Abstract
Regulating nucleo-cytoplasmic transport of RNA and protein is a key cellular control point. Perturbing the function of plant nuclear transport components can cause significant developmental defects and in this report we add an important line to this evidence. Overexpression of AtRAN1 or AtNUP62 in Nicotiana benthamiana causes significant damage to leaf tissue. This demonstrates that the precise control of nuclear transport is an important aspect of maintaining tissue integrity.
Keywords: RAN1, nicotiana benthamiana, nucleus, nuclear transport, nucleoporin, NUP62
Nucleo-cytoplasmic transport of RNA and proteins is a critical point of cellular regulation. Cargos are transported between compartments by the activity of karyopherin proteins that bind cargoes with the appropriate signal sequence.1 This process has many layers of regulation, firstly at the level of cargo recognition. This can occur via canonical or non-canonical NLS or NES signals that have such wide variety that identification of definitive transport motifs is a difficult task. Secondly, the karyopherin:cargo interacts with the GTPase RAN1 in either its GTP or GDP bound form. The gradient of RAN1 across the nuclear envelope (NE) is critical for driving nuclear transport so without the activity of nucleus or cytoplasm-specific enzymes that alter the guanine-nucleotide-binding of RAN1, the process of nuclear transport breaks down.2 Finally the karyopherin:cargo:RAN1 grouping passes through the nuclear pore complex (NPC), a process thought to be mediated by the interaction with hydrophobic FG repeat motifs contained within the secondary structure of certain nucleoporins.3 This entire process is well characterized in yeast and metazoan cell lines and in recent times a growing portfolio of evidence has described the action of members of the plant nuclear transport apparatus. This includes the proteomic identification of the plant orthologs of metazoan and yeast NUPs as well as genetic characterization of a range of nup mutants in Arabidopsis, tobacco and in legumes.3,4
Study of nuclear transport in plants has relied upon the identification of plants with a genetic defect in your protein of interest, be it a nucleoporin, a karyopherin or an enzyme involved in the RAN1.2,4 Many nuclear transport mutants show significant defects during gametogenesis or embryogenesis, resulting in a failure to produce viable offspring. However, given the fundamental importance of nuclear transport, a surprising number of mutants have been isolated that display mild or no developmental phenotypes. The mild phenotypes include reduced stature throughout the lifecycle and early flowering, which usually coincides with alterations in bulk mRNA export.4 Although the majority of this work has been performed in Arabidopsis, a number of studies have used the Nicotiana benthamiana expression system to investigate the activity of certain NUPs. VIGS-mediated down-regulation of NbNUP75, NbNUP88 and NbSeh1 causes an increase in susceptibility to the fungal pathogen Phytophora infestans5 while targeted downregulation of NbRAE1 causes a broader variety of growth phenotypes.6
Over-expression of proteins in N.benthamiana acts as an excellent experimental system to study the effect of pathogens on plant development. Therefore the initial aim of our study was to assess the effect of altering nuclear transport (NT) components on the infectivity of plant viruses. We aimed to over-express certain NT components in the presence of the Geminivirus Tomato Yellow Leave Curl Virus (TYLCV) 7 and evaluate the resulting molecular and phenotypic changes. Although we successfully used agro-infiltration to correctly deliver the virus and cause appropriate disease symptoms, our initial experimental aims were thwarted by the phenotypic changes we report below. In summary we found that overexpression of certain NT components caused a significant increase in growth defects when expressed individually within tobacco leaves.
In order to overexpress Arabidopsis proteins in N.benthamiana we utilised the CPMV-HT expression system.8 The gene of interest is cloned into the pEAQ plasmid that contains the P19 suppressor of silencing and the UTR sequence from Cowpea Mosaic Virus (CPMV). The CPMV sequence contains cis-acting elements that promote transcription of an associated gene without producing any infectious viral particles. Importantly we used the pEAQ-DEST3 vector that allowed attachment of a 6xHis-tag to either AtRAN1 or AtNUP62, which facilitated downstream analysis of gene expression.
Following inoculation we used a 6-point scale (0–5) to quantify the severity of symptoms in tobacco leaves that had been agro-infiltrated with either pEAQ-DEST3 (hereafter pEAQ), pEAQ-AtRAN1 or pEAQ-AtNUP62. We assessed symptom severity (Fig. 1) following each of these treatments for >20 days post infection (dpi) and observed an expected increase in severity over the time course (Fig. 3). It was not surprising to observe an increase in symptoms in leaves inoculated with pEAQ since in proof-of-concept experiments with the CPMV-HT system, expression of the marker protein DsRed caused significant tissue damage even at 8dpi.9
Figure 1.

Phenotype of Nicotiana benthemiana leaves after inoculation. Tobacco leaves were inoculated with Agrobacterium containing the appropriate construct and the severity of tissue decay was assessed over 20+ days. Severity of symptoms was rated on a scale between 0–5, where a rating of 0 showed no effect and a rating of 5 showed a completely necrotic tissue.
Figure 3.
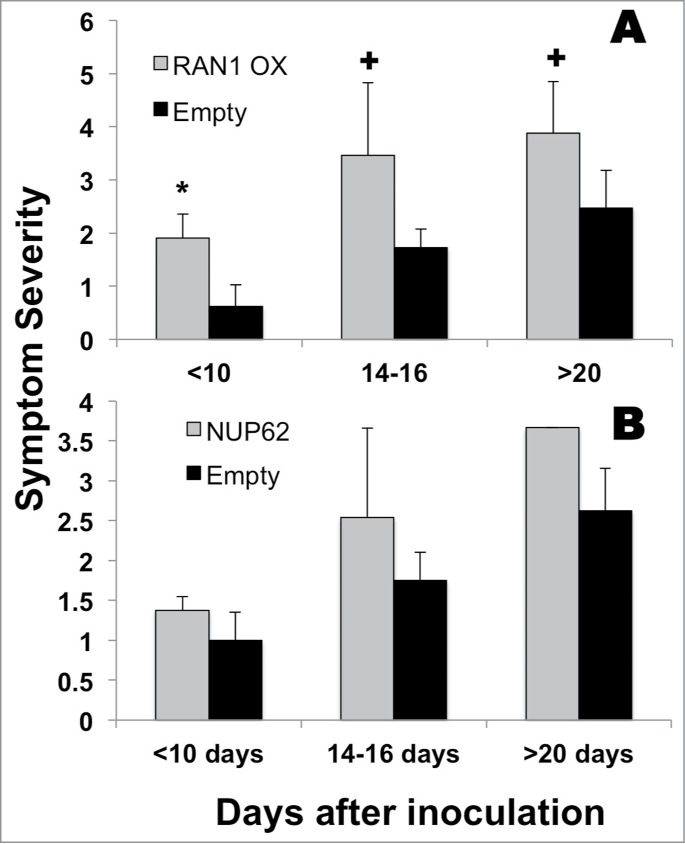
Overexpression of AtRAN1/AtNUP62 causes increased symptom severity. Symptom severity was assessed in plants inoculated with AtRAN1-His (A) or AtNUP62-His (B) and compared with those inoculated with the empty vector, pEAQ-DEST3. Data shown is representative of at least 2 repeats where in each experiment >3 leaves were assessed at each time point. Assignment of symptom severity scores is outlined in Figure. 1. Student Ttest values (P < 0.03=*, P < 0.15= +) comparing empty and AtRAN1 samples. Error bars represent StDev.
In order to verify that AtRAN1 and AtNUP62 were being successfully expressed we extracted RNA and protein from tobacco leaves at 2dpi (Fig. 2). Semi-quantitative RT-PCR using gene-specific primers and western blotting using an anti-His tag antibody were used to show that the genes were being expressed compared to pEAQ or no-plasmid controls (Fig. 2). Whereas the AtRAN1-His protein was expressed to a high level we were unable to visualize AtNUP62-His by protein gel blot even though the RT-PCR showed that the gene was expressed (Fig. 2). The reason for this difference is unclear. However we were unsurprised to be unable to detect the AtNUP62 protein, even in this expression system. While other nucleoporins have been expressed behind the CaMV 35S promotor in Arabidopsis, the expression levels are generally low with no resulting phenotypic change outside of complementing an appropriate mutant phenotype.10,11
Figure 2.
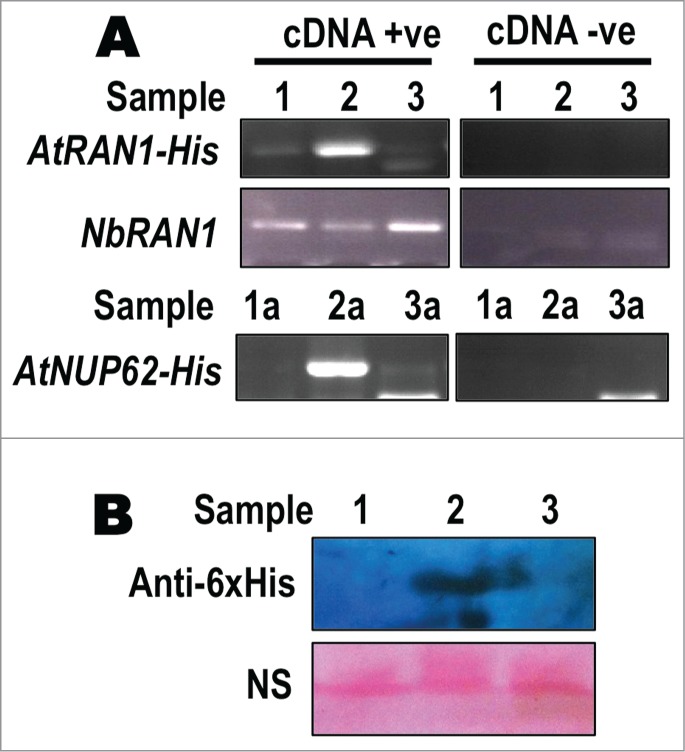
RAN1 and NUP62 expression in Nicotiana benthamiana leaves. RNA and protein was extracted from N.benthamiana leaves 2 days after inoculation with only agrobacteria or with agrobacteria containing an empty pEAQ-DEST3 plasmid, pEAQ-RAN1 plasmid or pEAQ-AtNUP62 plasmid (A) RT-PCR showing expression of AtRAN1-His, AtNUP62-His or endogenous NbRAN1. NbRAN1 was used as an internal control showing no genomic DNA contamination. (B) Western blot showing expression of 6xHis-tagged AtRAN1. NS- Non-specific band visualised by Ponceau Red staining. Sample numbers: 1- No plasmid 2- pEAQ-AtRAN1 3- pEAQ-DEST3. 1a- No plasmid 2a- pEAQ-AtNUP62 3a- pEAQ-DEST3. cDNA +ve/-ve represents presence of RT enzyme in cDNA preparation mix.
When assessing the symptoms generated following inoculation with pEAQ-AtRAN1 or pEAQ-AtNUP62 we showed an increase in severity at the earliest time point that continued to the end of the experimental time period (Fig. 3). This effect was more pronounced in leaves inoculated with AtRAN1. Inoculation with AtNUP62 caused a robust response but the small number of samples prevented the data obtaining good significance values. Although the control leaves also show an increase in symptom severity, the level remained <3 in both experiments. The difference between the ranking of 3 and 4 is notable as it represented a change from leaf discolouration to apparent necrosis of the leaf tissue. Overexpression of either AtRAN1 or AtNUP62 caused a severe phenotype demonstrating that continued growth of the leaf is compromised when these proteins are over-expressed.
It is not without precedent that altering the levels of either RAN1 or NUP62 has phenotypic consequences. Overexpression of AtNUP62 in Arabidopsis caused co-suppression of the endogenous NUP62 gene 12 while Arabidopsis mutants with a truncated NUP62 have reduced stature and early flowering.13,14 Furthermore it is impossible to identify homozygous null nup62 mutants since these are embryo lethal.14,15 Therefore it is unsurprising that overexpression of NUP62 can cause a severe phenotype in tobacco leaves. The precise mechanism by which this phenotype is generated is unknown since the functional significance of plant NUP62 still remains opaque. Surprisingly it was recently found that viable Arabidopsis nup62 mutants do not exhibit an alteration in mRNA export, which is usually a common molecular phenotype observed in other nup mutants.14 Therefore we have significant work still to do to elucidate why significantly altering AtNUP62 levels can cause severe phenotypes even though we verify here that it is an important regulator of cell growth.
In Arabidopsis there are 3 paralogs of the RAN protein, RAN1-3. However there are no available T-DNA null mutants of any of these proteins but it is expected that there is redundancy in their activity and that they have an essential function. Ran1 knockdown mutants have a mild embryo phenotype but in an interesting set of reciprocal crosses the authors show that altering the ratio of maternally and paternally derived RAN1 in a developing embryo has severe repercussions resulting in a high level of embryo abortion.16 Use of RNAi to knockdown expression of RAN1 in rice cause significant growth changes including reduced viability, changes in the response to ABA/salt stress and cell cycle defects.17,18 Opposingly, when RAN from rice or wheat is overexpressed either in its native plant or in Arabidopsis it causes a range of phenotypic changes. Overexpression of OsRAN1, TaRAN1 or OsRAN2 in rice causes resistance to cold stress,18,19 while overexpression of TaRAN1 or OsRAN1 in Arabidopsis alters apical dominance and plant stature.19,20 Therefore given this breadth of information it is not surprising that when AtRAN1 is overexpressed in tobacco there is such a severe phenotype. Clearly and as expected, RAN1 plays a significant role in maintaining normal cell growth so altering expressing levels as in AtRAN1 inoculated leaves prevents normal growth of that tissue.
Overall we have shown that overexpression of components of the nuclear transport apparatus in tobacco causes severe phenotypic change resulting in tissue necrosis. This highlights the importance of nuclear transport and the challenge is now to identify which molecules are differentially expressed and/or localized in these tissues. Gaining a better understanding of this process will provide future guidance as to how nuclear transport might be altered to influence plant growth responses.
Materials and Methods
Construction of clones
We amplified AtRAN1 and AtNUP62 from Arabidopsis cDNA prepared from 7do seedlings using the Bioline Tetro kit and the following primers: AtRAN1F, caccATGGCTCTACCTAACCAGCAAACCG; AtRAN1R, CTCAAAGATATCATCATCG; AtNUP62F, caccATGTCGGGGTTTCCATTTGGTC; AtNUP62R, AGACATCCAGTGCTTTGGAGC. These PCR products were cloned into pEAQ-DEST3 using standard Gateway cloning techniques (LifeTech).8 Agrobacterium strain GV3101 was then transformed with pEAQ-DEST clones using the freeze-thaw method.
Tobacco leaf inoculation
Transformed agrobacterium were grown overnight in LB media containing Kanamycin and Rifamycin (both 50μg/ml) and then resuspended in Induction buffer (10 mM Mes, pH 5.6, 10 mM MgCl2 and 150 μM acetosyringone) at OD600 ∼0.1 for 2hr at RT. Leaves from ∼1 mth old tobacco plants were inoculated and symptoms were assessed over the following 20+ days.
Expression analysis
Two days after agro-infiltration, a representative leaf was removed and flash-frozen. Total RNA was extracted using a Sigma Spectrum kit and cDNA prepared as above. Gene expression was analyzed using Bioline BioRed Taq polymerase with the following primers: AtRAN1F; AtNUP62F; 6xHisTag, CTAGTGATGGTGATGGTGATGCCC; NbRAN1F, ATGGCAGCACCACCGGCAAGAGC; NbRAN1R, CCATCAAGCTCAATGGTCCG. We performed a crude protein preparation in Extraction Buffer (100mM Tris-HCl pH 7.5, 100mM NaCl, 10% Glycerol, 2mM DTT, 1mM PMSF, 1/300 Plant Protease Inhibitor Cocktail (Sigma) 0.1% Triton). We then separated samples using PAGE, blotted onto PVDF membrane, probed with 1/2000 anti-6xHis (Abcam) and 1/1000 anti-rabbit antibodies (LifeTech) and visualised with ECL reagents (LifeTech) all with standard techniques.
Acknowledgments
We would also like to thank George Lomonossoff for the gift of the pEAQ plasmids.
Funding
We would like to thank the British Society of Plant Pathology for summer studentship awards to CK and AC as well as a Marie Curie Career Integration Grant (FP7-PEOPLE-2011-CIG) for support purchasing consumable items.
References
- 1. Tamura K, Hara-Nishimura I. Functional insights of nucleocytoplasmic transport in plants. Front Plant Sci 2014; 5:118; PMID:24765097; http://dx.doi.org/ 10.3389/fpls.2014.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meier I, Somers DE. Regulation of nucleocytoplasmic trafficking in plants. Curr Opin Plant Biol 2011; 14:538-46; PMID:21764628; http://dx.doi.org/ 10.1016/j.pbi.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 3. Tamura K, Hara-Nishimura I. The molecular architecture of the plant nuclear pore complex. J Exp Bot 2013; 64:823-32; PMID:22987840; http://dx.doi.org/ 10.1093/jxb/ers258 [DOI] [PubMed] [Google Scholar]
- 4. Parry G. Assessing the function of the plant nuclear pore complex and the search for specificity. J Exp Bot 2013; 64:833-45; PMID:23077202; http://dx.doi.org/ 10.1093/jxb/ers289 [DOI] [PubMed] [Google Scholar]
- 5. Ohtsu M, Shibata Y, Ojika M, Tamura K, Hara-Nishimura I, Mori H, Kawakita K, Takemoto D. Nucleoporin 75 is involved in the ethylene-mediated production of phytoalexin for the resistance of Nicotiana benthamiana to Phytophthora infestans. Mol Plant Microbe Interact 2014; PMID:25122483 [DOI] [PubMed] [Google Scholar]
- 6. Lee JY, Lee HS, Wi SJ, Park KY, Schmit AC, Pai HS. Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J 2009; 59:278-91; PMID:19392703; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03869.x [DOI] [PubMed] [Google Scholar]
- 7. Reyes MI, Nash TE, Dallas MM, Ascencio-Ibanez JT, Hanley-Bowdoin L. Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to tomato yellow leaf curl virus and tomato mottle virus infection in tomato. J Virol 2013; 87:9691-706; PMID:23824791; http://dx.doi.org/ 10.1128/JVI.01095-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peyret H, Lomonossoff GP. The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol Biol 2013; 83:51-8; PMID:23479085; http://dx.doi.org/ 10.1007/s11103-013-0036-1 [DOI] [PubMed] [Google Scholar]
- 9. Sainsbury F, Lomonossoff GP. Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol 2008; 148:1212-8; PMID:18775971; http://dx.doi.org/ 10.1104/pp.108.126284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 2006; 18:1590-603; PMID:16751346; http://dx.doi.org/ 10.1105/tpc.106.041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and Characterization of Nuclear Pore Complex Components in Arabidopsis thaliana. Plant Cell 2010; 22:4084-97; PMID:21189294; http://dx.doi.org/ 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao Q, Meier I. Identification and characterization of the Arabidopsis FG-repeat nucleoporin Nup62. Plant Signal Behav 2011; 6:330-4; PMID:21673506; http://dx.doi.org/ 10.4161/psb.6.3.13402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrandez-Ayela A, Alonso-Peral MM, Sanchez-Garcia AB, Micol-Ponce R, Perez-Perez JM, Micol JL, Ponce MR. Arabidopsis TRANSCURVATA1 encodes NUP58, a component of the nucleopore central channel. PLoS One 2013; 8:e67661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parry G. Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J Exp Bot 2014; 65:6057-67; PMID:25165147; http://dx.doi.org/ 10.1093/jxb/eru346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meinke D, Muralla R, Sweeney C, Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 2008; 13:483-91; PMID:18684657; http://dx.doi.org/ 10.1016/j.tplants.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 16. Liu P, Qi M, Wang Y, Chang M, Liu C, Sun M, Yang W, Ren H. Arabidopsis RAN1 mediates seed development through its parental ratio by affecting the onset of endosperm cellularization. Molecular plant 2014; 7:1316-28; PMID:24719465; http://dx.doi.org/ 10.1093/mp/ssu041 [DOI] [PubMed] [Google Scholar]
- 17. Zang A, Xu X, Neill S, Cai W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J Exp Bot 2010; 61:777-89; PMID:20018899; http://dx.doi.org/ 10.1093/jxb/erp341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen N, Xu Y, Wang X, Du C, Du J, Yuan M, Xu Z, Chong K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ 2011; 34:52-64; PMID:20825577; http://dx.doi.org/ 10.1111/j.1365-3040.2010.02225.x [DOI] [PubMed] [Google Scholar]
- 19. Xu P, Cai W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J Exp Bot 2014; 65:3277-87; PMID:24790113; http://dx.doi.org/ 10.1093/jxb/eru178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, Xu Z, Chong K. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol 2006; 140:91-101; PMID:16361516; http://dx.doi.org/ 10.1104/pp.105.071670 [DOI] [PMC free article] [PubMed] [Google Scholar]


