of a signal peptide to form complex β-barrels that span the OM as outer membrane proteins (Omps) (Koebnik et al., 2000). The characteristic β-barrel of Omp is comprised of amphipathic β-strands (with alternating hydrophobic amino acid residues) ensuring their solubility during secretion through the periplasm (Rosenbusch, 1974; Nakamura and Mizushima, 1976; Pugsley, 1993). Lipoproteins are adept at anchoring to either the IM or OM by means of the acyl moieties (hydrophobic interactions) in both gram negative and gram positive bacteria (Braun and Rehn, 1969; Narita, 2011). This review will focus upon lipoproteins in gram negative bacteria, which are exported to the surface of the OM in certain bacterial species. This provides insight to Brucella lipoproteins and the reason for their selection as vaccine candidates for treatment against brucellosis.
The General Secretory Twin-Arginine Translocation Systems: Inner Membrane To Periplasm
In gram negative bacteria, proteins destined for the OM are exported from the IM to the OM via the type I secretion system (T1SS), type III secretion system (T3SS), type IV secretion system (T4SS), or type VI secretion system (T6SS) in a single, direct step. Other proteins, mostly in gram negative bacteria that translocate to the OM, can initially be exported from the IM into the periplasm by means of the general secretory (Sec) or twin-arginine translocation (Tat) systems as protein precursors and thereafter translocated to the OM surface by other systems in a double reaction process. In most gram positive bacteria, translocation of the proteins to the cell surface is simpler and occurs via the Sec or Tat systems, with exception of mycobacteria. Components of the Sec system recognize a hydrophobic N-terminal sequence of the proteins destined for secretion and translocation of the protein occurs in an unfolded state, coupled with ATP hydrolysis and a proton gradient as the energy source (Sjostrom et al., 1987; Bibi, 1998; Papanikou et al., 2007). On the other hand, the Tat system identifies a sequence composed of basic amino acid residues (Ser- Arg- Arg- x- Phe- Leu -Lys) at the N-terminal end of the protein and protein export occurs in a folded state making use of a proton gradient (Pugsley, 1993; Brink et al., 1998; Müller, 2005).
Protein Secretion Pathways For General Secretory/Twin-Arginine Translocation Systems- Intermediates
Secretion can be further branched to the translocation of Sec/Tat-transported intermediates from the periplasm to external milieu through the chaperone-usher (CU) pilli pathway required for the formation of fimbrial adhesion involved in host attachment and invasion, biofilm formation, cell motility as well as protein and DNA transport export (Hultgren et al., 1991; Hal Jones et al., 1997). The other translocation pathways include the autotransporter (T5SS, type V secretion system); the type II secretion system (T2SS) and the type VII secretion system (T7SS), which form a channel utilized for the translocation of proteins across both the hydrophobic membrane and the cell in gram positive mycobacteria, and lastly the lipoprotein localization machinery (Lol) pathway. No stable intermediates have been identified for T1SS, T3SS, T4SS, and T6SS in the periplasm. Furthermore, the T3SS, T4SS, and T6SS pathways make use of a conducting channel capable of extending from the bacterial membranes to the host cell membrane specifically allowing for the secretion of bacterial virulence factors. Established pathways of lipoprotein secretion from the IM through the OM involve either the T2SS, T5SS, or Lol, or a model employed by bacteria of the class spirochetes referred as the spirochetal model (Pugsley, 1993). The protein secretion pathways extending beyond the scope of this review, i.e., lipoprotein secretion in gram negative bacteria, will not be discussed in further detail.
Bacterial Lipoproteins Function, Modification, And Localization
Lipoproteins have the ability to localize to diverse regions in the cell. In gram negative bacteria, mature lipoproteins anchor to the IM, the extracellular surface or the peripheral region of the OM, depending upon the distinct function performed in the cell (Wu and Tokunaga, 1986; Narita et al., 2004). Lipoproteins that are anchored peripherally to the OM play an important role in bacterial physiology including envelope stability, cell division, sporulation, conjugation, nutrient acquisition, signal transduction, transport, and protein folding, and in bacterial pathogenic processes for example adhesion, colonization, invasion, and persistence through immune evasion (Mathiopoulos et al., 1991; Sutcliffe and Russell, 1995; Khandavilli et al., 2008; Hutchings et al., 2009; Kovacs-Simon et al., 2011). The specific localization of lipoproteins is dependent on an effective lipoprotein modification and transport system, and a precise lipoprotein sorting mechanism (Hantke and Braun, 1973). In gram positive bacteria lipoproteins are exported through the IM and acylated to ensure secure localization on the bacterial cell surface. However, in gram negative bacteria lipoproteins destined for the OM must seek guidance from a lipoprotein-specific chaperoned pathway (Sutcliffe and Russell, 1995; Narita et al., 2004; Zückert, 2014). It would be naive to mold OM-associated proteins into a specific group due to their diverse nature and function, however, both the β-barrel structure of Omps and lipid modification of lipoproteins are characteristic of OM-associated proteins. Lipoproteins are generally hydrophilic, however, due to the N-terminal lipid moiety are hydrophobic. For this reason the release of lipoproteins does not occur spontaneously and must overcome the energetically unfavorable reaction at which they become detached from the IM, and are transported across the hydrophilic periplasm to reach the OM. The modification of lipoproteins by attachment of the hydrophobic lipid moiety occurs in the hydrophilic periplasm. This alteration does not impede their export from the periplasm to the surface of the OM (Pugsley, 1993; Tokuda and Matsuyama, 2004). Unlike Imps, which share α-helical membrane spanning domains, and integral Omps, which share the characteristic β-barrel domains, lipoproteins are structurally diverse and share only the N-terminal lipid moiety (MacIntyre et al., 1988; Bernstein, 2011; Narita, 2011).
Lipoproteins And Their “Lipobox”
The first component of lipoprotein secretion translocates preprolipoprotein precursors from the cytoplasm (where they are synthesized) through the IM and into the periplasm via the Sec or Tat systems (Berks et al., 2005; Driessen and Nouwen, 2008; Zückert, 2014). Lipoprotein genes of bacterial origin contain a C-terminal lipobox. It is this four-amino-acid motif, which shapes the molecular platform for in silico lipoprotein prediction algorithms (Inouye et al., 1977). With an increase in the number of bacterial genomes sequenced, more lipoproteins continue to be identified resulting in the degeneration of the original lipobox. At present the only conserved residue of the canonical lipobox is cysteine, which forms the target of acylation and is the new N-terminus amino acid (residue at position +1) of the mature lipoprotein (Seydel et al., 1999; Babu et al., 2006; Setubal et al., 2006). This clearly necessitates the development of new algorithms to identify lipoproteins. Following the +1 cysteine, peptide residues comprising the “tether” (second domain) are occasionally disordered, lacking observed secondary structure and vary in terms of length. This “tether” domain not only contains a lipoprotein sorting sequence but connects the lipid anchor to the third domain, thereby positioning lipoproteins in the cell envelope for optimal protein function (Seydel et al., 1999; Schulze and Zückert, 2006; Zückert, 2013, 2014).
Preprolipoprotein Diacylglyceryl Transferase, Lipoprotein Signal Peptidase, Lipoprotein N’N-acyl Transferase: Lipidation In The Periplasm
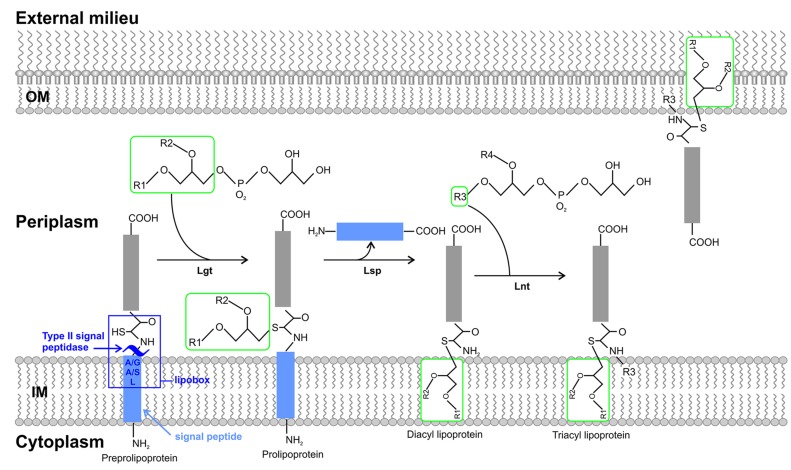
The second component of the biosynthetic pathway of bacterial lipoproteins yields a mature lipidated protein upon post translational modifications of prelipoproteins, catalyzed by three indispensable IM enzymes (Figure 1). In detail, preprolipoprotein diacylglyceryl transferase (Lgt) catalyzes diacylation (phosphatidylglycerol attachment to the thiol group) at the conserved cysteine residue of the lipoprotein via a thioester bond. This modification process occurs either within the IM or at the cytoplasmic surface (Tokunaga et al., 1982; Sankaran and Wu, 1994; Selvan and Sankaran, 2008). The signal peptide is then cleaved N-terminally of the +1 cysteine residue by a signal peptidase II enzyme, lipoprotein signal peptidase (Lsp) at the interface of the IM (gram negative bacteria) upon recognition of the lipobox (Tokunaga et al., 1982; Munoa et al., 1991; Vidal-Ingigliardi et al., 2007). The cleavage process avails the N-terminal amine group of the +1 cysteine residue for modification with a third acyl chain via an amide linkage by lipoprotein N’N-acyl transferase (Lnt) thereby completing the membrane anchor in gram negative bacteria (Gupta et al., 1993). This last process of the pathway is expendable in certain gram positive bacteria (Jackowski and Rock, 1986; Vidal-Ingigliardi et al., 2007; Tschumi et al., 2009; Hillmann et al., 2011). IM lipoproteins in gram negative bacteria do not interact with downstream pathways and are retained within the IM. Lipoproteins can localize either to the IM (toward the periplasm) as with gram positive bacteria or the OM as with gram negative bacteria. When translocated to the OM, lipoproteins are anchored to the OM either facing the periplasm, embedded in the inner leaflet of the OM (integral membrane proteins, displaying the C-terminal end to the external surface), anchored to the OM facing the external milieu or cleaved from the OM and released to the external milieu (Pugsley et al., 1986; Schulze and Zückert, 2006; Magnet et al., 2007; Roussel-Jazede et al., 2010; Bernstein, 2011; Zückert, 2014). In gram negative bacteria, OM lipoprotein translocation was first speculated to occur by means of localized fusion of the periplasm (hydrophilic) in between the IM and OM via Bayer’s junctions due to their hydrophobic properties (Bayer, 1968; Bayer, 1991). However, studies conducted by (Tokuda and Matsuyama, 2004), revealed the existence of the Lol pathway. The Lol pathway, T2SS, T5SS, and another pathway well characterized in spirochetes, the spirochetal OM model (involving a specialized flippase complex) are pathways associated with the final process of lipoprotein secretion to the OM (Schulze et al., 2010; Chen and Zuckert, 2011; Chen et al., 2011; Zückert, 2014). These are established pathways, which diversify among the subclasses of Proteobacteria. Examples of various types of surface exposed lipoproteins will be further expounded upon to illustrate these proposed models.
FIGURE 1.
The biosynthesis and sorting of bacterial lipoproteins. Prelipoproteins synthesized in the cytoplasm are translocated to the outer surface of the IM by the general secretory (Sec), two-arginine (Tat) pathways. Prelipoproteins possess a consensus motif termed the lipobox, which is a component of the N-terminal signal peptide cleavage site. It is the +1 cysteine residue of the lipobox that undergoes lipid modification in a sequential process catalyzed by three periplasmic enzymes to yield the mature lipoproteins. The first enzyme, preprolipoprotein diacylglyceryl transferase (Lgt) generates a thioether linkage by transferring a diacylglyceryl molecule to the sulfhydryl group of the +1 cysteine. The second enzyme, lipoprotein signal peptidase (Lsp) catalyzes the cleavage of the N-terminal signal peptide at +1 S-diacylglyceryl cysteine. Lastly, lipoprotein N’N-acyl transferase (Lnt) catalyzes aminoacylation of the α-amino group of the S-diacylglyceryl cysteine, producing a mature triacylated lipoprotein. Depending upon the lipoprotein sorting signal the lipoprotein is then targeted for the inner membrane (IM) or the outer membrane (OM). Abbreviations used: inner membrane (IM), general secretory (Sec), two-arginine (Tat), preprolipoprotein diacylglyceryl transferase (Lgt), lipoprotein signal peptidase (Lsp), lipoprotein N’N-acyl transferase (Lnt), outer membrane (OM).
The Bacterial Lipoprotein Localization Machinery Pathway: Periplasmic Sorting To The Outer Membrane Of Gram Negative Bacteria
The periplasmic sorting pathway exports specific lipoproteins from the IM using chaperones, the ATP binding cassette (ABC) transporter (LolCDE) and energy generated from ATP hydrolysis. A carrier protein (LolA) translocates the lipoproteins through the periplasm, and finally insertion into the inner leaflet of the OM is mediated by an OM receptor (LolB). In E. coli, the Lol pathway comprises of LolCDE which is inserted in the IM; LolA, a soluble chaperone located in the periplasm; and LolB, an OM lipoprotein receptor anchored to the inner leaflet of the OM (Matsuyama et al., 1995, 1997; Yakushi et al., 2000). Lipoproteins that are triacylated (i.e., mature and catalyzed by Lnt) are identified by the LolCDE complex. Upon ATP hydrolysis they are transferred to LolA, forming a water soluble complex. Translocation from the periplasm to the OM then occurs by means of diffusion (Tajima et al., 1998; Narita et al., 2002). The lipoprotein-specific periplasmic carrier protein LolA was the first characterized constituent of the Lol pathway and is vital for lipoprotein export through the periplasm. By forming a 1:1 stoichiometric ratio of carrier to lipoprotein, LolA encases the hydrophobic lipid moiety of the lipoprotein (Matsuyama et al., 1995; Tajima et al., 1998). LolB, a lipoprotein itself is situated on the inner periphery of the OM, directing the final step of lipoprotein sorting by accepting lipoproteins from LolA and anchoring them to the OM. As with LolA, knockout gene studies using lolB resulted in the accumulation of lipoprotein-LolA complex within the periplasm of E. coli (Matsuyama et al., 1997; Tanaka et al., 2001). A high degree of structural homology exists between LolA and LolB based on x-ray crystallography, even though a low degree of identity exists between the primary sequences (Takeda et al., 2003). Both components possess a hydrophobic cavity offering a probable binding site for the acyl chain of the lipoproteins. However, differences in their primary sequences and secondary structure permit the transfer of lipoproteins irreversibly from LolA to LolB in the periplasm (Matsuyama et al., 1997). Hypotheses have been postulated for the interaction of LolA-LolB with lipoproteins. In Pseudomonas aeruginosa (γ-Proteobacteria) for example, one acyl chain of the lipoprotein is bound internally, whereas the other two chains are proposed to connect with the proteins using surface hydrophobic patches (Remans et al., 2010). It has been established that homologs for LolB are present β-, γ- and δ-Proteobacteria (absent in α-Proteobacteria). Brucellae are placed taxonomically in the α-2 subdivision of the class Proteobacteria, which could suggest that LolA plays a more extensive role in lipoprotein localization. In Brucella the mechanism of lipoprotein maturation and translocation has been assumed based on sequence similarity between species (??). Components for lipoprotein maturation (Lgt, Lsp, and Lnt) have been inferred by homology (orthologs closely related species) and components for lipoprotein translocation such as LolA and LolC/E have been predicted (in the absence of proof at protein, transcript, or homology levels), whereas LolD has been inferred by homology (DelVecchio et al., 2002; Halling et al., 2005). The Lol pathway is considered a general mechanism for lipoprotein transport given that the genes encoding Lol proteins are conserved in other gram negative Proteobacteria. It is hypothesized that an Omp or a lipoprotein with a sequence dissimilar to LolB yet still retaining its function could exist, or LolA could be dual functioning, both accepting and incorporating lipoproteins due to the absence of LolB in α-Proteobacteria (Sutcliffe et al., 2012). In Mycobacterium tuberculosis for example, a structural LolA/B homolog interacts with all three acyl chains of the lipoprotein. It was assumed that the LolA-B fold is flexible, allowing lipoproteins to encase presenting hydrophobic molecules (Drage et al., 2010).
Studies conducted by deleting LolA allowed for the identification of the LolCDE complex in E. coli, which releases lipoproteins into the periplasm. The LolCDE complex is an ABC transporter present in the IM, which initiates the coupling of a lipoprotein to LolA (Yakushi et al., 2000). LolC and LolE are integral Imps homologous to each other, whereas LolD is located within the cytoplasmic leaflet of the IM. LolD possesses ATP-binding domains (conserved Walker A and B motifs) and forms a 2:1:1 stoichiometric ratio with LolC-LolE (Yakushi et al., 1998; Yakushi et al., 2000; Ito et al., 2006). Studies have suggested OM lipoproteins are first retained by LolE causing allosteric modifications increasing LolD’s affinity for ATP. Meanwhile, LolC attaches to LolA and as ATP binds to LolD, lipoprotein interaction with LolE is reduced resulting in lipoprotein relocation to LolA upon ATP hydrolysis and finally the lipoprotein-LolA complex attaches to LolB (Ito et al., 2006; Okuda and Tokuda, 2009; Mizutani et al., 2013; Zückert, 2014).
Lipoprotein Sorting Within The Periplasm And The Evolution Of The “+2” Rule
The N-terminal end of the mature lipoprotein (triacylated) has a sorting signal, i.e., the suggested lipobox, which is recognized by the Lol pathway for export. In principle, the amino acid residue (i.e., side chain) at position two of the lipoprotein sequence determines membrane localization in E. coli. Aspartic acid at +2 functions as an IM retention signal whereas other amino acids at this position function as an OM targeting signal. The aspartic acid functions as a LolCDE avoidance signal for lipoproteins by interfering with the interaction between LolCDE and cysteine (at +1 position) (Yamaguchi et al., 1988; Gennity and Inouye, 1991; Seydel et al., 1999; Terada et al., 2001). Studies involving substituting large non-protein molecules to cysteine at position +2 revealed that LolCDE recognizes the N-acyl S-diacylglyceryl of cysteine and this appears to be the sole structural requirement for LolCDE recognition (Masuda et al., 2002; Hara et al., 2003). Additionally, histidine and lysine at position +3 reduced the stringency of IM retention by the +2 aspartic acid residue. These studies among others rendered the +2 rule a guiding standard instead of a universal rule for lipoprotein sorting by the Lol pathway (Gennity and Inouye, 1991; Zückert, 2014).
Brucella Pathogenesis And Outer Membrane Biogenesis
According to the Food and Agriculture Organization (FAO) and World Health Organization (WHO), brucellosis is one of the most widespread zoonotic diseases in the world (Lopes et al., 2010). The disease is caused by bacteria from the genus Brucella. The main economically important species include B. abortus (in cattle), B. melitensis (in sheep and goats), and B. suis (in swine) (Bruce, 1887; Bang, 1897; Traum, 1914). The internalization of Brucella within the host cell is a regulated system involving interactions between host cell surface factors and pathogen molecular factors. As part of the intracellular trafficking in host mammalian cells, protein substrates for the T4SS are transported to the host cell cytosol where they allegedly assist translocation and interactions with the endoplasmic reticulum to reach the replicative vacuole. In fact Brucella virulence is dependent upon the ability to survive and replicate in the vacuolar phagocytic compartments of macrophages (Arenas et al., 2000; Naroeni et al., 2001). T4SS is categorized as one of the known bacterial secretion systems (discussed previously) and is one of the most important virulence systems identified as crucial in the intracellular processing of Brucella within macrophages, which ultimately controls the biogenesis of the intracellular compartment where Brucella will finally take residence (Hong et al., 2000; Xiang et al., 2006; de Jong et al., 2008). Similarly, the two-component regulatory system BvrR/BvrS (TCS BvrRS) is another crucial component of the virulence system. The BvrRS is composed of a histidine kinase sensor located in the IM (BvrS) and a cytoplasmic regulator (BvrR). This system controls the expression of a T4SS component, participates in the homeostasis of the OM by controlling the structure of the LPS and the expression of periplasmic proteins and Omps (Omp25 and Omp22) (Guzman-Verri et al., 2002; Manterola et al., 2005; Lamontagne et al., 2007). The interaction between T4SS and BvrRS may epitomize an evolutionarily conserved survival tactic used by the α-Proteobacteria subclass (Comerci et al., 2001; Delrue et al., 2001; Martinez-Nunez et al., 2010). Furthermore, gene expression profiling used to define Brucella pathogenesis identified the Omps, lipoproteins, stress response proteins, chaperones, flagellar genes, ABC transport proteins, and genes for LPS and fatty acid biosynthesis as key components for cell envelope or OM biogenesis (Guzman-Verri et al., 2002; Lamontagne et al., 2007; Manterola et al., 2007; Rossetti et al., 2009; Viadas et al., 2009). Immunogenic proteins (particularly Omp25, Omp31, Omp2b) found on the bacterial cell surface are superior vaccine candidates as they establish the initial contact site between the pathogen and the host (Delvecchio et al., 2006). Ultimately the knowledge gained on pathogen-associated factors (with a particular interest to Brucella Omps and lipoproteins) have been used for the application of these factors as antigens in vaccine formulation and studies have confirmed their ability to confer protective immunity against Brucella and other intracellular pathogens (Fu et al., 2012).
Brucella Outer Membrane Versus Other Gram Negative Bacteria Outer Membrane
What is known about the Brucella cell envelope is limited and indirect but does, however, prove the plasticity of its cell envelope (Moriyon and Lopez-Goni, 1998). Subtle yet noticeable differences exist between the cell envelopes of Brucella and those of other gram negative Proteobacteria used as model organisms. Studies have verified the failure of using standard bacterial cell fractionation methods for the Brucella cell envelope extraction and they also exhibit sensitivity to media classically used in other gram negative bacteria. Interestingly Brucella cells cultured in media and within other cells spontaneously release OM components lavish with LPS, native hapten polysaccharide, phosphatidylcholine, Omps, and other proteins maintaining cell viability (Gamazo and Moriyon, 1987). Some of these components inhibit the fusion of phagosomes with lysosomes thereby enabling virulence (Moreno et al., 2004). This shedding is associated with the blebbing of Brucella OM and capture of surface and periplasmic constituents (Zhou et al., 1998). The Brucella OM tends to form steadier bilayers compared to other gram negative bacteria, owing to the major presence of phosphatidylcholine (lipid) in the OM in contrast to phosphatidylethanolamine in other gram negative bacteria (Moriyon and Lopez-Goni, 1998). The presence of neutral lipids in Brucella OM suggests the interaction of positively charged ornithine lipids with the negatively charged phosphates of the lipid A (Martin and Hancock, 1990). The fatty acids in Brucella tend to possess longer acyl chains proposing a greater membrane span compared to the model gram negative bacteria (Moriyon and Lopez-Goni, 1998). Contrary to other gram negative bacteria, stronger hydrophobic interactions exist between the Brucella Omps and other OM components. The Brucella Omps also have a hydrophilic domain enabling interaction with the peptidoglycan layer and surface exposure (Dubray, 1976). However, based on Omp extraction procedures and similarity to Rhizobium leguminosarum (a member of the α-2 Proteobacteria subdivision), a portion of Omp could be covalently attached to the peptidoglycan layer (Roest et al., 1995; Vizcaino et al., 1996). Previous studies have shown heat inactivation causes the IM of Brucella to collapse, however, the OM maintains morphology, suggesting a greater OM stiffness in Brucella compared to other gram negative bacteria (Rubbi, 1991). The anchorage of the lipid (acyl chains) component of Brucella lipoproteins is suggested to be stronger too, based on other lipid components in the OM (Moriyon and Lopez-Goni, 1998). Another difference proven is that hydrophobic substrates readily permeate the OM of Brucella compared to other gram negative bacteria due to characteristics of the LPS (Douglas et al., 1984; Martinez de Tejada and Moriyon, 1993). Moreover, resistance to the bactericidal oxygen-independent systems of phagocytes characterizes the Brucella OM as a result of the resistance the LPS have to polycations (Martin and Hancock, 1990; Martinez de Tejada et al., 1995). The characteristics of the Brucella cell envelope define its pathogenicity and resistance. These findings will provide crucial insight into developing an understanding of Brucella lipoproteins localization within the Brucella OM given their (Brucella vs. other gram negative bacteria) variation in OM components.
Outer Membrane Proteins Accommodated In The Brucella Outer Membrane
The Brucella cell envelope is constituted of an IM, periplasm (peptidoglycan with soluble components) and OM like other gram negative bacteria (Dubray, 1976; Moriyon and Lopez-Goni, 1998). The OM of Brucella is riddled with Omps (comparable with other gram negative bacteria), which were first isolated during the extraction of the cell envelope in the 1980s. These Omps were categorized based upon their molecular mass into group 1 (94 or 88 kDa), group 2 (36–38 kDa), and group 3 (31–34 and 25–27 kDa) with several of them forming tight complex connections with the LPS and peptidoglycan layer (Winter, 1987). Group 2 Omp were classified as porins and lipoproteins bearing similarity to E. coli Braun’s lipoprotein (Lpp) (Douglas et al., 1984). Further classification of the group 2 (Omp2a and Omp2b) and group 3 proteins (Omp25 for 25–27 kDa and Omp31 for the 31–34 kDa) designated them as major Omps (Verstreate et al., 1982). Studies have confirmed the surface exposure of the major Omps based upon experiments with the use of monoclonal antibodies (MAbs) (Cloeckaert et al., 1990, 1991; Bowden et al., 1995). The ability of anti-Omp MAbs to bind to the OM surface of Brucella Rough (R) and Smooth (S) strains was assessed using ELISA. Labeling with MAb specific for the Brucella Omps revealed these Omps have epitopes that are exposed to the OM surface in S-Brucella cells. Moreover, this exposure is greater in R-Brucella and varies at species level. In order to understand the situation in the OM of the S-Brucella cells, it is thus suggested that the LPS with shorter O-chains are favored spatially relative to certain Omps, and/or the native hapten polysaccharides are irregularly dispersed on the OM surface (Gómez-Miguel et al., 1987; Cloeckaert et al., 1990; Bowden et al., 1995). Previous studies performed in vitro proposed the ability of antibodies to assist macrophage (cell mediated immune response) opsonization of Brucella (Ralston and Elberg, 1969). In mice, high protection was conferred by polyclonal immune sera that was raised against the peptidoglycan layer (reacted with Omps and S-LPS using immunoblots). However, protection conferred by the anti-Omp MAbs was lower in comparison to that conferred by the anti-S-LPS MAbs. Lower protection could be correlated to weaker opsonization as a result of low antigen-antibody accessibility and avidity (Cloeckaert et al., 1991). These investigations formed the basis of using Brucella Omps to further probe the effect these Omps would have on Brucella virulence.
Brucella Porins (Group 2 Outer Membrane Proteins)
Omp2a forms a monomeric pore in the OM, while Omp2b forms a trimeric channel composed of oligomers (in the presence of SDS, it is resistant to heat denaturation), both typical of some bacterial porins (Marquis and Ficht, 1993). Structural differences exist between the two Omps but do not differentiate which is more efficient or demonstrates better activity (Cloeckaert et al., 2002). However, the expression of Omp2a during intracellular growth of Brucella species has not been confirmed to date. Secondary structure prediction methods revealed both these porins have a 16 stranded β-barrel domain with eight large surface loop regions that are exposed to the external milieu (Mobasheri et al., 1997; Paquet et al., 2001). Major differences in the porins are two insertions and/or deletions which exist at the surface exposed loop 3 and loop 5, which vary between the Brucella species. It is suggested that pore constriction of Brucella porins is most likely exerted by loop 5, instead of the loop 3 (Paquet et al., 2001). Loop 6, loop 7, and loop 8 were identified as epitopes based on analysis with other chimeric porins and studies conducted with MAbs indicated a strong surface exposure of these loops, which is in agreement with the proposed role in bacterial adhesion of the Arg- Gly- Asp (RGD)-containing L6 loop (Cloeckaert et al., 1990; Campbell et al., 1994; Bogdan and Apicella, 1995; Bowden et al., 1995).
Brucella Omp25/Omp31 Family Similarity And Differences
At the level of DNA, a high degree of similarity is observed among the Brucella species. However, differences in pathogenicity and host preference that are noted could relate to OM composition. The Omp25/Omp31 family of Brucella is composed of seven homologous Omps (Omp22, Omp25b, Omp25c, Omp25d, Omp31b, Omp31, and Omp25). Some of these Omps may be involved in virulence with the encoding genes displaying DNA polymorphisms (Edmonds et al., 2002; Salhi et al., 2003; Vizcaíno et al., 2004; Martín-Martín et al., 2008). It was speculated that Brucella Omp25 shares identity with E. coli OmpA and even though they are not homologous and differ in size, it is suggested that these β-barrel proteins play a similar function in the OM (Moriyon and Lopez-Goni, 1998). Topology predictions suggest Omp25 is composed of eight stranded transmembrane β domains with large surface-exposed loops. A variation observed in the binding patterns of anti-Omp25 MAbs to B. ovis Omp25 suggests an antigenic shift exists due to a 36 bp deletion localized to the last surface-exposed loop at the C-terminal end (Cloeckaert et al., 2002). Like E. coli OmpA, Brucella Omp25 binds detergents and LPS, causing a characteristic (of β sheets) heat-modifiable electrophoretic migration (Moriyon and Berman, 1983; Moriyon and Lopez-Goni, 1998).
Interspecies differences are described for Omp31. Omp31 is absent in B. abortus and a nine nucleotide substitution is observed between B. ovis in comparison to B. melitensis. This substitution results in a difference in antigenicity between B. ovis and B. melitensis (Vizcaino et al., 2001b; Cloeckaert et al., 2002). Structural differences in the processing of Omps between gram negative bacteria are evident; an example is the heterologous expression of B. melitensis Omp31 in E. coli. In contrast to B. melitensis, in E. coli Omp31 does not seem to interact with the peptidoglycan. This suggests that the interaction of the peptidoglycan to the OM allows a greater “OM stiffness” in Brucella compared to other gram negative bacteria (Vizcaino et al., 1996). In other Brucella species Omp31 forms oligomers, which are resistant to denaturation by sodium dodecyl sulfate at low temperatures and as such is classified as a porin (since it displays porin characteristics) in Brucella species with lower omp2 gene expression (i.e., Omp31 compensates for Omp2). Functional studies are required to explain these porin characteristics of Brucella Omp since in E. coli porins are homologous, but Brucella Omp2b and Omp31 are divergent (Cloeckaert et al., 1996; Moriyon and Lopez-Goni, 1998). If porin activity occurs as a result of Omp31, its absence in B. abortus should have an effect on cell viability. This function could be compensated for by a similar protein, which may suggest the high level of Omp2b in B. abortus (Salhi et al., 2003; Caro-Hernandez et al., 2007; Martín-Martín et al., 2009). Based on topology predictions, Omp31, like Omp25, possesses an eight stranded β-barrel domain, however, with much larger surface exposed loops. Three amino acid residue substitutions form part of an immunodominant region on the N-terminal end of surface exposed loop 1 explain the perceived antigenic variation between Omp31 in B. melitensis and B. ovis (Vizcaino et al., 2001a,b). Furthermore, Omp25 and Omp31 of Brucella reveal a significant level of identity to Omps of other members of α-2 Proteobacteria including Rhizobium, Sinorhizobium, Mesorhizobium, Agrobacterium, Bartonella, and Rickettsia species (Moreno and Moriyon, 2002). Also, even though Omp31 is a major Omp, its absence has a compensatory effect on the Omp25/31 family that does not influence virulence. Omp31b shares 61% identity with Omp31, however, is absent in B. melitensis but identified in B. abortus and B. suis (yet to be identified in B. ovis) using antibody reactivity and protein detection techniques (Salhi et al., 2003; Martín-Martín et al., 2009). Mutant BvrRS Brucella strains are avirulent in the mouse model, which is indicated by decreased cell invasion, inhibition of intracellular replication and lysosome fusion and the inability to replicate intracellularly by Brucella (Sola-Landa et al., 1998). Studies involving the disruption of BvrR or BvrS resulted in a reduction of Brucella Omp2b, Omp22, Omp25, Omp25c, and Omp31b implicating the Omps in Brucella virulence. No reduction in Omp25d was noticeable, suggesting omp25d was mostly likely differentially expressed and thus detected at very low levels (Lamontagne et al., 2007). Even though group 3 Omps are suggested as crucial for the integrity of Brucella OM owing to their close association with LPS, no fluctuations were observed (Edmonds et al., 2001; López-Goni et al., 2004). The homology shared between the Omp25/Omp31 family could suggest structural and functional interchangeability, maintaining OM integrity and virulence in Brucella. Due to this redundancy, investigating single Omp mutants of the Omp25/Omp31 family would be misleading (Salhi et al., 2003; Caro-Hernandez et al., 2007; Martín-Martín et al., 2009). These investigations with the Omp25/Omp31 family and the BvrRS reveal a compensatory mechanism among the Brucella species and insight into the combined effort of all OM components of Brucella to maintain OM homeostasis, which could be speculated to affect lipoprotein localization.
Lipoproteins Accommodated In The Brucella Outer Membrane
The B. abortus genome contains approximately 80 genes encoding putative lipoproteins (Chain et al., 2005). Nonetheless, Omp10, Omp16, Omp19, and Omp89 are the four minor Omps previously identified by the use of MAbs that have been examined in much greater detail compared to the other putative lipoproteins (Cloeckaert et al., 1990). Three of the four minor Omps (Omp19, Omp16.5, and Omp10) are low molecular weight proteins, display poor polymorphisms and are expressed in all six Brucella species including their biovars (Thirkell et al., 1991; Tibor et al., 1996; Vizcaíno et al., 2000). Based on their amino acid sequences, they are mainly hydrophilic and display similarity to the peptidoglycan-associated lipoproteins (Pal) of other gram negative bacteria hence their association to the peptidoglycan (Moriyon and Lopez-Goni, 1998). Like E. coli Lpp, they are lipoproteins (however, Omp89 is not characterized as a lipoprotein) predicted to have three fatty acids connected to an N-terminus glycerylcysteine and the N-terminal signal peptides contain a tetrapeptide with high similarity to the consensus sequence, i.e., the lipobox (Tibor et al., 1999). It is through these lipid moieties that they are periplasmically linked to the OM and it is through these longer acyl chains that these lipoproteins are more strongly anchored to the Brucella OM in comparison to other Pal of gram negative bacteria (Moriyon and Lopez-Goni, 1998). Studies performed revealed a similar pathogenic mechanism was observed in macrophages when the lipidated Omp19 (but not the unlipidated form or LPSs) stimulated dendritic cell maturation thereby producing a Th1 type immune response (Giambartolomei et al., 2004; Zwerdling et al., 2008). It is this lipid moiety that is shared by all bacterial lipoproteins that display the same ability to down-regulate antigen capture and increase T-cell response (Chain et al., 2005; Zwerdling et al., 2008). This will be discussed further in the review with regards to Toll-like receptors (TLRs).
Brucella Phenotypic Studies Using Phagocytic Cells To Determine The Effect Of Lipoprotein Gene Mutations
The characteristic and hence pathogenic function of Brucella lipoproteins has been assessed by introducing mutations in the genes to determine the resultant phenotype. The model system used to assess the effect of attenuation caused by Brucella mutant strains is growth within professional and non-professional phagocytic cells (bovine macrophage cells and human epithelial cells), where its replicative niche is established (Stabel and Stabel, 1995; Delrue et al., 2001). Virulent Brucella fail to fuse with lysosomes; by contrast avirulent Brucella co-localize with the endosome post-infection (Arenas et al., 2000). By mimicking in vitro host intracellular defences, such as treatment with a polycationic peptide (polymyxin B) or with a reactive oxidative agent (H2O2), resistance conferred can be extrapolated to the characteristic phenotype of the Brucella cell envelope (Martinez de Tejada et al., 1995; Freer et al., 1996; Sola-Landa et al., 1998). Furthermore, virulence in mice, sensitivity to bovine complement-mediated lysis (extracellular killing) and growth in minimal media should be assessed in conjunction with in vitro studies (Corbeil et al., 1988). Deletions of the immunoreactive lipoproteins, Brucella omp19 and omp10 genes were assessed previously (Tibor et al., 2002). The omp10 mutant resulted in decreased survival rates in mice, and defective growth in media. However, neither in vitro growth nor OM characteristics were compromised. Thus it is suggested that replication is impeded in phagocytes during the intracellular stage with the omp10 mutant. With the omp19 mutant no splenomegaly (enlarged spleen, clinical symptom) was observed in mice, a lower growth rate was observed in vitro suggesting sensitivity to conditions within phagosomes before growth in an intracellular replication compartment. An increase in sensitivity to bovine serum complement was also observed for the mutant (Tibor et al., 2002). Additionally, omp19 deletion in the R-Brucella strain did not impact the survival of mice (Vemulapalli et al., 2000). These observations indirectly suggest Omp19 maintains interactions between OM components and hence the LPS lattice, thereby affecting the extracellular and intracellular survival of Brucella (Freer et al., 1996). Brucella lipoproteins continue to be identified and similar results were obtained with a conserved 20 kDa lipoprotein mutant (Kim et al., 2013). Mutations were introduced using a transposon mutagenesis screen (Kim et al., 2012). The Brucella mutant strain depicted a lower rate of intracellular replication in comparison to the wild type strain in human epithelial cells and macrophages. Moreover, fewer mutants were recovered from the spleen of infected mice compared to infection with the wild type strain (Kim et al., 2013). These experiments provide evidence on the possible use of Brucella lipoproteins as candidate vaccines; however, results in the natural host would be crucial. Studies involving the disruption of BvrR or BvrS resulted in a substantial increase in periplasmic proteins, chaperones, and transporters (for, e.g., the inorganic ABC transporter family) with a reduction in Omp25/Omp31 family. The unexpected increase occurred as a result of nutritional stress displaying no auxotrophic defects. A substantial increase in Omp19 was observed. However, Omp10 was mostly likely differentially expressed at levels too low to be detected (Lamontagne et al., 2007). This implies that Brucella virulence should be examined as a multidimensional component where compounds play a coordinated role to adapt to various environments.
The Topology Of Brucella Lipoproteins In The Outer Membrane
The Omp10, Omp19, and Omp16 Brucella lipoproteins were also suggested to be localized to both sides of the membrane based on antibody accessibility studies (Gómez-Miguel et al., 1987; Cloeckaert et al., 1990; Bowden et al., 1995). This unifies strongly with the existence of dual topology proteins (Lpp, Pal, and P6) identified in other gram negative bacteria and will be expounded on further (Bernstein, 2011; Cowles et al., 2011; Michel et al., 2013, 2015). Omp16 is homologous to the Pals of other gram negative bacteria, however, no homology has been found for Omp10 and Omp19 with other bacteria (Tibor et al., 1994, 1996). Brucella Omp10, Omp16, and Omp19 satisfy the characteristics of lipoproteins. Studies using an inhibitor of signal peptidase II resulted in the accumulation of the lipoprotein precursors and immunoprecipitation studies using anti-Omp MAbs confirm the lipid modification of these minor Omp. These studies have provided insight on a proposed topology model for Omp10 and Omp19 where the lipid moiety attached to the lipoprotein N-terminal end is inserted in the external surface of the OM and these lipoproteins would be localized at interface of the OM and the external milieu (Tibor et al., 1999).
Dual Topology Proteins In Gram Negative Bacteria
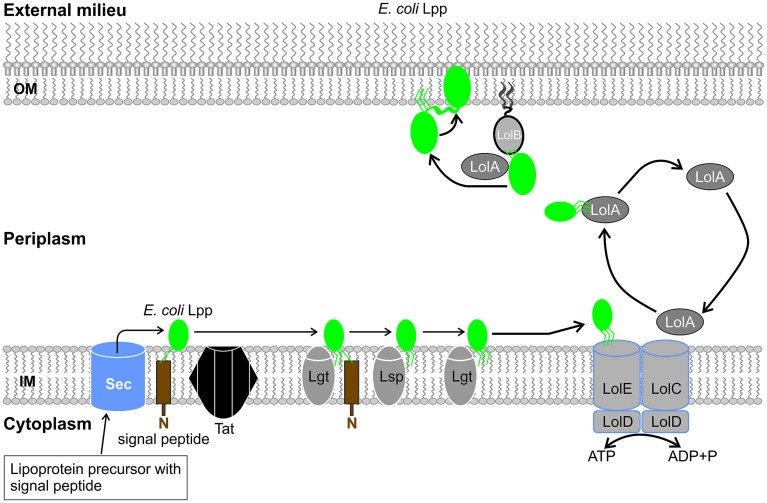
It is accepted that E. coli Lpp monomers are anchored to the inner surface of the OM (with the N-terminal lipid moiety embedded in the OM) (Yokota et al., 1999). Previous studies suggested that epitopes for both E. coli Lpp and Pal are exposed on the surface of E. coli (Hellman et al., 1997). These findings were further developed when dual topology characteristics were observed for these E. coli lipoproteins. E. coli Lpp exists in two distinct forms: a bound form, which resides in the periplasm with its C-terminal covalently attached to the peptidoglycan by transpeptidases, and a free form which spans the OM and is surface-exposed (Magnet et al., 2007; Cowles et al., 2011). Figure 2 provides insight to a speculated pathway for Lpp translocation in E. coli. Approximately 500 000 (of 750 000) copies of Lpp exist in free form in an E. coli cell, proving the importance of these structures to the E. coli OM bilayer (Braun et al., 1970; Inouye et al., 1972). Crystallographic studies of the soluble recombinant protein resolved a parallel three-stranded coiled-coil structure. The bound form exists as a homotrimer anchored to the OM by its lipid moieties due to the attachment of three lysine residues to the peptidoglycan layer. In comparison, the free form, which also exists as a homotrimer, extends over the OM to form a helix bundle, bearing similarity to transmembrane proteins (Bowie, 1999; Shu et al., 2000).
FIGURE 2.
The proposed translocation pathway for the dual topology E. coli Braun’s lipoprotein (Lpp). After generating the mature Lpp (green structure), the lipoprotein in the inner membrane (IM) interacts with the lipoprotein localization machinery (Lol) located in the periplasm. Adenosine triphosphate (ATP) hydrolysis, an ABC transporter (LolCDE), a carrier protein (LolA) and outer membrane (OM) lipoprotein receptor (LolB) mediates the “mouth-to-mouth” the transfer of Lpp from the IM to the OM. Lpp may reside in the periplasm anchored to the inner leaflet of the OM (bound form) or spans the OM resulting in the surface exposure of its C terminus (free form). Abbreviations used: Braun’s lipoprotein (Lpp), inner membrane (IM), lipoprotein localization machinery (Lol), adenosine triphosphate (ATP), outer membrane (OM), ATP binding cassette (ABC).
H. influenza P6 is more complex compared to E. coli Lpp in terms of structure. It is composed of a mesh of α-helices, loops, and a single β-sheet (Parsons et al., 2006). It is the second lipoprotein (next to E. coli Lpp) identified to have dual orientation characteristic in γ-Proteobacteria class of gram negative bacteria. P6 was recognized as a vaccine candidate for the treatment of non-typeable H. influenzae infections in humans due to its location on the surface of the OM; the gene is also well conserved among pathogenic strains and was identified as immunogenic (Murphy et al., 1985; Nelson et al., 1988; Bogdan and Apicella, 1995; DeMaria et al., 1996). Since P6 is recognized as one of the Pal, it localizes primarily to the periplasm (Murphy et al., 2006; Shaw et al., 2013; Shaw et al., 2014). This implies that, in contrast to E. coli Lpp where more copies of the surface exposed form exist, a greater portion of P6 was exported to the inner periphery of the OM (Michel et al., 2013).
The Lol pathway also exports E. coli Pal from the IM into the OM (Liang et al., 2005; Godlewska et al., 2009). The N-terminal end of E. coli Pal is suggested to interact with the inner surface of the OM via the lipid moieties and the C-terminal end binds to the peptidoglycan layer, thereby allowing Pal to be located in OM orientated toward the inner surface of the OM (Cascales and Lloubès, 2004; Parsons et al., 2006; Godlewska et al., 2009). The E. coli Pal lipoprotein shares similarity in terms of sequence and structure to P6, which would suggest that the lipoproteins possibly share function, protein interaction, and hence localization (Parsons et al., 2006; Godlewska et al., 2009). Pal is localized like P6 mainly to the periplasm, associated with the C-terminal domains of OmpA, TolB (periplasmic protein), TolA, and the peptidoglycan layer (Clavel et al., 1998; Godlewska et al., 2009). E. coli Pal and H. influenza P6 both form a monomeric α/β sandwich with the secondary structures (α-helices, loops, and a β-sheet) and have a high (50%) sequence identity (Chen and Henning, 1987; Parsons et al., 2006). Recently evidence of a dual topology role of Pal was established (Michel et al., 2015). Approximately 25% of the lipoprotein is exposed to the OM surface (similar to H. influenza P6). The dual orientation was further implicated in a pathological mechanism since the surface-exposed Pal may be released from the OM during sepsis (Hellman et al., 2000).
Brucella Lipoproteins And Their Similarity To Other Gram Negative Dual Topology Proteins
Due to the similarity of Brucella Omp16 with other Pal of gram negative bacteria, this may imply that Brucella Omp16 may be composed of a complex folding involving α/β secondary structure and like H. influenza P6 it may have surface exposed regions or be completely exposed to the external milieu. Even though the idea of a dual orientation lipoprotein might be probable, immunological studies have not confirmed the existence thereof. Previous studies in other gram negative bacteria have proposed the dual orientation of the lipoproteins takes place as a result of gene duplication followed by divergent evolution of topology; this may also occur post protein synthesis in the endoplasmic reticulum (Rapp et al., 2006). The dual orientation of membrane proteins has been conserved through evolution signifying an importance for the double orientation of these proteins (Michel et al., 2013). It is believed that bacterial lipoproteins have a common post-translational modification pathway (Pugsley, 1993). Based on the successful modification of Brucella Omp10, Omp16, and Omp19 when expressed in E. coli, it can be assumed the pathway for lipoprotein maturation is functionally conserved between Brucella, E. coli and other related gram negative bacteria (Tibor et al., 1999). The integration of lipoproteins into the OM could indicate the existence of novel types of membrane channels. A translocase (or a folding catalyst) reserved for interconverting the two forms has been implicated (Cowles et al., 2011).
Dual Topology Brucella Lipoproteins And Using Experimental Models Described For E. coli Of Lol And Other Pathways
Surface proteolysis is most commonly used to validate the surface exposure of a protein experimentally yet some of these gram negative bacteria proteins are protease resistant. Brucella Omps are no exception. Group 3 Omps form tight interactions to LPS, conferring resistance of these Omps to protease digestion (Gamazo and Moriyon, 1987). Recently dual topology proteins were identified using a novel, protease independent method which involved labeling with the water-soluble biotin reagents succinimidyl-6′-(biotinamido)-6-hexanamido hexanoate (NHS-LC-LC-biotin) and sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropinate (Sulfo-NHS-SS-biotin), followed by cell disruption techniques to quantify the population of cell surface Omps versus periplasmic Omps. These compounds are impermeable to the E. coli OM due to their relative high molecular mass and hydrophobicity and therefore surface exposed E. coli Omps can be labeled (Cowles et al., 2011; Michel et al., 2015). In assessing the dual topology of Brucella lipoproteins on the Brucella OM this would be impractical since these biotin analogs establish hydrophobic aliphatic biotin modifications on the proteins. This may alter the characteristics of surface exposed Omps (in particular lipoproteins as they are regarded as hydrophilic with the hydrophobic moiety embedded in the OM) in Brucella since hydrophobic compounds spontaneously permeate the OM of Brucella (Douglas et al., 1984; Martinez de Tejada and Moriyon, 1993). Another labeling reagent, which could be of use in the Brucella OM, is polyethylene glycol (PEG)-NHS-biotin. This analog is a primary-amine reactive, due to the PEG backbone it is hydrophilic and its high molecular mass makes it inaccessible to the periplasm (Nakae and Nikaido, 1975; Decad and Nikaido, 1976). Alternately, indirect visualization of surface-exposed lipoproteins using confocal microscopy could allow detection using a MAb specific for the lipoprotein and a secondary antibody conjugated fluorophore. As a negative control, a lipoprotein mutant strain may be used (Michel et al., 2015).
Brucella Lipoproteins And The Lipoprotein Localization Machinery
Outer Membrane Lipoprotein Carrier, LolA and LolB?
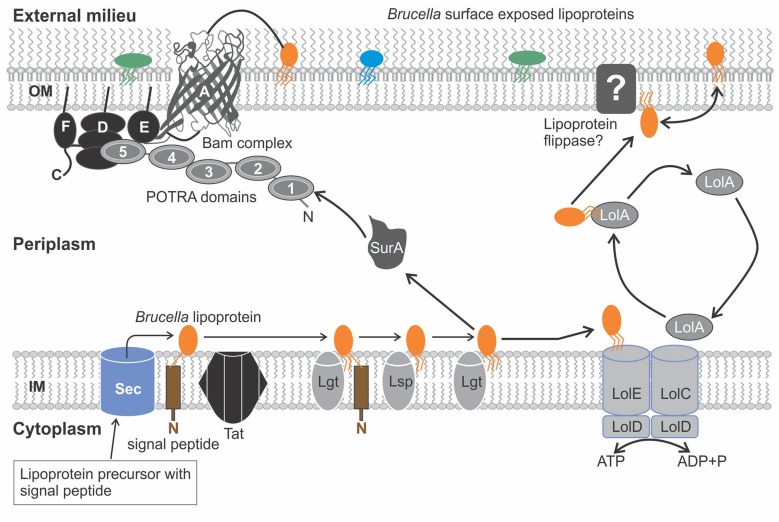
Brucella lipoproteins like those present in other gram negative bacteria are transported into the periplasm via the Sec or Tat pathways. They then undergo the process of lipidation catalyzed by the Imps, Lgt, Lsp, and Lnt (Figure 1). Depending on the function of the specific Brucella lipoprotein, the lipoprotein (like in other gram negative bacteria) is secreted to the OM (Figure 3). Components of the Lol pathway required for the modification and translocation of Brucella lipoproteins have been characterized (??). All components, except for LolB have been identified in Brucella since this gene is absent in α-Proteobacteria indicating the presence of a “dual functioning LolA” or another Lol component dissimilar to the conventional LolB as previously suggested (Narita, 2011; Sutcliffe et al., 2012). In other gram negative bacteria LolB is structurally similar to LolA and also possesses a hydrophobic cavity; however, LolB has a greater affinity for the lipid moiety due to the predominant presence of smaller hydrophobic amino acid residues such as leucine and isoleucine as opposed to bulky aromatic residues present in LolA. An efficient one-way lipoprotein transfer in the periplasm occurs as a result of the formation of a hydrophobic channel between LolA and LolB and the difference in affinity for lipoproteins, since energy sources are absent in the periplasm (Narita, 2011). If LolA plays a dual functional role in α-Proteobacteria, how does LolA translocate lipoproteins to the OM as ATP hydrolysis only fuels lipoprotein IM detachment and interaction of acyl chain to LolA? Furthermore, LolB is a lipoprotein anchored to the OM by a N-terminal acyl chain and LolA is not. This anchorage was identified as an essential component for incorporation of lipoproteins to the OM. The presence of extra C-terminal loop characteristic of LolA, confirms LolA cannot be targeted to membranes. It was therefore established that although LolA and LolB are structurally similar they are functionally distinct in the sorting of lipoproteins (Okuda et al., 2008; Tsukahara et al., 2009). This evidence suggests the possible existence of another protein dissimilar to LolB in sequence but which would perform the same function as LolB in α-Proteobacteria.
FIGURE 3.
The translocation pathways for the surface exposure of lipoproteins in Brucella. Prelipoproteins synthesized in the cytoplasm undergo lipid modification generating a mature, triacylated lipoprotein. Brucella lipoproteins (orange, green, and blue structures) may be released from the inner membrane (IM) to the outer membrane (OM) using the lipoprotein localization machinery (Lol) pathway, which is propelled by Adenosine triphosphate (ATP hydrolysis), an ATP binding cassette (ABC) transporter (LolCDE) and a carrier protein (LolA). LolB is absent in α-Proteobacteria, as such it is proposed lipoprotein flippase located in the OM transfers lipoproteins to the OM. Another proposed translocation pathway involves the export of the Brucella lipoproteins through the β- barrel domain of β- barrel assembly complex (Bam) complex embedded in the OM, assisted by the periplasmic chaperones (SurA) to the surface of the OM. The Brucella Bam complex lacks the BamB and BamC components found in other gram negative bacteria, however, a BamF component with a conserved sequence motif related to the BamC component may perform a similar role. Abbreviations used: inner membrane (IM), outer membrane (OM), lipoprotein localization machinery (Lol), adenosine triphosphate (ATP), ATP binding cassette (ABC), β- barrel assembly complex (Bam).
LolD-LolC/E Lipoprotein Releasing System
In Brucella, the lipoprotein releasing system is achieved by a “LolC/E” complex. This substantiates the likelihood of lipoprotein release by LolC–LolD heterodimer since LolE is only present in γ-Proteobacteria and not in the other subdivisions. Conformational changes in LolA are required to accommodate the acyl chains of lipoproteins. This change is achieved by interaction with LolC or LolE; however, LolC and not LolE cross-links with LolA suggesting the transfer of lipoproteins from LolA to LolC. This indicates that LolE is not important for lipoprotein transport in some bacteria since LolC interacts with LolA and would substantiate the absence of LolE in β-, δ-, and α-Proteobacteria (Narita, 2011).
Establishing The Existence Of The Lol Pathway Components For Brucella Lipoprotein Translocation Using Experimental Models
In E. coli, lipoprotein components were identified by generating spheroplasts and observing the effect that periplasmic (e.g., LolA) and OM components (e.g., a LolB homolog) have on the translocation of lipoproteins (Matsuyama et al., 1995, 1997; Yokota et al., 1999). Spheroplasts, in which the OM is disturbed, secrete proteins destined for the periplasm or OM. Brucella lipoproteins destined for the OM surface would therefore remain intact in the IM, as components for translocation are absent. In order to assess the periplasmic components required for lipoprotein translocation, an isolated periplasmic component (e.g., LolA) combined with the spheroplasts this would result in the secretion of Brucella lipoproteins. This periplasmic component would most likely be LolA as identified in other gram negative bacteria (Matsuyama et al., 1995). These secreted Brucella lipoproteins form a complex with LolA, which when isolated and incubated separately with the extracted Brucella OM component would presumably allow for the translocation and hence incorporation of the Brucella lipoproteins in the OM. This would suggest an Omp component is essential for lipoprotein incorporation in the OM. By solubilizing the Omp with detergents and addition of phospholipids a proteoliposome is generated which would determine the OM component required (since LolB is absent in Brucella) (Matsuyama et al., 1997; Yokota et al., 1999). Similarly the LolD-LolC/E component located in the IM of Brucella may be identified.
The Brucella lipoproteins characterized to date have a peripheral location with the anchorage of the lipid moiety to either the outer surface or the periplasmic leaflet of the OM as alluded to with Ab accessibility and similarity with E. coli PAL and Lpp. If they are anchored to the periplasmic leaflet, it is likely that the lipoprotein adopts an integral membrane protein conformation resulting in the surface exposure of its C-terminal end (like E. coli Lpp). However, it may also be exported to both the inner leaflet and outer leaflet of the OM (like E. coli Pal) (Michel et al., 2015). Studies conducted on the orientation of a Pal present in H. influenza P6, implies the existence of an energy driven reaction (in order to translocate across the cell envelope) catalyzed by a “flippase” enzyme to chaperone the “flipping” of the lipoprotein to the OM outer surface. The orientation of P6 could possibly occur via the Lol system during or after translocation into the OM (Michel et al., 2013). These suggestions seem most likely to exist for the translocation of Brucella lipoproteins since they are similar to these dual topology lipoproteins. In order to identify the OM component (an Omp, or a specific translocase) involved in “flipping” lipoproteins to the surface of the OM, Brucella knockout mutants (single mutations) may be created for the OM component. It would be essential to monitor the integrity of the Brucella cell envelope (additionally, IM marker detection would be useful). These results may also vary between using the R- and S-Brucella strains as previously suggested (Cloeckaert et al., 1990, 1996). Based on these knockout strains, components essential for Brucella lipoprotein surface exposure would result in no surface detection using the hydrophilic biotin analog and immunofluorescence techniques described previously (Michel et al., 2015).
Brucella Lipoproteins And The β-Barrel Assembly Complex
The involvement of the Bam (β-assembly machine, consisting of BamA; BamB; BamC; BamD; and BamE) complex (which translocates β-barrel proteins in the OM) could perhaps exist in lipoprotein translocation too (Ruiz et al., 2006; Bernstein, 2011). This was observed for the Neisseria gonorrhoeae NalP which is an autotransporter permitting the export of immunogenic proteins into the extracellular milieu by means of its protease domain and can itself be exported from the OM after further N-terminal processing. It is also branded as a lipoprotein with a characteristic C-terminal lipobox within the signal sequence (Van Ulsen et al., 2003). In fact, lipidation was found to be important for NalP’s autotransporter function (Van Ulsen et al., 2003; Roussel-Jazede et al., 2013). The export of NalP in an unfolded state is mediated by the T5SS pathway and includes the Bam complex (lodged in OM) and the integral membrane protein chaperones Skp, SurA, and DegP (located in the periplasm). These chaperones enable NalP’s translocator domain (β-barrel with a narrow hydrophilic pore containing an N-terminal α-helix) to insert into the OM, followed by translocation of its N-terminal passenger domain through the OM and finally its autolytic cleavage. It is suggested that the lipid anchor is incorporated on the OM surface since the N-terminal passenger domain is surface localized; however, the anchor topology is unknown (Oomen et al., 2004; Zückert, 2014). Gram negative bacteria autotransporters are integrated in the OM by a common machinery due to a consensus C-terminal sequence recognized by Omp85, which in turn is responsible for the translocation of other Omp (Robert et al., 2006). In general, three possible post-translocations can occur for the secreted proteins. The protein can remain covalently attached to the translocator domain, can be cleaved yet still remain connected to the OM or, as with NalP, it can be released into the external milieu (Van Ulsen et al., 2003; Zückert, 2014).
Would it be possible for the Bam complex, which mediates translocation of Brucella Omps to export Brucella lipoproteins resembling a variation of the T5SS model for Neisseria gonorrhoeae NalP? Moreover, certain components of the Bam complex are lipoproteins and in Neisseria meningitidis for example BamC is regarded a prospective vaccine candidate (surface antigen). The concept of lipoprotein translocation via the Bam complex cannot be considered far-fetched as the functions and interrelation of lipoproteins in the OM of gram negative bacteria are vast (Pajon et al., 2009). Analysis of bacterial genome sequences suggests the OM assembly machinery might be similar in gram negative bacteria (Gatsos et al., 2008). ?? consists of the Brucella equivalent components of the β-barrel assembly pathway that are present in other gram negative bacteria and have been demonstrated in Brucella based upon sequence similarity. All components and processes are similar in Brucella compared to other gram negative bacteria, preceding delivery of protein substrates to the Bam complex (Figure 3). SurA is a periplasmic chaperone mediating the translocation of proteins across the periplasm to the OM. In Brucella and certain other α-Proteobacteria, SurA is diminished lacking a periplasmic peptidyl-prolyl isomerase domain indicating these domains are not essential for binding protein substrates (Alcock et al., 2008). The main component of the Bam complex, BamA/Omp85 consists of a N-terminal end comprised of multiple polypeptide-transport-associated (POTRA) domains and a C-terminal β-barrel domain embedded in the OM. Structural analysis of the POTRA domains reveal a promiscuous binding capability to diverse peptide substrates. In E. coli BamA forms a complex with other lipoproteins namely BamB, BamC, BamD, and BamE. The second, third, and fourth POTRA domains are essential to facilitate interactions with BamB and the fifth POTRA domain with BamD and BamE (Kim et al., 2007; Misra, 2007). Interestingly enough, the Brucella BamA homolog possesses all five POTRA domains, although in Neisseria meningitides BamA is comprised only of the fifth POTRA domain and maintains its function like the other homologs (Robert et al., 2006; Bos et al., 2007). The lipoprotein components of BamA are assembled and anchored by the lipid moiety in the inner or outer surface of the OM as discussed previously. A BamC homolog could not be inferred in α-Proteobacteria, however, studies conducted using Caulobacter crescentus (an α-Proteobacterium) identified a novel BamF component with a conserved sequence motif related to those present in BamC. BamF is found exclusively in α-Proteobacteria, hence also in Brucella and is described as having evolved like BamC from the ancestral Bam complex (Anwari et al., 2012). Although BamB is present in some species of α-Proteobacteria, Brucella like Neisseria lacks a homolog for this component (Bos et al., 2007). It was established that certain protein substrates interact with BamB whereas with others, SurA carrying other protein substrates interacts directly with BamA compensating for the lack of BamB (Vuong et al., 2008). The complete activity and diversity of the Bam complex is largely unknown and could perhaps assist lipoprotein translocation too (Ruiz et al., 2006; Bernstein, 2011).
Brucella Lipoproteins And Other Possible Models
Various studies have established the existence of different models for lipoprotein translocation, which directly relates to the function of the specific lipoprotein. One such example is the pathogenic bacterial species of the spirochete class Borrelia, which possess many surface exposed lipoproteins functioning as major antigens, some being vital in the pathogenesis of Lyme disease (zoonotic disease) and relapsing fever. Secretion of the lipoproteins to the external surface is proposed to occur in an unfolded monomeric state (which can occur at the C-terminal end) and once at the surface it assembles into a quaternary structure (Kumru et al., 2011). This development paved the way for another hypothesis on the existence of a periplasmic “holding” chaperone, which interacted with the unfolded lipoproteins as they surface from the IM Sec system into the periplasm. This chaperone then delivers the lipoprotein to a “flippase” complex, which in turn exports it to the OM surface with N-terminal anchored into the OM (Schulze et al., 2010; Chen and Zuckert, 2011; Chen et al., 2011). LolA may function similarly to the “holding” chaperone; however, the involvement of the Lol pathway in this model remains to be resolved (Zückert, 2014). Other studies further implicate the role of the Bam complex in translocation of Borrelia burgdorferi lipoproteins (Lenhart and Akins, 2010). The spirochetal model could seem plausible since Brucella is absent of LolB, this would suggest the transfer of the mature lipoprotein from LolA to the holding chaperone and “flipping” to the OM surface. Furthermore, the T2SS model has been described for Caulobacter crescentus (classified as an α-Proteobacterium) under phosphate starvation conditions (Le Blastier et al., 2010). The T2SS in gram negative bacteria is a double-step mechanism, allowing for the secretion of folded and/or oligomeric proteins to the cell’s external milieu (Filloux et al., 1990). The proteins are translocated across the IM via the Sec or Tat systems thereafter the T2SS secreton, which is composed of three sub-complexes (GspDQ secretin, T2SS proteins and pseudopilus), recognizes and exports the proteins across the OM (Pugsley, 1993). Lipoproteins do not possess a predetermined, universal structure. They vary, from consisting of a large globular domain (E. coli BamC) to having a translocator domain (Neisseria autotransporter NalP) to a parallel three-stranded coiled-coil structure (E. coli Lpp). Barely any detail on the mechanism of lipoprotein translocation to the OM is available due to the variation in biological activity elicited by the lipoproteins and hence the complex machinery associated with lipoprotein transport in these bacteria. However, the mechanisms and intricacies illustrated in other gram negative bacteria have provided a stepping-stone in understanding the translocation of Brucella lipoproteins. Different models have been suggested for the translocation of lipoproteins to the OM of gram negative bacteria. These models interlink in mechanism. The Lol pathway could integrate the spirochetal model by use of a “flippase” complex in species devoid of LolB and the spirochetal model may engage LolA of the Lol pathway or a BamA homolog used by the T5SS model. Using these and the T2SS models, a relative understanding and a model for the translocation of Brucella lipoproteins will be established. To establish whether the Bam complex or any other model for lipoprotein translocation may potentially be used by Brucella instead of the Lol pathway, a knockout mutant Brucella strain of all components of the Lol pathway should be created. If lipoprotein translocation and surface exposure occurs without the Lol pathway this would implicate the role of another translocation pathway. The interlinking of various pathways may also be established by the formation of proteoliposomes and including required IM and periplasmic components (for, e.g., LolD–LolC/E and LolA) and OM components (e.g., the Bam complex) for Brucella lipoprotein translocation.
Brucella Lipoproteins Alone Are Toll-Like Receptor 2 Agonists As Vaccine Antigens
Pathogens, in particular bacteria, display molecular motifs known as pathogens-associated molecular patterns (PAMPs) that are recognized by the host’s innate immune system, thereby protecting the host from infection. The most common PAMPs are nucleic acids derived from pathogens and molecular components exposed on the surface, which include LPS, peptidoglycan, lipoproteins, and flagellin. Pathogens express a set of PAMPs that interact with multiple host pattern-recognition receptors (PRR). These receptors are grouped into secreted molecules and surface receptors for pathogen engulfment (on phagocytic cells) or the release of cytokines. After the PAMP-PRR interaction, signal transduction causes the activation of transcription factors that ultimately proceeds to the expression of inflammatory cytokines, type I interferon (IFN), chemokines and other compounds requires to elicit an immune response. Bacterial lipoproteins, i.e., Brucella Omp16 and Omp19, being lipoproteins are potent activators of the transmembrane proteins, TLRs (a group of PRRs) and in particular the cell surface TLR2, which complexes as a heterodimer with TLR1 or TLR6. Studies were performed using crystal structures of TLR2/1 and TLR2/6 (from model bacteria) heterodimerized with synthetic triacyl and diacyl lipopeptide (modified +1 cysteine residue of the lipoprotein). Crystallography revealed the triacylated lipopeptides/lipoproteins activate via the TLR2/1 heterodimer and diacylated lipopeptides/lipoproteins via the TLR2/6 heterodimer (Takeuchi et al., 2001, 2002; Takeda et al., 2002). Furthermore, it was speculated that the TLR2 bonds with the O-esterified fatty acids, the glyceryl group and the thioether moiety (S-glycerylcysteine residue of triacyl and diacyl lipopeptides) and that the TLR1 uses it hydrophobic cavity to distinguish the amide-linked fatty acid of the triacyl lipopeptide. TLR6, however, does not possess a distinguishing cavity. Other studies have elaborated upon this model suggesting lipidation alone is not solely responsible for TLR2 heterodimer selectivity, but also the amino acid residues after the +1 cysteine (lipidated) residue hence acyl chains position and protein/lipid compositions (Buwitt-Beckmann et al., 2005, 2006; Omueti et al., 2005; Kurokawa et al., 2009). However, TLR2 alone is not involved in controlling in vivo Brucella infection. Other PAMPs, such as B. abortus DNA was recognized as a TLR9 activator and LPS, and unlipidated Omp16, a TLR4 activator. Furthermore, lipoproteins instead of LPS were identified as the stimulators of the inflammatory response caused by B. abortus (Giambartolomei et al., 2004; Kang et al., 2009; Pasquevich et al., 2010; Pasquevich et al., 2011; Delpino et al., 2012; Gomes et al., 2012). Therefore, it is speculated that the use of multiple PAMPs to elicit various components of the immune system should be investigated further in an effort to design an effective vaccine that would target Brucella given that the pathogen already establishes a replicative niche within the host. At present the superiority of live-attenuated B. abortus vaccines (e.g., S19 and RB51) to stimulate an effective immune response (particularly a T-cell response) supersedes currently developed vaccines against brucellosis, yet at the expense of a constant serological response (Schurig et al., 2002). This review has focused upon experimentally characterized and studied Brucella lipoproteins, which have been regarded as protective, surface antigens. However, with the advancement of modern technology such as reverse vaccinology, which allows for the identification of protective antigens that have specific structural, immunogenic and functional qualities based on in silico genome analysis, many more OM constituents and their biosynthetic pathways will be documented in the future for the development of a safe and effective Brucella vaccine.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ABBREVIATIONS
- B. melitensis
Brucella melitensis
- B. ovis
Brucella ovis
- B. suis
Brucella suis
- CU
chaperone-usher
- E. coli
Escherichia coli
- H. influenza
Haemophilus influenza
- IM
inner membrane
- Imp
inner membrane protein
- Lgt
preprolipoprotein diacylglyceryl transferase
- Lsp
lipoprotein signal peptidase
- Lnt
lipoprotein N’N-acyl transferase
- Lol
lipoprotein localization machinery
- Lpp
Braun’s lipoprotein
- LPS
lipopolysaccharides
- MAb
monoclonal antibodies
- OM
outer membrane
- Omp
outer membrane protein
- PAMPs
pathogens-associated molecular patterns
- Pal
peptidoglycan-associated lipoproteins
- POTRA
polypeptide-transport-associated
- PRR
pattern-recognition receptors
- RGD
Arg- Gly- Asp
- Sec, system
general secretory
- Tat system
twin-arginine translocation
- T1SS
type I secretion system
- T2SS
type II secretion system
- T3SS
type III secretion system
- T4SS
type IV secretion system
- T5SS
type V secretion system
- T6SS
type VI secretion system
- T7SS
type VII secretion system
- TCS BvrRS
two-component regulatory system BvrR/BvrS
- TLRs
toll-like receptors
Supplementary Material
The Suplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01189
References
- Alcock F., Grossmann J., Gentle I., LikiC V., Lithgow T., Tokatlidis K. (2008). Conserved substrate binding by chaperones in the bacterial periplasm and the mitochondrial intermembrane space. Biochem. J. 409 377–387. 10.1042/BJ20070877 [DOI] [PubMed] [Google Scholar]
- Anwari K., Webb C. T., Poggio S., Perry A. J., Belousoff M., Celik N., et al. (2012). The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol. Microbiol. 84 832–844. 10.1111/j.1365-2958.2012.08059.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas G. N., Staskevich A. S., Aballay A., Mayorga L. S. (2000). Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 68 4255–4263. 10.1128/IAI.68.7.4255-4263.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M. M., Priya M. L., Selvan A. T., Madera M., Gough J., Aravind L., et al. (2006). A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188 2761–2773. 10.1128/JB.188.8.2761-2773.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang B. (1897). The etiology of epizootic abortion. J. Comp. Pathol. Ther. 10 125–IN2 10.1016/S0368-1742(97)80014-8 [DOI] [Google Scholar]
- Bayer M. (1991). Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J. Struct. Biol. 107 268–280. 10.1016/1047-8477(91)90052-X [DOI] [PubMed] [Google Scholar]
- Bayer M. E. (1968). Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 53 395–404. 10.1099/00221287-53-3-395 [DOI] [PubMed] [Google Scholar]
- Berks B. C., Palmer T., Sargent F. (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8 174–181. 10.1016/j.mib.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Bernstein H. D. (2011). The double life of a bacterial lipoprotein. Mol. Microbiol. 79 1128–1131. 10.1111/j.1365-2958.2011.07538.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi E. (1998). The role of the ribosome-translocon complex in translation and assembly of polytopic membrane proteins. Trends Biochem. Sci. 23 51–55. 10.1016/S0968-0004(97)01134-1 [DOI] [PubMed] [Google Scholar]
- Bladen H. A., Mergenhagen S. E. (1964). Ultrastructure of Veillonella and morphological correlation of an outer membrane with particles associated with endotoxic activity. J. Bacteriol. 88 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan J. A., Jr., Apicella M. A. (1995). Mapping of a surface-exposed, conformational epitope of the P6 protein of Haemophilus influenzae. Infect. Immun. 63 4395–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos M. P., Robert V., Tommassen J. (2007). Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61 191–214. 10.1146/annurev.micro.61.080706.093245 [DOI] [PubMed] [Google Scholar]
- Bowden R. A., Cloeckaert A., Zygmunt M. S., Bernard S., Dubray G. (1995). Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect. Immun. 63 3945–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U. (1999). Helix-bundle membrane protein fold templates. Protein Sci. 8 2711–2719. 10.1110/ps.8.12.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V., Rehn K. (1969). Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. Eur. J. Biochem. 10 426–438. 10.1111/j.1432-1033.1969.tb00707.x [DOI] [PubMed] [Google Scholar]
- Braun V., Rehn K., Wolff H. (1970). Supramolecular structure of the rigid layer of the cell wall of Salmonella, Serratia, Proteus, and Pseudomonas fluorescens. Number of lipoprotein molecules in a membrane layer. Biochemistry (N. Y.) 9 5041–5049. 10.1021/bi00828a001 [DOI] [PubMed] [Google Scholar]
- Brink S., Bogsch E. G., Edwards W. R., Hynds P. J., Robinson C. (1998). Targeting of thylakoid proteins by the ΔpH-driven twin-arginine translocation pathway requires a specific signal in the hydrophobic domain in conjunction with the twin-arginine motif. FEBS Lett. 434 425–430. 10.1016/S0014-5793(98)01028-X [DOI] [PubMed] [Google Scholar]
- Bruce S. D. (1887). Note on the discovery of a microorganism in Malta fever. Practitioner 39 161. [Google Scholar]
- Buwitt-Beckmann U., Heine H., Wiesmuller K. H., Jung G., Brock R., Akira S., et al. (2006). TLR1- and TLR6-independent recognition of bacterial lipopeptides. J. Biol. Chem. 281 9049–9057. 10.1074/jbc.M512525200 [DOI] [PubMed] [Google Scholar]
- Buwitt-Beckmann U., Heine H., Wiesmüller K., Jung G., Brock R., Akira S., et al. (2005). Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur. J. Immunol. 35 282–289. 10.1002/eji.200424955 [DOI] [PubMed] [Google Scholar]
- Campbell G., Adams L., Sowa B. (1994). Mechanisms of binding of Brucella abortus to mononuclear phagocytes from cows naturally resistant or susceptible to brucellosis. Vet. Immunol. Immunopathol. 41 295–306. 10.1016/0165-2427(94)90103-1 [DOI] [PubMed] [Google Scholar]
- Caro-Hernandez P., Fernandez-Lago L., de Miguel M. J., Martin-Martin A. I., Cloeckaert A., Grillo M. J., et al. (2007). Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect. Immun. 75 4050–4061. 10.1128/IAI.00486-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Lloubès R. (2004). Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 51 873–885. 10.1046/j.1365-2958.2003.03881.x [DOI] [PubMed] [Google Scholar]
- Chain P. S., Comerci D. J., Tolmasky M. E., Larimer F. W., Malfatti S. A., Vergez L. M., et al. (2005). Whole-genome analyses of speciation events in pathogenic Brucellae. Infect. Immun. 73 8353–8361. 10.1128/IAI.73.12.8353-8361.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Henning U. (1987). Nucleotide sequence of the gene for the peptidoglycan-associated lipoprotein of Escherichia coli K12. Eur. J. Biochem. 163 73–77. 10.1111/j.1432-1033.1987.tb10738.x [DOI] [PubMed] [Google Scholar]
- Chen S., Kumru O. S., Zuckert W. R. (2011). Determination of Borrelia surface lipoprotein anchor topology by surface proteolysis. J. Bacteriol. 193 6379–6383. 10.1128/JB.05849-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zuckert W. R. (2011). Probing the Borrelia burgdorferi surface lipoprotein secretion pathway using a conditionally folding protein domain. J. Bacteriol. 193 6724–6732. 10.1128/JB.06042-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T., Germon P., Vianney A., Portalier R., Lazzaroni J. C. (1998). TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29 359–367. 10.1046/j.1365-2958.1998.00945.x [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., de Wergifosse P., Dubray G., Limet J. N. (1990). Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect. Immun. 58 3980–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloeckaert A., Jacques I., Bosseray N., Limet J. N., Bowden R., Dubray G., et al. (1991). Protection conferred on mice by monoclonal antibodies directed against outer-membrane-protein antigens of Brucella. J. Med. Microbiol. 34 175–180. 10.1099/00222615-34-3-175 [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Verger J., Grayon M., Vizcaíno N. (1996). Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol. Lett. 145 1–8. 10.1111/j.1574-6968.1996.tb08547.x [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Vizcaíno N., Paquet J., Bowden R. A., Elzer P. H. (2002). Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90 229–247. 10.1016/S0378-1135(02)00211-0 [DOI] [PubMed] [Google Scholar]
- Comerci D. J., Martinez-Lorenzo M. J., Sieira R., Gorvel J. P., Ugalde R. A. (2001). Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3 159–168. 10.1046/j.1462-5822.2001.00102.x [DOI] [PubMed] [Google Scholar]
- Corbeil L. B., Blau K., Inzana T. J., Nielsen K. H., Jacobson R. H., Corbeil R. R., et al. (1988). Killing of Brucella abortus by bovine serum. Infect. Immun. 56 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. E., Li Y., Semmelhack M. F., Cristea I. M., Silhavy T. J. (2011). The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79 1168–1181. 10.1111/j.1365-2958.2011.07539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decad G. M., Nikaido H. (1976). Outer membrane of gram-negative bacteria. XII. Molecular-sieving function of cell wall. J. Bacteriol. 128 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M., Sun Y., den Hartigh A., van Dijl J., Tsolis R. (2008). Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 70 1378–1396. 10.1111/j.1365-2958.2008.06487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpino M. V., Barrionuevo P., Macedo G. C., Oliveira S. C., Genaro S. D., Scian R., et al. (2012). Macrophage-elicited osteoclastogenesis in response to Brucella abortus infection requires TLR2/MyD88-dependent TNF-alpha production. J. Leukoc. Biol. 91 285–298. 10.1189/jlb.04111185 [DOI] [PubMed] [Google Scholar]
- Delrue R., Martinez-Lorenzo M., Lestrate P., Danese I., Bielarz V., Fonck P., et al. (2001). Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3 487–497. 10.1046/j.1462-5822.2001.00131.x [DOI] [PubMed] [Google Scholar]
- Delvecchio V. G., Alefantis T., Ugalde R. A., Comerci D., Marchesini M. I., Khan A., et al. (2006). Identification of protein candidates for developing bacterial ghost vaccines against Brucella. Methods Biochem. Anal. 49 363–377. [PubMed] [Google Scholar]
- DelVecchio V. G., Kapatral V., Redkar R. J., Patra G., Mujer C., Los T., et al. (2002). The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. U.S.A. 99 443–448. 10.1073/pnas.221575398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria T. F., Murwin D. M., Leake E. R. (1996). Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 64 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas J. T., Rosenberg E. Y., Nikaido H., Verstreate D. R., Winter A. J. (1984). Porins of Brucella species. Infect. Immun. 44 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drage M. G., Tsai H. C., Pecora N. D., Cheng T. Y., Arida A. R., Shukla S., et al. (2010). Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nat. Struct. Mol. Biol. 17 1088–1095. 10.1038/nsmb.1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen A. J., Nouwen N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77 643–667. 10.1146/annurev.biochem.77.061606.160747 [DOI] [PubMed] [Google Scholar]
- Dubray G. (1976). Localisation cellulaire des polyosides des bacteries des genres Brucella et Escherichia en phase lisse (S) ou rugueuse (R). Ann. Microbiol. 127 133–149. [PubMed] [Google Scholar]
- Edmonds M. D., Cloeckaert A., Booth N. J., Fulton W. T., Hagius S. D., Walker J. V., et al. (2001). Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am. J. Vet. Res. 62 1461–1466. 10.2460/ajvr.2001.62.1461 [DOI] [PubMed] [Google Scholar]
- Edmonds M. D., Cloeckaert A., Elzer P. H. (2002). Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 88 205–221. 10.1016/S0378-1135(02)00110-4 [DOI] [PubMed] [Google Scholar]
- Filloux A., Bally M., Ball G., Akrim M., Tommassen J., Lazdunski A. (1990). Protein secretion in gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 9 4323–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer E., Moreno E., Moriyon I., Pizarro-Cerda J., Weintraub A., Gorvel J. P. (1996). Brucella-Salmonella lipopolysaccharide chimeras are less permeable to hydrophobic probes and more sensitive to cationic peptides and EDTA than are their native Brucella sp. counterparts. J. Bacteriol. 178 5867–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Xu J., Li X., Xie Y., Qiu Y., Kwaik Y. A. (2012). Immunization of mice with recombinant protein CobB or AsnC confers protection against Brucella abortus infection. PLoS ONE 7:e29552 10.1371/journal.pone.0029552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazo C., Moriyon I. (1987). Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect. Immun. 55 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsos X., Perry A. J., Anwari K., Dolezal P., Wolynec P. P., Likic V. A., et al. (2008). Protein secretion and outer membrane assembly in Alpha Proteobacteria. FEMS Microbiol. Rev. 32 995–1009. 10.1111/j.1574-6976.2008.00130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennity J. M., Inouye M. (1991). The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 266 16458–16464. [PubMed] [Google Scholar]
- Giambartolomei G. H., Zwerdling A., Cassataro J., Bruno L., Fossati C. A., Philipp M. T. (2004). Lipoproteins, not lipopolysaccharide, are the key mediators of the proinflammatory response elicited by heat-killed Brucella abortus. J. Immunol. 173 4635–4642. 10.4049/jimmunol.173.7.4635 [DOI] [PubMed] [Google Scholar]
- Godlewska R., Wisniewska K., Pietras Z., Jagusztyn-Krynicka E. K. (2009). Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298 1–11. 10.1111/j.1574-6968.2009.01659.x [DOI] [PubMed] [Google Scholar]
- Gomes M. T. R., Campos P. C., de Almeida L. A., Oliveira F. S., Costa M., Marim F. M., et al. (2012). The role of innate immune signals in immunity to Brucella abortus. Front. Cell. Infect. Microbiol. 2:130 10.3389/fcimb.2012.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Miguel M. J., Moriyón I., López J. (1987). Brucella outer membrane lipoprotein shares antigenic determinants with Escherichia coli Braun lipoprotein and is exposed on the cell surface. Infect. Immun. 55 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. D., Gan K., Schmid M. B., Wu H. C. (1993). Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J. Biol. Chem. 268 16551–16556. [PubMed] [Google Scholar]
- Guzman-Verri C., Manterola L., Sola-Landa A., Parra A., Cloeckaert A., Garin J., et al. (2002). The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U.S.A. 99 12375–12380. 10.1073/pnas.192439399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hal Jones C., Danese P. N., Pinkner J. S., Silhavy T. J., Hultgren S. J. (1997). The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16 6394–6406. 10.1093/emboj/16.21.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Peterson-Burch B. D., Bricker B. J., Zuerner R. L., Qing Z., Li L. L., et al. (2005). Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 187 2715–2726. 10.1128/JB.187.8.2715-2726.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K., Braun V. (1973). Covalent binding of lipid to protein. Eur. J. Biochem. 34 284–296. 10.1111/j.1432-1033.1973.tb02757.x [DOI] [PubMed] [Google Scholar]
- Hara T., Matsuyama S., Tokuda H. (2003). Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J. Biol. Chem. 278 40408–40414. 10.1074/jbc.M307836200 [DOI] [PubMed] [Google Scholar]
- Hellman J., Loiselle P. M., Tehan M. M., Allaire J. E., Boyle L. A., Kurnick J. T., et al. (2000). Outer membrane protein A, peptidoglycan-associated lipoprotein, and murein lipoprotein are released by Escherichia coli bacteria into serum. Infect. Immun. 68 2566–2572. 10.1128/IAI.68.5.2566-2572.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman J., Zanzot E. M., Loiselle P. M., Amato S. F., Black K. M., Ge Y., et al. (1997). Antiserum against Escherichia coli J5 contains antibodies reactive with outer membrane proteins of heterologous gram-negative bacteria. J. Infect. Dis. 176 1260–1268. 10.1086/514121 [DOI] [PubMed] [Google Scholar]
- Hillmann F., Argentini M., Buddelmeijer N. (2011). Kinetics and phospholipid specificity of apolipoprotein N-acyltransferase. J. Biol. Chem. 286 27936–27946. 10.1074/jbc.M111.243519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P. C., Tsolis R. M., Ficht T. A. (2000). Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68 4102–4107. 10.1128/IAI.68.7.4102-4107.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren S. J., Normark S., Abraham S. N. (1991). Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45 383–415. 10.1146/annurev.mi.45.100191.002123 [DOI] [PubMed] [Google Scholar]
- Hutchings M. I., Palmer T., Harrington D. J., Sutcliffe I. C. (2009). Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol. 17 13–21. 10.1016/j.tim.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Inouye M., Shaw J., Shen C. (1972). The assembly of a structural lipoprotein in the envelope of Escherichia coli. J. Biol. Chem. 247 8154–8159. [PubMed] [Google Scholar]
- Inouye S., Wang S., Sekizawa J., Halegoua S., Inouye M. (1977). Amino acid sequence for the peptide extension on the prolipoprotein of the Escherichia coli outer membrane. Proc. Natl. Acad. Sci. U.S.A. 74 1004–1008. 10.1073/pnas.74.3.1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Matsuzawa H., Matsuyama S., Narita S., Tokuda H. (2006). Genetic analysis of the mode of interplay between an ATPase subunit and membrane subunits of the lipoprotein-releasing ATP-binding cassette transporter LolCDE. J. Bacteriol. 188 2856–2864. 10.1128/JB.188.8.2856-2864.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S., Rock C. O. (1986). Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J. Biol. Chem. 261 11328–11333. [PubMed] [Google Scholar]
- Kang J. Y., Nan X., Jin M. S., Youn S., Ryu Y. H., Mah S., et al. (2009). Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31 873–884. 10.1016/j.immuni.2009.09.018 [DOI] [PubMed] [Google Scholar]
- Khandavilli S., Homer K. A., Yuste J., Basavanna S., Mitchell T., Brown J. S. (2008). Maturation of Streptococcus pneumoniae lipoproteins by a type II signal peptidase is required for ABC transporter function and full virulence. Mol. Microbiol. 67 541–557. 10.1111/j.1365-2958.2007.06065.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Lim J. J., Lee J. J., Kim D. G., Lee H. J., Min W., et al. (2012). Identification of genes contributing to the intracellular replication of Brucella abortus within HeLa and RAW 264.7 cells. Vet. Microbiol. 158 322–328. 10.1016/j.vetmic.2012.02.019 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Son B. G., Lim J. J., Lee J. J., Kim D. G., Lee H. J., et al. (2013). The role of a Brucella abortus lipoprotein in intracellular replication and pathogenicity in experimentally infected mice. Microb. Pathog. 54 34–39. 10.1016/j.micpath.2012.09.002 [DOI] [PubMed] [Google Scholar]
- Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., Kahne D. (2007). Structure and function of an essential component of the outer membrane protein assembly machine. Science 317 961–964. 10.1126/science.1143993 [DOI] [PubMed] [Google Scholar]
- Koebnik R., Locher K. P., Van Gelder P. (2000). Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37 239–253. 10.1046/j.1365-2958.2000.01983.x [DOI] [PubMed] [Google Scholar]
- Kovacs-Simon A., Titball R., Michell S. L. (2011). Lipoproteins of bacterial pathogens. Infect. Immun. 79 548–561. 10.1128/IAI.00682-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru O. S., Schulze R. J., Rodnin M. V., Ladokhin A. S., Zuckert W. R. (2011). Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J. Bacteriol. 193 2814–2825. 10.1128/JB.00015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Lee H., Roh K. B., Asanuma M., Kim Y. S., Nakayama H., et al. (2009). The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for toll-like receptor 2. J. Biol. Chem. 284 8406–8411. 10.1074/jbc.M809618200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne J., Butler H., Chaves-Olarte E., Hunter J., Schirm M., Paquet C., et al. (2007). Extensive cell envelope modulation is associated with virulence in Brucella abortus. J. Proteome Res. 6 1519–1529. 10.1021/pr060636a [DOI] [PubMed] [Google Scholar]
- Le Blastier S., Hamels A., Cabeen M., Schille L., Tilquin F., Herman C. (2010). Phosphate starvation triggers production and secretion of an extracellular lipoprotein in Caulobacter crescentus. PLoS ONE 5:e14198 10.1371/journal.pone.0014198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart T. R., Akins D. R. (2010). Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol. Microbiol. 75 692–709. 10.1111/j.1365-2958.2009.07015.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M. D., Bagchi A., Warren H. S., Tehan M. M., Trigilio J. A., Beasley-Topliffe L. K., et al. (2005). Bacterial peptidoglycan-associated lipoprotein: a naturally occurring toll-like receptor 2 agonist that is shed into serum and has synergy with lipopolysaccharide. J. Infect. Dis. 191 939–948. 10.1086/427815 [DOI] [PubMed] [Google Scholar]
- Lopes L., Nicolino R., Haddad J. (2010). Brucellosis– risk factors and prevalence: a review. Open Vet. Sci. J. 4 72–84. 10.2174/1874318801004010072 [DOI] [Google Scholar]
- López-Goni I., Manterola L., Pan S., Moriyón I. (2004). “The Brucella BvrS/BvrR and related two-component regulatory systems of the α-2 proteobacteria: common regulatory strategies of animal and plant pathogens and endosymbionts,” in Brucella: Molecular and Cellular Biology, eds López-Goñi I., Moriyón I. (Norwich: Horizon Bioscience; ), 213–230. [Google Scholar]
- MacIntyre S., Freudl R., Eschbach M. L., Henning U. (1988). An artificial hydrophobic sequence functions as either an anchor or a signal sequence at only one of two positions within the Escherichia coli outer membrane protein OmpA. J. Biol. Chem. 263 19053–19059. [PubMed] [Google Scholar]
- Magnet S., Bellais S., Dubost L., Fourgeaud M., Mainardi J. L., Petit-Frere S., et al. (2007). Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J. Bacteriol. 189 3927–3931. 10.1128/JB.00084-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola L., Guzman-Verri C., Chaves-Olarte E., Barquero-Calvo E., de Miguel M. J., Moriyon I., et al. (2007). BvrR/BvrS-controlled outer membrane proteins Omp3a and Omp3b are not essential for Brucella abortus virulence. Infect. Immun. 75 4867–4874. 10.1128/IAI.00439-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola L., Moriyon I., Moreno E., Sola-Landa A., Weiss D. S., Koch M. H., et al. (2005). The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol. 187 5631–5639. 10.1128/JB.187.16.5631-5639.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis H., Ficht T. A. (1993). The omp2 gene locus of Brucella abortus encodes two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect. Immun. 61 3785–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N., Hancock R. (1990). “Function and structure of the major components of the outer membrane of gram-negative bacteria,” in Advances in Brucellosis Research, ed. Adams L. G. (College Station, TX: Texas A&M University Press; ), 55–75. [Google Scholar]
- Martinez de Tejada G., Moriyon I. (1993). The outer membranes of Brucella spp. are not barriers to hydrophobic permeants. J. Bacteriol. 175 5273–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Tejada G., Pizarro-Cerda J., Moreno E., Moriyon I. (1995). The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun. 63 3054–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Nunez C., Altamirano-Silva P., Alvarado-Guillen F., Moreno E., Guzman-Verri C., Chaves-Olarte E. (2010). The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J. Bacteriol. 192 5603–5608. 10.1128/JB.00567-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Martín A. I., Caro-Hernández P., Orduña A., Vizcaíno N., Fernández-Lago L. (2008). Importance of the Omp25/Omp31 family in the internalization and intracellular replication of virulent B. ovis in murine macrophages and HeLa cells. Microb. Infect. 10 706–710. 10.1016/j.micinf.2008.02.013 [DOI] [PubMed] [Google Scholar]
- Martín-Martín A. I., Caro-Hernández P., Sancho P., Tejedor C., Cloeckaert A., Fernández-Lago L., et al. (2009). Analysis of the occurrence and distribution of the Omp25/Omp31 family of surface proteins in the six classical Brucella species. Vet. Microbiol. 137 74–82. 10.1016/j.vetmic.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Masuda K., Matsuyama S., Tokuda H. (2002). Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. U.S.A. 99 7390–7395. 10.1073/pnas.112085599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiopoulos C., Mueller J., Slack F., Murphy C., Patankar S., Bukusoglu G., et al. (1991). A Bacillus subtilis dipeptide transport system expressed early during sporulation. Mol. Microbiol. 5 1903–1913. 10.1111/j.1365-2958.1991.tb00814.x [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Tajima T., Tokuda H. (1995). A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14 3365–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Yokota N., Tokuda H. (1997). A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16 6947–6955. 10.1093/emboj/16.23.6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L. V., Shaw J., MacPherson V., Barnard D., Bettinger J., D’Arcy B., et al. (2015). Dual orientation of the outer membrane lipoprotein Pal in Escherichia coli. Microbiology 161 1251–1259. 10.1099/mic.0.000084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L. V., Snyder J., Schmidt R., Milillo J., Grimaldi K., Kalmeta B., et al. (2013). Dual orientation of the outer membrane lipoprotein P6 of nontypeable Haemophilus influenzae. J. Bacteriol. 195 3252–3259. 10.1128/JB.00185-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R. (2007). First glimpse of the crystal structure of YaeT’s POTRA domains. ACS Chem. Biol. 2 649–651. 10.1021/cb700212p [DOI] [PubMed] [Google Scholar]
- Mitchell P. (1961). Approaches to the analysis of specific membrane transport. Biol. Struct. Funct. 2 581–599. [Google Scholar]
- Mizutani M., Mukaiyama K., Xiao J., Mori M., Satou R., Narita S., et al. (2013). Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex. FEBS Lett. 587 23–29. 10.1016/j.febslet.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Mobasheri H., Ficht T., Marquis H., Lea E., Lakey J. (1997). Brucella Omp2a and Omp2b porins: single channel measurements and topology prediction. FEMS Microbiol. Lett. 155 23–30. 10.1111/j.1574-6968.1997.tb12681.x [DOI] [PubMed] [Google Scholar]
- Moreno E., Gorvel J., López-Goñi I., Moriyón I. (2004). “Invasion, intracellular trafficking and replication of Brucella organisms in professional and non-professional phagocytes,” in Brucella: Molecular And Cellular Biology, eds López-Goñi I., Moriyón I. (Norfolk: Horizon Scientific Press; ), 287–312. [Google Scholar]
- Moreno E., Moriyon I. (2002). Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. U.S.A. 99 1–3. 10.1073/pnas.022622699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyon I., Berman D. T. (1983). Isolation, purification, and partial characterization of Brucella abortus matrix protein. Infect. Immun. 39 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyon I., Lopez-Goni I. (1998). Structure and properties of the outer membranes of Brucella abortus and Brucella melitensis. Int. Microbiol. 1 19–26. [PubMed] [Google Scholar]
- Müller M. (2005). Twin-arginine-specific protein export in Escherichia coli. Res. Microbiol. 156 131–136. 10.1016/j.resmic.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Munoa F. J., Miller K. W., Beers R., Graham M., Wu H. C. (1991). Membrane topology of Escherichia coli prolipoprotein signal peptidase (signal peptidase II). J. Biol. Chem. 266 17667–17672. [PubMed] [Google Scholar]
- Murphy T. F., Kirkham C., Lesse A. J. (2006). Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect. Immun. 74 5169–5176. 10.1128/IAI.00692-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T. F., Nelson M. B., Dudas K. C., Mylotte J. M., Apicella M. A. (1985). Identification of a specific epitope of Haemophilus influenzae on a 16,600-dalton outer membrane protein. J. Infect. Dis. 152 1300–1307. 10.1093/infdis/152.6.1300 [DOI] [PubMed] [Google Scholar]
- Nakae T., Nikaido H. (1975). Outer membrane as a diffusion barrier in Salmonella typhimurium. Penetration of oligo– and polysaccharides into isolated outer membrane vesicles and cells with degraded peptidoglycan layer. J. Biol. Chem. 250 7359–7365. [PubMed] [Google Scholar]
- Nakamura K., Mizushima S. (1976). Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J. Biochem. 80 1411–1422. [DOI] [PubMed] [Google Scholar]
- Narita S. (2011). ABC transporters involved in the biogenesis of the outer membrane in gram-negative bacteria. Biosci. Biotechnol. Biochem. 75 1044–1054. 10.1271/bbb.110115 [DOI] [PubMed] [Google Scholar]
- Narita S., Matsuyama S., Tokuda H. (2004). Lipoprotein trafficking in Escherichia coli. Arch. Microbiol. 182 1–6. 10.1007/s00203-004-0682-4 [DOI] [PubMed] [Google Scholar]
- Narita S., Tanaka K., Matsuyama S., Tokuda H. (2002). Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J. Bacteriol. 184 1417–1422. 10.1128/JB.184.5.1417-1422.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naroeni A., Jouy N., Ouahrani-Bettache S., Liautard J. P., Porte F. (2001). Brucella suis-impaired specific recognition of phagosomes by lysosomes due to phagosomal membrane modifications. Infect. Immun. 69 486–493. 10.1128/IAI.69.1.486-493.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. B., Murphy T. F., van Keulen H., Rekosh D., Apicella M. A. (1988). Studies on P6, an important outer-membrane protein antigen of Haemophilus influenzae. Rev. Infect. Dis. 10 S331–S336. 10.1093/cid/10.Supplement_2.S331 [DOI] [PubMed] [Google Scholar]
- Okuda S., Tokuda H. (2009). Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl. Acad. Sci. U.S.A. 106 5877–5882. 10.1073/pnas.0900896106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., Watanabe S., Tokuda H. (2008). A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins. FEBS Lett. 582 2247–2251. 10.1016/j.febslet.2008.05.022 [DOI] [PubMed] [Google Scholar]
- Omueti K. O., Beyer J. M., Johnson C. M., Lyle E. A., Tapping R. I. (2005). Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J. Biol. Chem. 280 36616–36625. 10.1074/jbc.M504320200 [DOI] [PubMed] [Google Scholar]
- Oomen C. J., van Ulsen P., van Gelder P., Feijen M., Tommassen J., Gros P. (2004). Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23 1257–1266. 10.1038/sj.emboj.7600148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajon R., Yero D., Niebla O., Climent Y., Sardiñas G., García D., et al. (2009). Identification of new meningococcal serogroup B surface antigens through a systematic analysis of Neisserial genomes. Vaccine 28 532–541. 10.1016/j.vaccine.2009.09.128 [DOI] [PubMed] [Google Scholar]
- Papanikou E., Karamanou S., Economou A. (2007). Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5 839–851. 10.1038/nrmicro1771 [DOI] [PubMed] [Google Scholar]
- Paquet J. Y., Diaz M. A., Genevrois S., Grayon M., Verger J. M., de Bolle X., et al. (2001). Molecular, antigenic, and functional analyses of Omp2b porin size variants of Brucella spp. J. Bacteriol. 183 4839–4847. 10.1128/JB.183.16.4839-4847.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L. M., Lin F., Orban J. (2006). Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry (N. Y.) 45 2122–2128. 10.1021/bi052227i [DOI] [PubMed] [Google Scholar]
- Pasquevich K. A., Ibañez A. E., Coria L. M., Samartino C. G., Estein S. M., Zwerdling A., et al. (2011). An oral vaccine based on U-Omp19 induces protection against Brucella abortus mucosal challenge by inducing an adaptive IL-17 immune response in mice. PLoS ONE 6:e16203 10.1371/journal.pone.0016203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquevich K. A., Samartino C. G., Coria L. M., Estein S. M., Zwerdling A., Ibañez A. E., et al. (2010). The protein moiety of Brucella abortus outer membrane protein 16 is a new bacterial pathogen-associated molecular pattern that activates dendritic cells in vivo, induces a Th1 immune response, and is a promising self-adjuvanting vaccine against systemic and oral acquired brucellosis. J. Immunol. 184 5200–5212. 10.4049/jimmunol.0902209 [DOI] [PubMed] [Google Scholar]
- Paulsen I. T., Seshadri R., Nelson K. E., Eisen J. A., Heidelberg J. F., Read T. D., et al. (2002). The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. U.S.A. 99 13148–13153. 10.1073/pnas.192319099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P. (1993). The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P., Chapon C., Schwartz M. (1986). Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J. Bacteriol. 166 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston D. J., Elberg S. S. (1969). Serum-mediated immune cellular responses to Brucella melitensis REV 1. II. Restriction of Brucella by immune sera and macrophages. J. Reticuloendothel. Soc. 6 109–139. [PubMed] [Google Scholar]
- Rapp M., Granseth E., Seppälä S., Von Heijne G. (2006). Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 13 112–116. 10.1038/nsmb1057 [DOI] [PubMed] [Google Scholar]
- Remans K., Pauwels K., van Ulsen P., Buts L., Cornelis P., Tommassen J., et al. (2010). Hydrophobic surface patches on LolA of Pseudomonas aeruginosa are essential for lipoprotein binding. J. Mol. Biol. 401 921–930. 10.1016/j.jmb.2010.06.067 [DOI] [PubMed] [Google Scholar]
- Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., Tommassen J. (2006). Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:e377 10.1371/journal.pbio.0040377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest H. P., Bloemendaal C. J., Wijffelman C. A., Lugtenberg B. J. (1995). Isolation and characterization of ropA homologous genes from Rhizobium leguminosarum biovars viciae and trifolii. J. Bacteriol. 177 4985–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbusch J. P. (1974). Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J. Biol. Chem. 249 8019–8029. [PubMed] [Google Scholar]
- Rossetti C. A., Galindo C. L., Lawhon S. D., Garner H. R., Adams L. G. (2009). Brucella melitensis global gene expression study provides novel information on growth phase-specific gene regulation with potential insights for understanding Brucella: host initial interactions. BMC Microbiol. 9:81 10.1186/1471-2180-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel-Jazede V., Grijpstra J., van Dam V., Tommassen J., van Ulsen P. (2013). Lipidation of the autotransporter NalP of Neisseria meningitidis is required for its function in the release of cell-surface-exposed proteins. Microbiology 159 286–295. 10.1099/mic.0.063982-0 [DOI] [PubMed] [Google Scholar]
- Roussel-Jazede V., Jongerius I., Bos M. P., Tommassen J., van Ulsen P. (2010). NalP-mediated proteolytic release of lactoferrin-binding protein B from the meningococcal cell surface. Infect. Immun. 78 3083–3089. 10.1128/IAI.01193-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi C. (1991). Antígenos Proteicos de Brucella abortus: Aislamiento y Estudio de su Asociación al LPS y de Factores que Afectan a la Producción de Anticuerpos Monoclonales Contra los Mismos. Ph.D. thesis, Facultad de Ciencias Exactas, Universidad Nacional de la Plata, La Plata, 1–137. [Google Scholar]
- Ruiz N., Kahne D., Silhavy T. J. (2006). Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4 57–66. 10.1038/nrmicro1322 [DOI] [PubMed] [Google Scholar]
- Salhi I., Boigegrain R. A., Machold J., Weise C., Cloeckaert A., Rouot B. (2003). Characterization of new members of the group 3 outer membrane protein family of Brucella spp. Infect. Immun. 71 4326–4332. 10.1128/IAI.71.8.4326-4332.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran K., Wu H. C. (1994). Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269 19701–19706. [PubMed] [Google Scholar]
- Schulze R. J., Chen S., Kumru O. S., Zuckert W. R. (2010). Translocation of Borrelia burgdorferi surface lipoprotein OspA through the outer membrane requires an unfolded conformation and can initiate at the C-terminus. Mol. Microbiol. 76 1266–1278. 10.1111/j.1365-2958.2010.07172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R. J., Zückert W. R. (2006). Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 59 1473–1484. 10.1111/j.1365-2958.2006.05039.x [DOI] [PubMed] [Google Scholar]
- Schurig G. G., Sriranganathan N., Corbel M. J. (2002). Brucellosis vaccines: past, present and future. Vet. Microbiol. 90 479–496. 10.1016/S0378-1135(02)00255-9 [DOI] [PubMed] [Google Scholar]
- Selvan A. T., Sankaran K. (2008). Localization and characterization of prolipoprotein diacylglyceryl transferase (Lgt) critical in bacterial lipoprotein biosynthesis. Biochimie 90 1647–1655. 10.1016/j.biochi.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Setubal J. C., Reis M., Matsunaga J., Haake D. A. (2006). Lipoprotein computational prediction in Spirochaetal genomes. Microbiology 152 113–121. 10.1099/mic.0.28317-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel A., Gounon P., Pugsley A. P. (1999). Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34 810–821. 10.1046/j.1365-2958.1999.01647.x [DOI] [PubMed] [Google Scholar]
- Shaw J., Schmidt R., MacPherson V., Pichichero M., Michel L. (2014). Quantifying the two populations of dual oriented P6 in nontypable Haemophilus influenzae and Pal in Escherichia coli (948.6). FASEB J. 28 948.6. [Google Scholar]
- Shaw J., Schmidt R., Snyder J., Pichichero M., Michel L. V. (2013). Using biotinylation to determine the orientations of Pal in Escherichia coli. FASEB J. 27 790.8. [Google Scholar]
- Shu W., Liu J., Ji H., Lu M. (2000). Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 å resolution. J. Mol. Biol. 299 1101–1112. 10.1006/jmbi.2000.3776 [DOI] [PubMed] [Google Scholar]
- Sjostrom M., Wold S., Wieslander A., Rilfors L. (1987). Signal peptide amino acid sequences in Escherichia coli contain information related to final protein localization. A multivariate data analysis. EMBO J. 6 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola-Landa A., Pizarro-Cerda J., Grillo M., Moreno E., Moriyón I., Blasco J., et al. (1998). A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29 125–138. 10.1046/j.1365-2958.1998.00913.x [DOI] [PubMed] [Google Scholar]
- Stabel J. R., Stabel T. J. (1995). Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 45 211–220. 10.1016/0165-2427(94)05348-V [DOI] [PubMed] [Google Scholar]
- Sutcliffe I. C., Harrington D. J., Hutchings M. I. (2012). A phylum level analysis reveals lipoprotein biosynthesis to be a fundamental property of bacteria. Protein Cell 3 163–170. 10.1007/s13238-012-2023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe I. C., Russell R. R. (1995). Lipoproteins of gram-positive bacteria. J. Bacteriol. 177 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima T., Yokota N., Matsuyama S., Tokuda H. (1998). Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 439 51–54. 10.1016/S0014-5793(98)01334-9 [DOI] [PubMed] [Google Scholar]
- Takeda K., Miyatake H., Yokota N., Matsuyama S., Tokuda H., Miki K. (2003). Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22 3199–3209. 10.1093/emboj/cdg324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Takeuchi O., Akira S. (2002). Recognition of lipopeptides by Toll-like receptors. J. Endotoxin Res. 8 459–463. 10.1177/09680519020080060101 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Kawai T., Muhlradt P. F., Morr M., Radolf J. D., Zychlinsky A., et al. (2001). Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13 933–940. 10.1093/intimm/13.7.933 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., et al. (2002). Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169 10–14. 10.4049/jimmunol.169.1.10 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Matsuyama S. I., Tokuda H. (2001). Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J. Bacteriol. 183 6538–6542. 10.1128/JB.183.22.6538-6542.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada M., Kuroda T., Matsuyama S. I., Tokuda H. (2001). Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276 47690–47694. 10.1074/jbc.M109307200 [DOI] [PubMed] [Google Scholar]
- Thirkell D., Myles A. D., Russell W. C. (1991). Palmitoylated proteins in urea plasma urealyticum. Infect. Immun. 59 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Decelle B., Letesson J. (1999). Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect. Immun. 67 4960–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Saman E., de Wergifosse P., Cloeckaert A., Limet J. N., Letesson J. (1996). Molecular characterization, occurrence, and immunogenicity in infected sheep and cattle of two minor outer membrane proteins of Brucella abortus. Infect. Immun. 64 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Wansard V., Bielartz V., Delrue R. M., Danese I., Michel P., et al. (2002). Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 70 5540–5546. 10.1128/IAI.70.10.5540-5546.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Weynants V., Denoel P., Lichtfouse B., De Bolle X., Saman E., et al. (1994). Molecular cloning, nucleotide sequence, and occurrence of a 16.5-kilodalton outer membrane protein of Brucella abortus with similarity to pal lipoproteins. Infect. Immun. 62 3633–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda H., Matsuyama S. (2004). Sorting of lipoproteins to the outer membrane in Escherichia coli. Biochim. Biophys. Acta 1694 IN1–IN9. [PubMed] [Google Scholar]
- Tokunaga M., Tokunaga H., Wu H. C. (1982). Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl. Acad. Sci. U.S.A. 79 2255–2259. 10.1073/pnas.79.7.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traum J. (1914). Annual Report to the Chief of the Bureau of Animal Industry, Year Ending June 30 1914. Quezon City: USDA Bureau of Animal Industry, 30. [Google Scholar]
- Tschumi A., Nai C., Auchli Y., Hunziker P., Gehrig P., Keller P., et al. (2009). Identification of apolipoprotein N-acyltransferase (Lnt) in Mycobacteria. J. Biol. Chem. 284 27146–27156. 10.1074/jbc.M109.022715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara J., Mukaiyama K., Okuda S., Narita S., Tokuda H. (2009). Dissection of LolB function–lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 276 4496–4504. 10.1111/j.1742-4658.2009.07156.x [DOI] [PubMed] [Google Scholar]
- Van Ulsen P., Van Alphen L., Ten Hove J., Fransen F., Van Der Ley P., Tommassen J. (2003). A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50 1017–1030. 10.1046/j.1365-2958.2003.03773.x [DOI] [PubMed] [Google Scholar]
- Vemulapalli R., Cravero S., Calvert C. L., Toth T. E., Sriranganathan N., Boyle S. M., et al. (2000). Characterization of specific immune responses of mice inoculated with recombinant vaccinia virus expressing an 18-kilodalton outer membrane protein of Brucella abortus. Clin. Diagn. Lab. Immunol. 7 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreate D. R., Creasy M. T., Caveney N. T., Baldwin C. L., Blab M. W., Winter A. J. (1982). Outer membrane proteins of Brucella abortus: isolation and characterization. Infect. Immun. 35 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viadas C., Rodriguez M., Garcia-Lobo J., Sangari F., Lopez-Goni I. (2009). Construction and evaluation of an ORFeome-based Brucella whole-genome DNA microarray. Microb. Pathog. 47 189–195. 10.1016/j.micpath.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Vidal-Ingigliardi D., Lewenza S., Buddelmeijer N. (2007). Identification of essential residues in apolipoprotein N-acyl transferase, a member of the CN hydrolase family. J. Bacteriol. 189 4456–4464. 10.1128/JB.00099-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno N., Caro-Hernández P., Cloeckaert A., Fernández-Lago L. (2004). DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microb. Infect. 6 821–834. 10.1016/j.micinf.2004.04.009 [DOI] [PubMed] [Google Scholar]
- Vizcaíno N., Cloeckaert A., Verger J., Grayon M., Fernández-Lago L. (2000). DNA polymorphism in the genus Brucella. Microb. Infect. 2 1089–1100. 10.1016/S1286-4579(00)01263-6 [DOI] [PubMed] [Google Scholar]
- Vizcaino N., Cloeckaert A., Zygmunt M. S., Dubray G. (1996). Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect. Immun. 64 3744–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino N., Cloeckaert A., Zygmunt M. S., Fernandez-Lago L. (2001a). Characterization of a Brucella species 25-kilobase DNA fragment deleted from Brucella abortus reveals a large gene cluster related to the synthesis of a polysaccharide. Infect. Immun. 69 6738–6748. 10.1128/IAI.69.11.6738-6748.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino N., Kittelberger R., Cloeckaert A., Marin C. M., Fernandez-Lago L. (2001b). Minor nucleotide substitutions in the omp31 gene of Brucella ovis result in antigenic differences in the major outer membrane protein that it encodes compared to those of the other Brucella species. Infect. Immun. 69 7020–7028. 10.1128/IAI.69.11.7020-7028.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong P., Bennion D., Mantei J., Frost D., Misra R. (2008). Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 190 1507–1517. 10.1128/JB.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A. (1987). Outer membrane proteins of Brucella. Ann. Inst. Pasteur Microbiol. 138 87–89. 10.1016/0769-2609(87)90081-0 [DOI] [PubMed] [Google Scholar]
- Wu H., Tokunaga M. (1986). “Biogenesis of lipoproteins in bacteria,” in Protein Secretion and Export in Bacteria Anonymous, eds Wu H. C., Tai P. C. (Berlin: Springer; ), 127–157. [DOI] [PubMed] [Google Scholar]
- Xiang Z., Zheng W., He Y. (2006). BBP: Brucella genome annotation with literature mining and curation. BMC Bioinform. 7:347 10.1186/1471-2105-7-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushi T., Masuda K., Narita S., Matsuyama S., Tokuda H. (2000). A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2 212–218. 10.1038/35008635 [DOI] [PubMed] [Google Scholar]
- Yakushi T., Yokota N., Matsuyama S., Tokuda H. (1998). LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J. Biol. Chem. 273 32576–32581. 10.1074/jbc.273.49.32576 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yu F., Inouye M. (1988). A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53 423–432. 10.1016/0092-8674(88)90162-6 [DOI] [PubMed] [Google Scholar]
- Yokota N., Kuroda T., Matsuyama S., Tokuda H. (1999). Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274 30995–30999. 10.1074/jbc.274.43.30995 [DOI] [PubMed] [Google Scholar]
- Zhou L., Srisatjaluk R., Justus D. E., Doyle R. J. (1998). On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol. Lett. 163 223–228. 10.1111/j.1574-6968.1998.tb13049.x [DOI] [PubMed] [Google Scholar]
- Zückert W. R. (2013). A call to order at the Spirochaetal host-pathogen interface. Mol. Microbiol. 89 207–211. 10.1111/mmi.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zckert W. R. (2014). Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim. Biophys. Acta 1843 1509–1516. 10.1016/j.bbamcr.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerdling A., Delpino M., Barrionuevo P., Cassataro J., Pasquevich K. A., García Samartino C., et al. (2008). Brucella lipoproteins mimic dendritic cell maturation induced by Brucella abortus. Microb. Infect. 10 1346–1354. 10.1016/j.micinf.2008.07.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.