Abstract
Sunflower seedlings subjected to 120 mM NaCl stress exhibit high total peroxidase activity, differential expression of its isoforms and accumulation of lipid hydroperoxides. This coincides with high specific activity of phospholipid hydroperoxide glutathione peroxidase (PHGPX) in the 10,000g supernatant from the homogenates of 2–6 d old seedling cotyledons. An upregulation of PHGPX activity by NaCl is evident from Western blot analysis. Confocal laser scanning microscopic (CLSM) analysis of sections of cotyledons incubated with anti-GPX4 (PHGPX) antibody highlights an enhanced cytosolic accumulation of PHGPX, particularly around the secretory canals. Present work, thus, highlights sensing of NaCl stress in sunflower seedlings in relation with lipid hydroperoxide accumulation and its scavenging through an upregulation of PHGPX activity in the cotyledons.
Keywords: glutathione peroxidase, lipid hydroperoxides, phospholipid hydroperoxide glutathione peroxidase, sunflower, seedling cotyledons, salt stress
Abbreviations
- CLSM
confocal laser scanning microscopy
- GPX
glutathione peroxidase
- LHPO
lipid hydroperoxide
- POD
peroxidase
- PHGPX
phospholipid hydroperoxide glutathione peroxidase
- ROS
reactive oxygen species
Reactive oxygen species (ROS) are produced as byproducts of metabolic pathways in different cellular compartments.1 At moderate concentrations, ROS act as signaling mediators for various cellular responses or they may prove to be toxic at high concentrations in a cell.2 A sudden increase in ROS concentration, referred as “oxidative burst," leads to oxidative stress in the cells.3 Detoxification of ROS is essential for the protection of plant cells against oxidative damage.4 An important enzymatic system responsible for protecting cells against oxidative damage is glutathione peroxidase (GPX; EC 1.11.1.9)5 which catalyzes the reduction of hydrogen peroxide, organic hydroperoxides or lipid hydroperoxides to water and corresponding alcohol using reduced glutathione (GSH) as a reductant.
At least 5 GPX isozymes have been identified among mammals.6,7 Out of these, phospholipid hydroperoxide glutathione peroxidase (PHGPX or GPX4) is the principal enzymatic defense against oxidative destruction of biomembranes. Due to relatively high activities of ascorbate peroxidase and catalase in plants, it was earlier thought that PHGPX is either absent in plants or its presence is probably not physiologically important. Role of plant PHGPX has so far remained largely unexplored. Investigations on their possible induction under a wide range of abiotic and biotic stresses is likely to highlight their specific role in ROS scavenging, such as the removal of lipid hydroperoxides.
Lipid peroxidation of cell membranes is a natural metabolic process under normal (aerobic) conditions and is also an effect of oxidative stress caused by ROS in the cells. Lipid hydroperoxides are toxic compounds formed as a result of attack of hydroxyl radicals and singlet oxygen on the methylene groups of polyunsaturated fatty acids in the membranes, and they cause a decrease in membrane fluidity, increase the leakiness of membranes and damage membrane proteins, thereby inactivating receptors, enzymes, and ion channels. PHGPX is a major candidate for scavenging cellular lipid hydroperoxides. Under stress conditions, H2O2 concentration increases drastically, leading to an increase in the activity of antioxidative enzymes (APX, CAT) involved in its scavenging.5 Activity of these enzymes, however, becomes insufficient to eliminate all the H2O2 and so, the excess of H2O2 undergoes Fenton reaction with Fe2+, leading to the formation of lipid hydroperoxides. Thus, PHGPX becomes active to reduce these lipid hydroperoxides and protect the cell from oxidative damage.
Oilseeds, such as sunflower, exhibit an abundance of intracellular biomembranes, principally oil body membranes, whose systematic proteolytic degradation is prerequisite for the mobilization of triacylglycerides in their matrix through lipase action.8,9 Sensing of salt stress by seedlings is likely to affect oil body mobilization through lipid peroxidation of the oil body membranes. Present investigations focus on the modulation of PHGPX activity in sunflower seedling cotyledons in response to salt stress. After analyzing cytosolic peroxidase activity in relation with NaCl stress at the 3 developmental stages of seedlings, main focus of this work has been to monitor qualitative, quantitative and spatial analysis of PHGPX in the cotyledons. Analysis of cotyledons from 2 day-old etiolated seedlings highlights the signaling of NaCl stress from roots to cotyledons and accumulation of lipid hydroperoxides formed in cells as a result of salinity stress.
POD activity is maximum in 4-day-old seedling cotyledons. A peak of POD activity is evident in cotyledons of 4-day-old seedling, both in the absence or presence of 120 mM NaCl and it is marginally lowered in seedlings subjected to NaCl stress (Fig. 1A). A 95 kDa POD isoform is maximally intense in 4-day-old seedling cotyledons (both in control and in presence of NaCl) and in 6-day-old seedling cotyledons (control) (Fig. 1B). Western blot analysis using anti-peroxidase antibody, however, shows 3 isoforms of POD at 36, 40 and 52 kDa, which exhibit quantitative differences in their expression with respect to development. A gradual decline in the activity of all 3 isoforms is evident with the passage of seedling development. Their expression is affected by NaCl stress at all the stages of development and gets differentially reduced at later stages of seedling development in response to NaCl stress. No new isoforms are expressed in response to NaCl stress. Thus, all the POD isoforms contribute to the observed differences.
Figure 1.
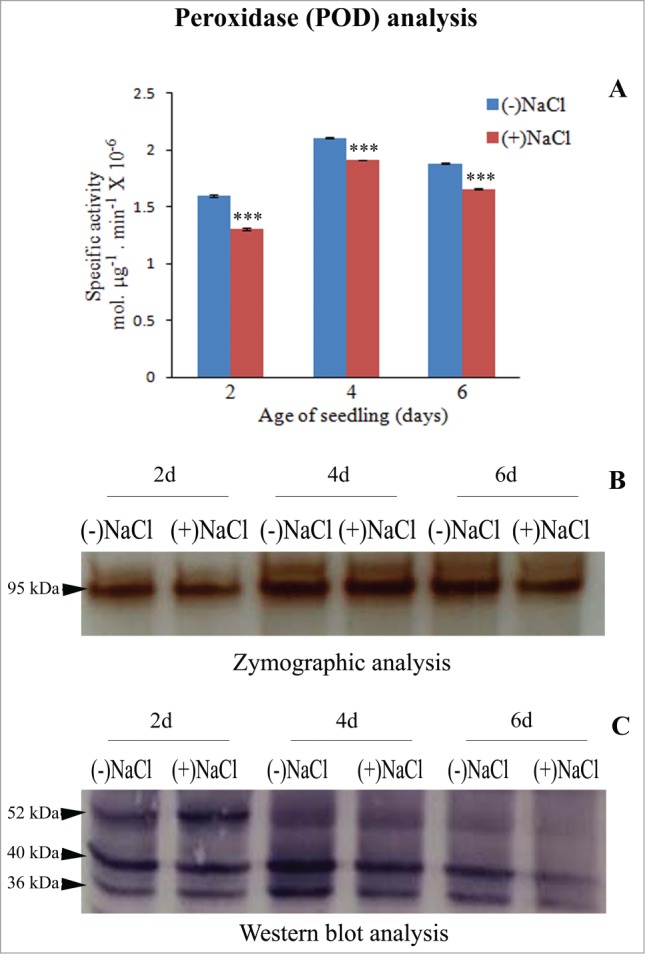
Analysis of peroxidase (POD) activity in total soluble protein (10,000g supernatant) from dark-grown sunflower seedling cotyledons grown in the absence or presence of 120 mM NaCl. (A) Spectrophotometric determination of specific activity of POD. NaCl-stress induced changes in enzyme activity were analyzed by one-way ANOVA using SPSS 22.0 and were found to be statistically significant (***P < 0.001) in comparison to control [(-)NaCl]. Bars represent SE (n = 3). (B) Zymographic detection of POD isoforms. (C) Western blot analysis of POD using anti-peroxidase antibody.
The lipid hydroperoxide (LHPO) content increased by 40% with the passage of seedling development (Fig. 2A). The differences were, however, not so significant in the absence or presence of NaCl stress. PHGPX activity was highest in 2-day-old sunflower seedling cotyledons subjected to 120 mM NaCl stress (Fig. 2B and C). The enzyme activity exhibited a 50% increase in response to NaCl stress in 4-day-old seedling cotyledons although the activity remained lower than that observed in 2-day-old respective samples. An enhancement of PHGPX activity in response to NaCl-stress with respect to controls at the respective stages of development implies increased detoxification of lipid hydroperoxides formed as a result of NaCl stress. Lowest PHGPX activity was evident in 6-day-old seedling cotyledons which correlates with elevated LHPO content. CLSM analysis revealed highest expression of PHGPX in the cytosol of the specialized epidermal cells enclosing the secretory canal in the neighborhood of vascular strands (Fig. 3). Significant differences highlighted the differential and elevated expression of PHGPX in response to NaCl stress. Present observations indicate maxima detoxifying action of PHGPX in the early stages of seedling growth i.e., 2-day-old seedling cotyledons, thus highlighting early signaling role of this enzyme in response to NaCl stress.
Figure 3.
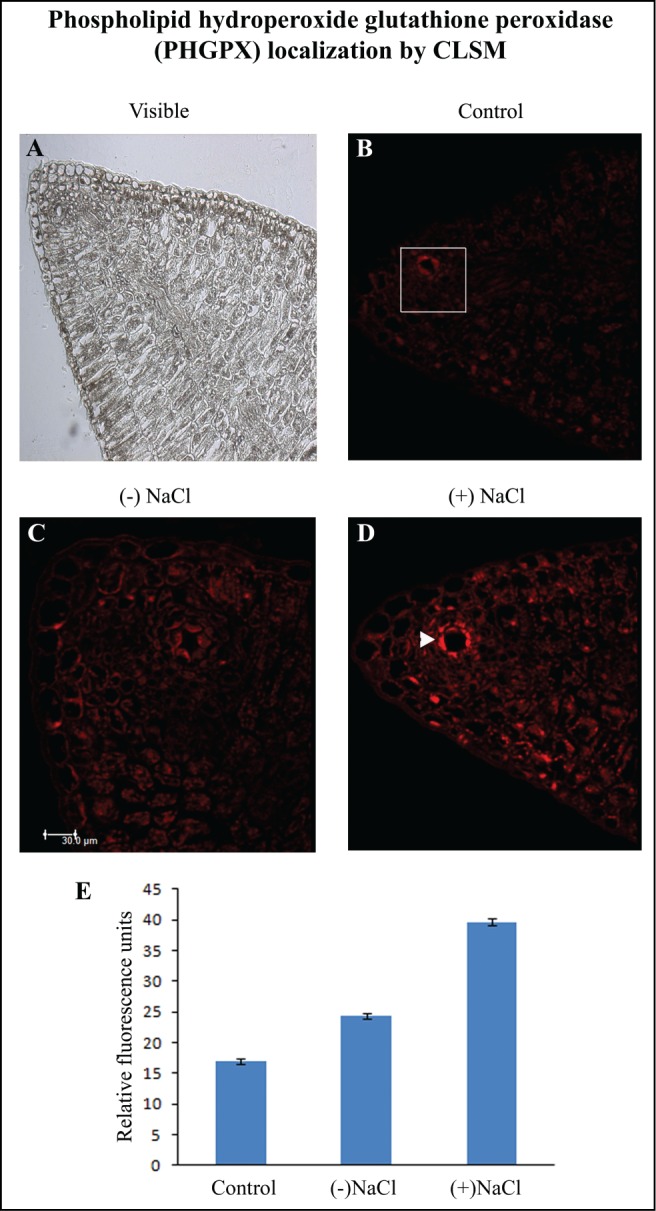
Localization of phospholipid hydroperoxide glutathione peroxidase (PHGPX) by CLSM imaging using anti-GPX4 (PHGPX) antibody. (A) Visible image of 7 μm thick transverse sections of cotyledon from 2 d-old etiolated seedlings. Fluorescence micrographs of cotyledon transverse sections in the absence of anti-GPX4 (PHGPX) antibody (B), in the presence of anti-GPX4 (PHGPX) antibody in cotyledons from (−) NaCl seedling (C) and cotyledons from (+) NaCl seedlings (D); ▸ indicates secretory canal. (E) Quantification of fluorescence from the indicated zone. Magnification: 200×.
Figure 2.
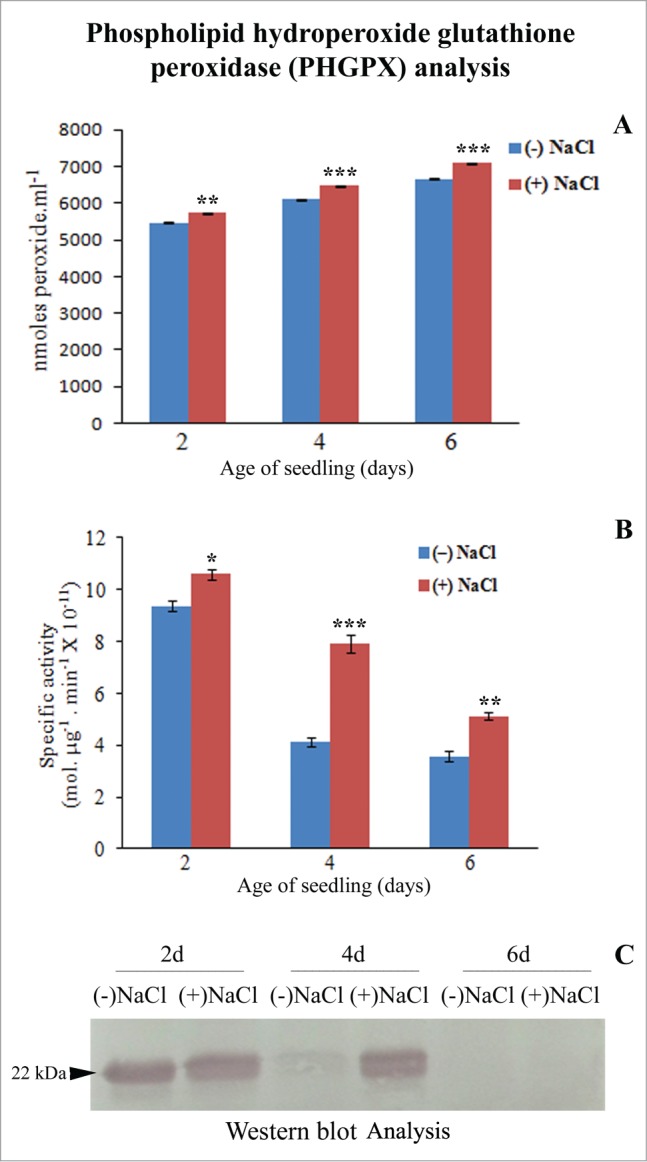
Analysis of phospholipid hydroperoxide glutathione peroxidase (PHGPX) activity in total soluble protein (10,000g supernatant) from dark-grown sunflower seedling cotyledons grown in the absence or presence of 120 mM NaCl. (A) Quantification of lipid hydroperoxide content using PeroxiDetect Kit (Sigma-Aldrich Chemicals Pvt. Ltd., USA). Changes in lipid hydroperoxide content were found to be statistically significant (**P < 0.01, ***P < 0.001), as analyzed by one-way ANOVA using SPSS 22.0. Bars represent SE (n = 3). (B) Spectrophotometric determination of specific activity of PHGPX. NaCl-stress induced changes in enzyme activity, were analyzed by one-way ANOVA using SPSS 22.0 and were found to be statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001) in comparison to control [(-)NaCl]. Bars represent SE (n = 3). (C) Western blot analysis of PHGPX using anti-GPX4 (PHGPX) antibody.
Analysis of GPXs in the plant kingdom started with the cloning of the first GPX cDNA from the leaves of wild tobacco (Nicotiana sylvestris), which showed homology to animal, selenium-dependent, PHGPX.10 Since then, more than 100 cDNAs encoding GPX proteins exhibiting high sequence similarity to animal GPXs have been isolated, cloned and characterized from various plant species.11-23 Most GPX genes isolated from plants so far have highest sequence homology to animal phospholipid hydroperoxide glutathione peroxidase (PHGPX). Cit-SAP was the first naturally isolated PHGPX protein, partially purified from salt-adapted citrus cells.24 Phospholipid hydroperoxide glutathione peroxidase (PHGPX, EC 1.11.1.12), a member of GPX family, is a monomer of 20–25 kDa and is involved in direct reduction of phospholipid hydroperoxides and complex hydroperoxy lipids.7,25 Earlier investigations have also shown that plant PHGPXs exhibit lower enzymatic activities than their animal counterparts because they contain Cys at the putative catalytic site rather than the selenocysteine, typical of animal GPXs.11,18,26,27 Yeast and plant GPXs could be part of an alternative pathway that scavenges peroxides, especially phospholipid hydroperoxides.5 GPX proteins (Gpx-1, Gpx-2) from Synechocystis PCC 6803 utilize NADPH instead of glutathione (GSH) as a reductant.28 In addition, there have been reports on GPX isoenzymes that can utilize both GSH and thioredoxin (Trx) as reducing agents in yeast,29 sunflower and tomato.27 Investigations in Arabidopsis30 and Chinese cabbage18 have revealed that GPX utilizes thioredoxin as the sole electron donor for the reduction of hydroperoxides.
The response of antioxidative enzymes to salinity stress differs depending on the species, tissues, and subcellular localization.31,32 Differential expression of POD isoforms is evident in sunflower seedling cotyledons with reference to age and absence or presence of NaCl stress (present work). PHGPX has also been reported to play a role in H2O2 scavenging, signaling events and photoprotection in some other plant systems.33,34 PHGPX cDNAs isolated from different plants is induced by salinity, heavy metal toxicity and infection with bacterial or viral pathogens. Isolation and characterization of PHGPX gene from rice has also been reported.34-36 Present findings highlight a role of PHGPX in combating hydroperoxide production as an early signaling response in 2-day-old seedlings.
The major function of plant PHGPXs appears to be the scavenging of phospholipid hydroperoxides, thereby protecting cell membranes from peroxidative damage.37 Gene expression analysis has shown an increase in the levels of PHGPX mRNA in several plant species undergoing different biotic and abiotic stresses, such as pathogen infections,10,14,38 high salt concentrations,39,22 exposure to heavy metals,40,41 mechanical stimulation,16 aluminum toxicity,42 seed germination,12 salt and osmotic stress,43 oxidative stress,39,35 chilling stress.22 PHGPXs may play dual roles as a redox transducer in addition to acting as a H2O2 scavenger under stress.33 Thus, PHGPX proteins may have different functions in plant cells, with some isoforms functioning in signal transduction pathway, while others are involved in catalyzing the reduction of harmful products formed by ROS. A signal transducing role for a GPX has also been demonstrated in yeast44 and Arabidopsis.33 The effect of various signaling molecules (salicylic acid, jasmonic acid, ABA, ethylene) and certain protein phosphatase inhibitors (cantharidin, endothall) on the expression of PHGPX gene in rice seedling leaves has demonstrated an upregulation of the mRNA levels.45 These data suggest the role of this enzyme in the induction of defense mechanisms in plant cells subjected to oxidative damage as a result of exposure to various environmental stresses, by reducing the phospholipid hydroperoxides formed in the biomembranes as a result of oxidative stress.
To sum up, etiolated sunflower seedlings exhibit enhanced total peroxidase (POD) activity and differential expression of its isoforms in the 10,000g supernatant of cotyledons in response to NaCl stress. Accumulation of lipid hydroperoxides coincides with an upregulation of PHGPX activity as an early event. NaCl stress elevates PHGPX activity in cotyledons all the more, at the respective stages of seedling growth. Spatial distribution of PHGPX also exhibits an overall elevated cytosolic distribution in response to NaCl stress and its concentration in the cells lining the secretory canals. A long distance sensing of NaCl stress in terms of upregulation of PHGPX activity from seedling roots to cotyledons is, thus proposed.
Seed germination, seedling growth and salt (120 mM) treatment were undertaken according to earlier reports from the author's laboratory.46 Peroxidase (POD) activity was estimated according to Alba et al.,47 with minor modifications. Protein obtained from cotyledons by homogenizing in 50 mM sodium acetate buffer (pH 4.0) and centrifugation at 10,000g was quantified by Bradford method48 and 25 μg protein was added to a 2.4 mL substrate solution (0.6 mM o-dianisidine, 8.8 mM H2O2 in 50 mM sodium phosphate buffer, pH-6). Absorbance was recorded at 460 nm for 5 min at an interval of 1 min. Enzyme activity from spectrophotometric analysis was expressed as specific activity of POD in Mol μg−1 min−1. For zymographic detection of peroxidase (POD) activity protein extracted and quantified as described above was constituted in Lamelli buffer without SDS. 25 μg protein was loaded for single dimension separation (vertical gel) on a 12% native-PAGE at 4°C (conditions: 75V for 0.5 h, 25 mA for 5 h) in Miniprotean Tetra Cell (Biorad, USA). Gel was then incubated in 0.2 M sodium acetate buffer (pH 5.0) containing 1.3 mM benzidine and 1.3 mM H2O2 till brown bands representing POD developed. Western blot detection of peroxidase (POD) isoforms was done on nitrocellulose membrane at a current of 400 mA for 1 h at 4°C to achieve complete transfer of proteins. Subsequently, the blot was incubated for overnight at 4°C with a rabbit polyclonal antibody against peroxidase (anti-peroxidase antibody obtained from Sigma-Aldrich Chemicals Pvt. Ltd., USA) in a dilution of 1:2000. Membrane was subsequently washed in wash buffer (0.2% Tween-20 in PBS pH 7.4) and incubated in secondary antibody (anti-rabbit IgG conjugated to alkaline phosphatase antibody obtained from Sigma-Aldrich Chemicals Pvt. Ltd., USA) (1:2500 in wash buffer) for 1 h at RT. Finally, the membrane was washed in wash buffer and developed using BCIP/NBT (1 Sigma Fast tablet dissolved in 10 mL distilled water) for 10–30 min for detection of alkaline phosphatase activity.
Lipid hydroperoxide content was measured using a PeroxiDetect Kit (Sigma-Aldrich Chemicals Pvt. Ltd., USA). A standard curve was generated using different concentrations of t-butyl hydroperoxide (provided in the kit). Lipid soluble fraction was isolated from tissue by chloroform-methanol extraction. This was then incubated with the color reagent (provided in the kit) for 30 mins at RT. Absorbance was recorded at 560 nm. Lipid hydroperoxide levels were calculated from the standard curve and expressed in nmoles. ml−1. PHGPX activity was estimated according to Yang et al.49 by detecting the oxidation of glutathione in a coupled assay. Cotyledons (500 mg) from sunflower seedlings grown in the absence or presence of NaCl (120 mM) were homogenized in 1 mL of extraction buffer (50 mM Tris-HCl, 0.4 M sucrose, 3 mM EDTA, 5 mM 2-mercaptoethanol, 1 mM PMSF, pH-7.5). PHGPX activity was measured in 12,000g supernatant by monitoring the decrease in absorbance at 340 nm due to oxidation of NADPH in the presence of GSH and GSH reductase. The reaction mixture contained 50 mM Tris-HCl, pH-8.0, 0.1 mM EDTA, 15 mM GSH, 0.12% Triton X-100, 3 units of glutathione reductase (Sigma-Aldrich Chemicals Pvt. Ltd., USA) and 0.25 mM NADPH. To this reaction mixture, 50 μg protein was added and preincubated at RT for 5–10 min. The reaction was then started by addition of 200 μM t-butyl hydroperoxide (Sigma-Aldrich Chemicals Pvt. Ltd., USA). Rate of NADPH consumption was corrected for hydroperoxide-independent, glutathione-independent and non-enzymatic oxidation of NADPH. Enzyme activity was calculated using an absorption coefficient of 6.22 mM−1 cm−1 for NADPH. Western blot analysis of separated bands for PHGPX was performed at a current of 400 mA for 1 h at 4°C. Subsequently the blot was incubated with a rabbit polyclonal antibody against glutathione peroxidase 4 [anti-GPX4 (PHGPX) antibody obtained from Abcam, UK] in a dilution of 1:1000 in blocking buffer, for overnight at 4°C. The membrane was washed in wash buffer (0.2% Tween-20 in PBS pH 7.4) and incubated in secondary antibody (anti-rabbit IgG conjugated to alkaline phosphatase antibody, Sigma-Aldrich Chemicals Pvt. Ltd., USA) (1:2500 in wash buffer) for 1 h at RT followed by development using BCIP/NBT (1 Sigma Fast tablet dissolved in 10 mL distilled water) for 10–30 min. Immunolocalization of PHGPX was done by CLSM using wax sections (7 μm thick) of cotyledons from 2 d old, dark-grown seedlings. Sections were incubated in BlockAid solution (Invitrogen, USA) for 30 min. After blocking and serial washings in PBS, the sections were incubated in anti-rabbit polyclonal primary antibody against PHGPX [anti-GPX4 (PHGPX) antibody obtained from Abcam, UK] at a dilution of 1:200 for 2 h at RT. Subsequently, sections were washed in PBS and incubated with Cy3-labeled anti-rabbit IgG (GE Life Sciences, England) diluted to 1:1500 in PBS. Sections without primary antibody served as control. The slides were mounted in PBS and viewed using CLSM (TCS SP5, Leica, Germany).
All experiments were undertaken at least thrice.
Funding Statement
Authors are grateful to University of Delhi and University Grants Commission for financial support.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 2010; 48:909-30; PMID:20870416; http://dx.doi.org/ 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- 2. Mittler R, Vanderauwera S, Gollery M, Breusegem FV. Reactive oxygen gene network of plants. Trends Plant Sci 2004; 9:490-8; PMID:15465684; http://dx.doi.org/ 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 3. Bhattacharjee S. Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. Curr Sci India 2005; 89(7): 1113-21. [Google Scholar]
- 4. Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 2002; 7:405-10; PMID:12234732; http://dx.doi.org/ 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- 5. Eshdat Y, Holland D, Zehava F, Ben-Hayyim G. Plant glutathione peroxidases. Physiol Plant 1997; 100:234-40; PMID:24843434; http://dx.doi.org/ 10.1111/j.1399-3054.1997.tb04779.x24843434 [DOI] [Google Scholar]
- 6. Herbette S, Roeckel-Drevet P, Drevet JR. Seleno-independent glutathione peroxidases more than simple antioxidant scavengers. FEBS J 2007; 274:2163-80; PMID:17419737; http://dx.doi.org/ 10.1111/j.1742-4658.2007.05774.x [DOI] [PubMed] [Google Scholar]
- 7. Ursini F, Mariorino M, Brigelius-Flohe R, Aumann KD, Roveri A, Schomburg D, Flohe L. Diversity of glutathione peroxidase. Method Enzymol 1995; 252:38-53; http://dx.doi.org/ 10.1016/0076-6879(95)52007-4 [DOI] [PubMed] [Google Scholar]
- 8. Sadeghipour HR, Bhatla SC. Diiferential sensitivity of oleosins to proteolysis during oil body mobilization in sunflower seedlings. Plant Cell Physiol 2002; 43(10): 1117-26; PMID:12407191; http://dx.doi.org/ 10.1093/pcp/pcf142 [DOI] [PubMed] [Google Scholar]
- 9. Sadeghipour HR, Bhatla SC. Light-enhanced oil body mobilization in sunflower seedlings accompanies faster protease action on oleosins. Plant Physiol Biochem 2003; 41:309-16; PMID:22705588; http://dx.doi.org/ 10.1016/S0981-9428(03)00024-X22705588 [DOI] [Google Scholar]
- 10. Criqui MC, Jamet E, Parmentier Y, Marbach J, Fleck J. Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol Biol 1992; 18:623-7; PMID:1536938; http://dx.doi.org/ 10.1007/BF00040684 [DOI] [PubMed] [Google Scholar]
- 11. Holland D, Ben-Hayyim G, Faltin Z. Molecular characterization of salt-stress associated protein in citrus: protein and cDNA sequence homology to mammalian glutathione peroxidases. Plant Mol Biol 1993; 21:923-7; PMID:8467085; http://dx.doi.org/ 10.1007/BF00027124 [DOI] [PubMed] [Google Scholar]
- 12. Johnson RR, Cranston HJ, Chaverra ME, Dyer WE. Characterization of cDNA clones for differentially expressed genes in embryos of dormant and nondormant Avena fatua L. caryopses. Plant Mol Biol 1995; 28:113-22; PMID:7787176; http://dx.doi.org/ 10.1007/BF00042043 [DOI] [PubMed] [Google Scholar]
- 13. Sugimoto M, Furui S, Suzuki Y. Molecular cloning and characterization of a cDNA encoding putative phospholipid hydroperoxide glutathione peroxidase from spinach. Biosci Biotech Bioch 1997; 61:1379-81; PMID:9301122; http://dx.doi.org/ 10.1271/bbb.61.1379 [DOI] [PubMed] [Google Scholar]
- 14. Roeckel-Drevet P, Gagne G, Labrouhe T, Dufaure JP, Nicolas P, Drever JR. Molecular characterization, organ distribution and stress-mediated induction of two glutathione peroxidase-encoding mRNAs in sunflower (Helianthus annuus). Physiol Plant 1998; 103:385-94; http://dx.doi.org/ 10.1034/j.1399-3054.1998.1030312.x [DOI] [Google Scholar]
- 15. Mullineaux PM, Karpinski S, Jimenez A, Cleary SP, Robinson C, Creissen GP. Identification of cDNA encoding plastid- targeted glutathione peroxidase. Plant J 1998; 13:375-9; PMID:9680987; http://dx.doi.org/ 10.1046/j.1365-313X.1998.00052.x [DOI] [PubMed] [Google Scholar]
- 16. Depege N, Drevet J, Boyer N. Molecular cloning and characterization of tomato cDNAs encoding glutathione peroxidase-like proteins. Eur J Biochem 1998; 253:445-51; PMID:9654095; http://dx.doi.org/ 10.1046/j.1432-1327.1998.2530445.x [DOI] [PubMed] [Google Scholar]
- 17. Churin Y, Schilling S, Borner T. A gene family encoding glutathione peroxidase homologues in Hordeum vulgare (barley). FEBS Lett 1999; 459:33-8; PMID:10508912; http://dx.doi.org/ 10.1016/S0014-5793(99)01208-9 [DOI] [PubMed] [Google Scholar]
- 18. Jung BG, Lee KO, Lee SS, Chi YH, Jang HH, Kang SS, Lee K, Lim D, Yoon SC, Yun DJ, et al. A chinese cabbage cDNA with high sequence identity to phospholipid hydroperoxide glutathione peroxidases encodes a novel isoform of thioredoxin-dependent peroxidase. J Biol Chem 2002; 277(15): 12572-8; PMID:11823460; http://dx.doi.org/ 10.1074/jbc.M110791200 [DOI] [PubMed] [Google Scholar]
- 19. Gaber A, Ogata T, Maruta T, Yoshimura K, Tamoi M, Shigeoka S. The involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol 2012; 53(9): 1596-606; PMID:22773682; http://dx.doi.org/ 10.1093/pcp/pcs100 [DOI] [PubMed] [Google Scholar]
- 20. Passaia G, Foninia LS, Caverzana A, Jardim-Messedera D, Christoff AP, Gaetac ML, Mariath JEA, Margis R, Margis-Pinheiroa M. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci 2013; 208:93-101; PMID:23683934; http://dx.doi.org/ 10.1016/j.plantsci.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 21. Zhai CZ, Zhao L, Yin LJ, Chen M, Wang QY, Li LC, Xu ZS, Ma YZ. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. Plos One 2013; 8(10):e73989; PMID:24098330; http://dx.doi.org/ 10.1371/journal.pone.0073989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim YJ, Jang MG, Noh HY, Lee HJ, Sukweenadhi J, Kim JH, Kim SY, Kwon WS, Yang DC. Molecular characterization of two glutathione peroxidase genes of Panax ginseng and their expression analysis against environmental stresses. Gene 2014; 535:33-41; PMID:24269671; http://dx.doi.org/ 10.1016/j.gene.2013.10.071 [DOI] [PubMed] [Google Scholar]
- 23. Fu JY. Cloning of a new glutathione peroxidase gene from tea plant (Camellia sinensis) and expression analysis under biotic and abiotic stresses. Bot Stud 2014; 55:7; http://dx.doi.org/ 10.1186/1999-3110-55-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beeor-Tzahar T, Ben-Hayyim G, Holland D, Faltin Z, Eshdat Y. A stress-associated citrus protein is a distinct plant phospholipid hydroperoxide glutathione peroxidase. FEBS Lett 1995; 366:151-5; PMID:7789534; http://dx.doi.org/ 10.1016/0014-5793(95)00521-A [DOI] [PubMed] [Google Scholar]
- 25. Faltin Z, Holland D, Velcheva M, Tsapoversky M, Roeckel-Drevet P, Handa AK, Abu-Abied M, Friedman-Einat M, Eshdat Y, Perl A. A glutathione peroxidase regulation of rective oxygen species level is crucial for in vitro plant differentiation. Plant Cell Physiol 2010; 51(7):1151-62; PMID:20530511; http://dx.doi.org/ 10.1093/pcp/pcq082 [DOI] [PubMed] [Google Scholar]
- 26. Faltin Z, Camoin L, Ben-Hayyim G, Perl A, Beeor-Tzahar T, Strosberg AD, Holland D, Eshdat Y. Cysteine is the presumed catalytic residue of Citrus sinensis phospholipid hydroperoxide glutathione peroxidase over-expressed under salt stress. Physiol Plant 1998; 104:741-6; http://dx.doi.org/ 10.1034/j.1399-3054.1998.1040432.x [DOI] [Google Scholar]
- 27. Herbette S, Lenne C, Leblanc N, Julien JL, Roeckel-Drevet J, Roeckel-Drevet P. Two GPX-like proteins from Lycopersicon esculentum and Helianthus annuus are antioxidant enzymes with phospholipid hydroperoxide glutathione peroxidase and thioredoxin peroxidase activities. Eur J Biochem 2002; 269:2414-20; PMID:11985625; http://dx.doi.org/ 10.1046/j.1432-1033.2002.02905.x [DOI] [PubMed] [Google Scholar]
- 28. Gaber A, Tamoi M, Takeda T, Nakano Y, Shigeoka S. NADPH-dependent glutathione peroxidase-like proteins (Gpx-1, Gpx-2) reduce unsaturated fatty acid hydroperoxides in Synechocystis PCC 6803. FEBS Lett 2001; 499:32-6; PMID:11418106; http://dx.doi.org/ 10.1016/S0014-5793(01)02517-0 [DOI] [PubMed] [Google Scholar]
- 29. Tanaka T, Izawa S, Inoue Y. GPX2, encoding a phospholipid hydroperoxide glutathione peroxidase homologue, codes for an atypical 2-Cys peroxiredoxin in Saccharomyces cerevisiae. J Biol Chem 2005; 280(51): 42078-87; PMID:16251189; http://dx.doi.org/ 10.1074/jbc.M508622200 [DOI] [PubMed] [Google Scholar]
- 30. Iqbal A, Yabuta Y, Takeda T, Nakano Y, Shigeoka S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J 2006; 273:5589-97; PMID:17096689; http://dx.doi.org/ 10.1111/j.1742-4658.2006.05548.x [DOI] [PubMed] [Google Scholar]
- 31. Sreenivasula N, Grimm B, Wobus U, Weschke W. Differential response of antioxidant compounds to salinity stress in salt-tolerant and salt-sensitive seedlings of foxtail millet (Setaria italica). Physiol Plant 2000; 109:435-41; http://dx.doi.org/ 10.1034/j.1399-3054.2000.100410.x [DOI] [Google Scholar]
- 32. Mittova V, Tal M, Volokita M, Guy M. Upregulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant Cell Environ 2003; 26:845-56; PMID:12803612; http://dx.doi.org/ 10.1046/j.1365-3040.2003.01016.x [DOI] [PubMed] [Google Scholar]
- 33. Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006; 18:2749-66; PMID:16998070; http://dx.doi.org/ 10.1105/tpc.106.044230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang CCC, Lesak IS, Jorda L, Sotnikov A, Melzer M, Miszalski Z, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol 2009; 150:670-83; PMID:19363092; http://dx.doi.org/ 10.1104/pp.109.135566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li WJ, Feng H, Fan JH, Zhang RQ, Zhao NM, Liu JY. Molecular cloning and expression of a phospholipid hydroperoxide glutathione peroxidase homolog in Oryza sativa. Biochim Biophys Acta 2000; 1493:225-30; PMID:10978528; http://dx.doi.org/ 10.1016/S0167-4781(00)00152-4 [DOI] [PubMed] [Google Scholar]
- 36. Chen S, Vaghchhipawala Z, Li W, Asard H, Dickman MB. Tomato phospholipids hydroperoxide glutathione peroxidase inhibits cell death induced by bax and oxidative stresses in yeast and plants. Plant Physiol 2004; 135:1630-41; PMID:15235116; http://dx.doi.org/ 10.1104/pp.103.038091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gueta-Dahan Y, Yaniv Z, Zilinskas B, Ben-Hayyim G. Salt and oxidative stress: similar and specific responses and their relation to salt tolerance in Citrus. Planta 1997; 203:460-9; PMID:9421931; http://dx.doi.org/ 10.1007/s004250050215 [DOI] [PubMed] [Google Scholar]
- 38. Herbette S, Lenne C, De Labrouhe DT, Drevet JR, Roeckel-Drevet P. Transcripts of sunflower antioxidant scavengers of the SOD and GPX families accumulate differentially in response to downy mildew infection, phytohormones, reactive oxygen species, nitric oxide, protein kinase and phosphatase inhibitors. Physiol Plant 2003; 119:418-28; http://dx.doi.org/ 10.1034/j.1399-3054.2003.00186.x [DOI] [Google Scholar]
- 39. Avsian-Kretchmer O, Eshdat Y, Gueta-Dahan Y, Ben-Hayyim G. Regulation of stress-induced phospholipid hydroperoxide glutathione peroxidase expression in citrus. Planta 1999; 209:469-77; PMID:10550628; http://dx.doi.org/ 10.1007/s004250050750 [DOI] [PubMed] [Google Scholar]
- 40. Sugimoto M, Sakamoto W. Putative phospholipid hydroperoxide glutathione peroxidase gene from Arabidopsis thaliana induced by oxidative stress. Genes Genet Syst 1997; 72:311-6; PMID:9511228; http://dx.doi.org/ 10.1266/ggs.72.311 [DOI] [PubMed] [Google Scholar]
- 41. Ramos J, Matamoros MA, Naya L, James EK, Rouhier N, Sato S, Tabata S, Becana M. The glutathione peroxidase gene family of Lotus japonicus: characterization of genomic clones, expression analyses and immunolocalization in legumes. New Phytol 2009; 181(1):103-14; PMID:18826485; http://dx.doi.org/ 10.1111/j.1469-8137.2008.02629.x [DOI] [PubMed] [Google Scholar]
- 42. Rodriguez-Milla MA, Butler E, Rodriguez-Huete A, Wilson CF, Anderson O, Gustafson JP. EST-based gene expression analysis under aluminium stress in rye (Secale cereale L.). Plant Physiol 2002; 130:1706-16; PMID:12481053; http://dx.doi.org/ 10.1104/pp.009969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao F, Chen J, Ma T, Li H, Wang N, Li Z, Zhang Z, Zhou Y. The glutathione peroxidase gene family in Thellungiella salsuginea: genome-wide identification, classification, and gene and protein expression analysis under stress conditions. Int J Mol Sci 2014; 15:3319-35; PMID:24566152; http://dx.doi.org/ 10.3390/ijms15023319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 2002; 111:471-81; PMID:12437921; http://dx.doi.org/ 10.1016/S0092-8674(02)01048-6 [DOI] [PubMed] [Google Scholar]
- 45. Agrawal GK, Rakwal R, Jwac NS, Agrawal VP. Effects of signaling molecules, protein phosphatase inhibitors and blast pathogen (Magnaporthe grisea) on the mRNA level of a rice (Oryza sativa L.) phospholipid hydroperoxide glutathione peroxidase (OsPHGPX) gene in seedling leaves. Gene 2002; 283:227-36; PMID:11867229; http://dx.doi.org/ 10.1016/S0378-1119(01)00854-X [DOI] [PubMed] [Google Scholar]
- 46. David A, Yadav S, Bhatla SC. Sodium chloride stress induces nitric oxide accumulation in root tips and oil body surface accompanying slower oleosin degradation in sunflower seelings. Physiol Plant 2010; 140:342-54; PMID:20738803; http://dx.doi.org/ 10.1111/j.1399-3054.2010.01408.x [DOI] [PubMed] [Google Scholar]
- 47. Alba CM, De Forchetti SM, Quesada MA, Valpuesta V, Tigier HA. Localization and general properties of developing peach seed coat and endosperm peroxidase isoenzymes. J Plant Growth Regul 1998; 17:7-11; http://dx.doi.org/ 10.1007/PL00007013 [DOI] [Google Scholar]
- 48. Bradford M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of dyebinding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/ 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 49. Yang XD, Dong CJ, Liu JY. A plant mitochondrial phosholipid hydroperoxide glutathione peroxidase: its precise localization and higher enzymatic activity. Plant Mol Biol 2006; 62:951-62; PMID:16944266; http://dx.doi.org/ 10.1007/s11103-006-9068-0 [DOI] [PubMed] [Google Scholar]


