Abstract
Membrane fusion is a tightly controlled process in all eukaryotic cell types. The SNARE family of proteins is required for fusion throughout the exocytic and endocytic trafficking pathways. SNAREs on a transport vesicle interact with the cognate SNAREs on the target membrane, forming an incredibly stable SNARE complex that provides energy for the membranes to fuse, although many aspects of the mechanism remain elusive. Recent advances in single-molecule and high-resolution structural methods provide exciting new insights into how SNARE complexes assemble, including measurements of assembly energetics and identification of intermediates in the assembly pathway. These techniques were also key in elucidating mechanistic details into how the SNARE complex is disassembled, including details of the energetics required for ATP-dependent α-SNAP/NSF-mediated SNARE complex disassembly, and the structural changes that accompany ATP hydrolysis by the disassembly machinery. Additionally, SNARE complex formation and disassembly are tightly regulated processes; innovative biochemical and biophysical characterization has deepened our understanding of how these regulators work to control membrane fusion and exocytosis.
Exocytosis and membrane fusion
Most eukaryotic cells carry out exocytosis for the delivery of lipids and proteins to facilitate cellular growth, as well as signaling to the cell’s exterior environment. Exocytic vesicle fusion allows the plasma membrane to expand, often in a specific location in the cell (i.e. polarized growth). Additionally, proteins that were embedded in the vesicle membrane are delivered to the plasma membrane to serve as signaling receptors, transport proteins, and channels. The soluble contents of the vesicles are also delivered to the extracellular space upon fusion. Exocytic vesicles originate from the trans-Golgi network or recycling endosomes, and are transported on the cytoskeleton to the plasma membrane. Upon arrival at the plasma membrane, SNARE (Soluble N-ethylmaleimide sensitive factor protein receptor) complexes form between the vesicle and the plasma membrane, leading to membrane fusion and cargo delivery.
Certain cell types have specialized forms of exocytosis that require additional regulation, such as neurotransmitter release in neurons, histamine release in mast cells, and transport of the glucose transporter GLUT4 to the plasma membrane in muscle and adipose cells. It is important to note that although regulation of these specific pathways is different from one another and from the growth pathway, they share many common core components—the different pathways use the cytoskeleton and motor proteins for transport of vesicles, and membrane fusion is carried out by the SNARE family of proteins [1–5]. This review will focus primarily on new information regarding the mechanism of SNARE-mediated membrane fusion at the plasma membrane and its regulation. More detailed reviews of the general steps and components of exocytosis and endocytosis can be found in [6–10] and references therein.
The fundamental machinery of trafficking
Exocytosis is a multi-step process that can be regulated at many points by many factors. Cargo transport begins when macromolecules are packaged into vesicles that bud from the trans-Golgi network. These vesicles are then carried by motor proteins of the myosin and kinesin families that walk towards the plasma membrane using actin filaments and microtubule tracks, and ATP as fuel [7]. The specificity of the cargo-motor protein interaction is controlled by the Rab family of small GTPases [10]. Once at the cell periphery, a multisubunit tethering complex called the exocyst facilitates SNARE-mediated membrane fusion. The exocyst has been suggested to tether the vesicle to the target membrane, bringing the SNARE proteins into close proximity. Additionally, the yeast exocyst plays a more direct role through its interaction with the SNARE subunit Sec9, the yeast homolog of SNAP-25 [11]. The exocyst does not appear to be required for synaptic vesicle fusion in neurons [12], but is essential in other cell types [13]. Other SNARE assembly and fusion regulators include the Sec1/Munc18 family [3,14], neuronal specific proteins including synaptotagmin and complexin (discussed below), and Munc13 [15]. After the membranes have been fused by the SNAREs, the SNARE complex must be either disassembled and recycled for another round of fusion, or degraded. Therefore, regulation of exocytosis occurs at the levels of cargo loading, vesicle transport, and membrane fusion. The studies highlighted below will focus specifically on the mechanisms of SNARE complex formation and disassembly, and how these processes can be regulated.
SNARE-mediated membrane fusion
Membrane fusion results from binding of the vesicle-associated (v-) SNARE VAMP (also called synaptobrevin) to the target membrane-associated (t-) SNAREs syntaxin and SNAP-25 [8]. Biochemical and biophysical characterization of these proteins indicated that their unassembled structures are comprised of random coils, which gain significant helicity when they form the assembled ternary complex (Fig. 1) [16]. The neuronal SNARE complex is very stable; it is resistant to both heat (with a melting temperature of greater than 90°C) and sodium dodecyl sulfate [16,17]. Analyses using both electron microscopy and spin labeling electron paramagnetic resonance spectroscopy showed that these helices formed a parallel coiled-coil, providing the first clue that these proteins might be not be antiparallel receptors for vesicle recognition, but rather “zipper” and pull the vesicle and target membranes together for fusion [18,19]. Moreover, the crystal structure established that the post-fusion SNARE complex is a parallel four-helix-bundle, with two helices contributed by SNAP-25 and one each by the membrane-embedded syntaxin and synaptobrevin [20]. This structure identified an ionic layer (three glutamine residues from SNAP-25 and syntaxin that interact with an arginine from synaptobrevin) buried at the interface of the complex and surrounded by hydrophobic interactions. These combined interactions may help to stabilize the ternary complex, properly align the register of the complex, and/or be important for complex disassembly, although this has been debated [20,21]. These data, together with the finding that SNARE complexes were necessary and sufficient to catalyze liposome fusion [22], support the hypothesis that v- and t-SNARE zippering from the N-termini to the membrane-embedded C-terminal helices [17,23,24] provides the energy for membrane fusion [25].
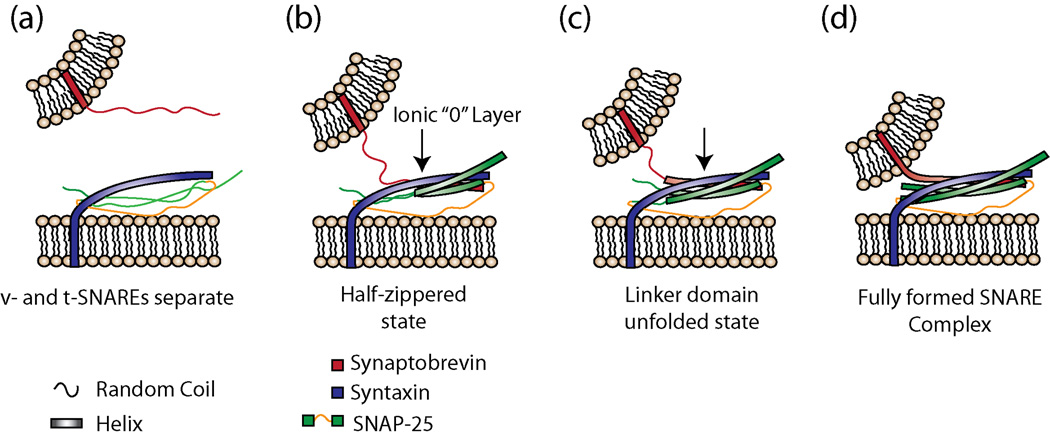
Fig 1.
SNARE complex structure formation during membrane fusion. (a) v- and t-SNAREs are separate. (b) Complex formation begins via association of the N-terminal regions of the individual SNARE proteins forming the half-zippered state [26,27**]. This association includes the ionic (0) layer. (c) A second intermediate state has been postulated [26], here called the partially unzipped state. (d) SNARE complex assembly and fusion complete. Here, the SNARE complex is drawn to reflect the conformation as observed in the crystal structure of the neuronal SNAREs [20]. It is still unclear at which point in these steps that membrane hemifusion and fusion occur.
SNARE complex formation originally appeared to be a simple two-state process: uncomplexed v- and t-SNAREs would directly form ternary complexes with no kinetic intermediates [25]. However, recent work challenged this idea, at least for neuronal SNAREs, but perhaps for others as well [26–29]; intermediates in the SNARE assembly pathway were identified that might be key to the regulation of assembly and fusion. The first line of evidence comes from experiments using optical tweezers, which allowed researchers to directly pull on the C-terminal regions (normally embedded in the membrane) of the complexed neuronal SNARES [26]. Gao et al. used this optical tweezers and Hidden Markov modeling to identify four distinct states: (1) ternary complex is completely formed; (2) application of force allows the SNAREs to partially unzip, oscillating between the partially unzipped form and the folded form; (3) increasing the application of force on the C-termini causes state 2 to further unfold to a half-zippered state; and (4) the half-zippered state crosses a threshold and completely unfolds. This process is shown in Figure 1, but in reverse order. When trap distances were analyzed, the half-zippered state corresponded to formation of the complex up to, and including, the ionic layer. An additional study by Min et al. using magnetic tweezers independently isolated this half-zippered state, but not the partially unzipped state 2 [27**]. Interestingly, both studies observed the same hysteresis behavior; this suggested that they may have been characterizing the same intermediate state, and was reminiscent of the assembly and disassembly hysteresis observed biochemically by Fasshauer et al. [30]. Taken together, these data suggest that neuronal SNARE complex assembly occurs with at least one intermediate.
Do these folding intermediates play a role in the regulation of SNARE complexes in different cell types? The authors of the studies above suggest that the half-zippered state is important for the fusion priming observed in neurons (see below), but its relevance in other cell types was not clear. To answer this, Zorman et al. compared the energetics for SNARE complex formation from four different origins: synaptic vesicle fusion in neurons, those involved in exocytosis of GLUT4 vesicles, endosomal SNAREs and the yeast exocytic SNAREs [28]. Using artificially concatenated SNARE proteins, the authors isolated the half-zippered state in all four SNAREs tested, although the energetics differed. The N-terminal half of the complexes all had similar folding energies. However, differences were discovered between the C-terminal halves of each complex. Additionally, formation of the C-terminal half of each complex was more favorable than their N-terminal halves, leading the authors to postulate that this evolved to counteract the increase in repulsive energies that arises as the membranes are moved closer together. Furthermore, the authors predict that these differences in energetics are not the key determinants in the regulatory mechanisms employed, although it was noted that the two SNAREs with the most favorable C-terminal domain folding energies (neuronal and GLUT4) are regulated by functionally similar fusion priming mechanisms.
Regulation of SNARE zippering
Neuronal fusion reactions are incredibly fast in vivo (on the order of ms); however, in vitro measurements previously reported a fusion rate of minutes [22]. A handful of more recent studies demonstrated fusion rates in the sub-second regime, but each of these studies have specific caveats, including a lack of dependence on a fully assembled t-SNARE [31,32]; the requirement for a polyethylene glycol (PEG) tether between the vesicle and target membrane [33]; or use of artificial PEGylated membranes [34]. A key difference between these in vitro studies and synaptic vesicle fusion in vivo is that the SNARE complexes likely exist in a primed pre-fusion state at presynaptic sites. Indeed, docked vesicles are directly observed in neurons prior to stimulation [35,36].
How does priming occur? Priming results from the cooperation between complexin and the Ca2+-sensor synaptotagmin to regulate neuronal SNAREs [25,37]. Complexin binds trans-SNARE complexes and appears to wedge a helix into the C-terminal region of the forming complex, thus sterically inhibiting the completion of SNARE folding [38,39]. Upon arrival of an action potential and Ca2+ influx, Ca2+ binds to synaptotagmin, a conformational change occurs that leads to the release of complexin, completion of SNARE assembly and fusion of the membranes. Recently, it was shown that synaptotagmin draws the opposing membranes together via electrostatic interactions while simultaneously removing complexin from blocking the completion of SNARE zippering [40*]. The exact nature of this synaptotagmin-induced structural change remains elusive. Additionally, crystallographic studies suggested that complexin might cross-link adjacent clamped SNARE complexes. Release of a single complexin may thus destabilize the remaining complexin-SNARE interactions, allowing for cooperative membrane fusion by the adjoining SNARE complexes [39,41]; while intriguing, this idea remains controversial [42].
SNARE disassembly
The post-fusion assembled SNARE complex is very stable. Given this stability, the complex will not spontaneously disassemble, which is required for the recycling and/or turnover of v- and t-SNAREs. As a result, the post-fusion SNARE complex must be mechanically disassembled. Two studies on the two proteins responsible for this disassembly reaction, the AAA+ ATPase N-ethylmaleimide sensitive factor (NSF) and α-SNAP (soluble NSF-attachment protein), provide exciting new insights into this mechanism [43**,44].
A combination of structural and single molecule microscopic experiments elucidate how post-fusion SNAREs are disassembled. High-resolution cryogenic electron microscopy was used by Zhao et al. to determine structures of the SNARE-αSNAP-NSF supercomplex in both the ADP- and ATP-bound states, resolving conformational changes between the two nucleotide states (Fig. 2) [44]. The structure reveals that NSF can adopt two states, an ATP-bound split washer conformation and an ADP-bound flat, open-washer conformation. This spring-loaded state change forces α-SNAP to alter its conformation as well, inducing a shearing motion in the SNARE complex. The authors propose that this motion induces SNARE unwinding and dissociation.
Fig 2.
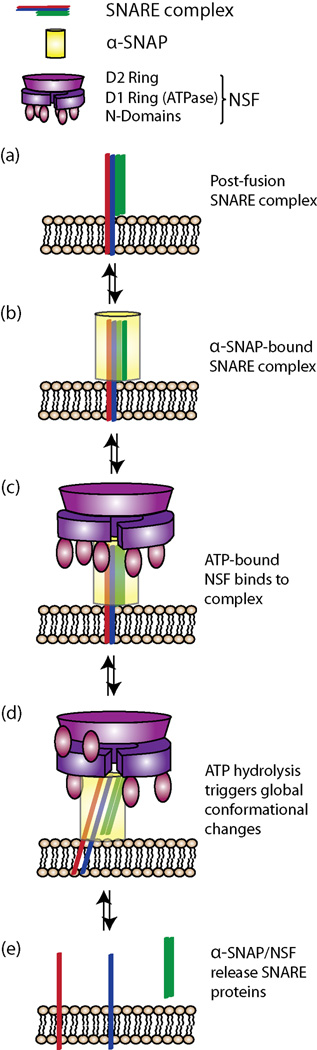
α-SNAP/NSF drive ATP-dependent cis-SNARE complex disassembly via shearing motions. (a) Post-fusion SNARE complex is shown as four parallel rods (omitting the SNAP-25 loop) for clarity of presentation. (b) Four α-SNAP monomers combine to form a collar around the SNARE complex [44**]. (c) ATP-bound NSF binds to the SNARE-α-SNAP complex. (d) A single round of ATP hydrolysis is sufficient to induce structural changes in NSF and α-SNAP, including two of the N-domains binding the D2 ring and the concomitant shifting of α-SNAP. These motions are sufficient to induce shearing and twisting motions in the SNARE complex. The shearing and twisting motions disrupt the inter-helical interactions and destabilize the four-helix bundle, driving disassembly (e). The exact order of release of the SNARE proteins remains unknown.
In a complementary study by Ryu et al., single molecule microscopy revealed that SNARE disassembly can be induced by a single round of ATP hydrolysis; the actual number of subunits that must hydrolyze ATP remains unclear [43**]. Another recent biochemical study suggests that hydrolysis of as few as 10 molecules of ATP are needed per disassembly reaction [45]. Additionally, single molecule Fӧrster Resonance Energy Transfer experiments on SNAREs during the α-SNAP/NSF-mediated disassembly reaction [43**] supported the shearing motion described in [44**]. A more detailed inspection of the kinetics from the single molecule experiments in [38] will be necessary to delve deeper into the mechanism, particularly with respect to conformations observed by Zhao et al [44**].
Disassembly regulation and moonlighting functions of the machinery
The requirement for α-SNAP/NSF in SNARE disassembly has been known for decades [46,47], but recent evidence suggests that their involvement in membrane fusion might be more complicated than originally proposed. Studies of the yeast homologs to α-SNAP and NSF (Sec17 and Sec18, respectively) demonstrated that Sec17 can associate with SNARE complexes and recruit Sec1/Munc18 regulatory proteins to inhibit disassembly by Sec18 [48]. Furthermore, bulk fusion assays demonstrated that Sec17 can enhance SNARE-mediated membrane fusion in the absence of Sec18 [49*]. Mutational analysis indicated that this stimulatory effect was dependent on a hydrophobic loop on Sec17, leading to the proposal that the effect was caused by a direct interaction of Sec17 with both target and vesicle membranes [40*]. The in vivo implications of this work are unknown, but it is clear that our understanding of the disassembly machinery remains incomplete.
Conclusions
Recent biophysical and structural characterizations of SNARE proteins and their regulators provided critical insights into how these molecules orchestrate the fundamental process of membrane fusion. Single molecule studies defined potentially important regulatory intermediates in the SNARE assembly process and showed that these intermediates are conserved in different cell types. Extension of these studies to include regulatory factors, such as synaptotagmin and complexin, will be important to our understanding of highly regulated membrane fusion reactions. Moreover, high-resolution structural electron cryomicroscopy and single molecule microscopy were integral in elucidating key aspects of the SNARE complex disassembly process. These studies revealed that a single round of ATP hydrolysis coupled to conformational changes in α-SNAP/NSF led to a shearing and twisting motion that could unravel and destabilize the incredibly stable SNARE complex. Surprisingly, biochemical characterization found moonlighting functions for the disassembly machinery: they appear to promote assembly under specific conditions. Further improvements in single molecule microscopy and electron cryomicroscopy, combined with careful quantitative analyses, will improve our understanding of the molecular mechanisms of the membrane fusion and SNARE complex disassembly machinery.
Highlights.
SNARE complex formation drives membrane fusion
Complex formation occurs with intermediates, potentially important for regulation
α-SNAP/NSF disassemble SNAREs by inducing shearing/rotation with ATP hydrolysis
Complexin/Synaptotagmin regulate vesicle release via conformational changes
Acknowledgments
We thank Michelle Dubuke, Margaret Heider, Anne Mirza, and Alexandra D’Ordine for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM068803 to M.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Rizo J, Sudhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- 4.Sudhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudhof TC. A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat Med. 2013;19:1227–1231. doi: 10.1038/nm.3338. [DOI] [PubMed] [Google Scholar]
- 6.Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 9.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 10.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 11.Morgera F, Sallah MR, Dubuke ML, Gandhi P, Brewer DN, Carr CM, Munson M. Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Mol Biol Cell. 2012;23:337–346. doi: 10.1091/mbc.E11-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murthy M, Garza D, Scheller RH, Schwarz TL. Mutations in the exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron. 2003;37:433–447. doi: 10.1016/s0896-6273(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 13.Inoue M, Chang L, Hwang J, Chiang SH, Saltiel AR. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- 14.Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22:488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Wang S, Sheng Y, Zhang M, Zou W, Wu L, Kang L, Rizo J, Zhang R, Xu T, et al. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol. 2015;22:547–554. doi: 10.1038/nsmb.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger AT. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- 17.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 19.Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 20.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 21.Lauer JM, Dalal S, Marz KE, Nonet ML, Hanson PI. SNARE complex zero layer residues are not critical for N-ethylmaleimide-sensitive factor-mediated disassembly. J Biol Chem. 2006;281:14823–14832. doi: 10.1074/jbc.M512706200. [DOI] [PubMed] [Google Scholar]
- 22.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 23.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 24.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 25.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Min D, Kim K, Hyeon C, Cho YH, Shin YK, Yoon TY. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism. Nat Commun. 2013;4:1705. doi: 10.1038/ncomms2692. Magnetic tweezer experiments demonstrate a single folding intermediate in SNARE complex assembly. This data corroborates earlier studies indicating an intermediate in neuronal SNARE complex formation that could be important for the regulation of SNARE complex formation in neurons (i.e. the insertion occlusion of complete formation of the four-helix bundle).
- 28.Zorman S, Rebane AA, Ma L, Yang G, Molski MA, Coleman J, Pincet F, Rothman JE, Zhang Y. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. Elife. 2014;3:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz ML, Merz AJ. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol. 2009;185:535–549. doi: 10.1083/jcb.200811082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fasshauer D, Antonin W, Subramaniam V, Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat Struct Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 31.Bowen ME, Weninger K, Brunger AT, Chu S. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs) Biophys J. 2004;87:3569–3584. doi: 10.1529/biophysj.104.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T, Tucker WC, Bhalla A, Chapman ER, Weisshaar JC. SNARE-driven, 25-millisecond vesicle fusion in vitro. Biophys J. 2005;89:2458–2472. doi: 10.1529/biophysj.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domanska MK, Kiessling V, Stein A, Fasshauer D, Tamm LK. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J Biol Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karatekin E, Di Giovanni J, Iborra C, Coleman J, O'Shaughnessy B, Seagar M, Rothman JE. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc Natl Acad Sci U S A. 2010;107:3517–3521. doi: 10.1073/pnas.0914723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nofal S, Becherer U, Hof D, Matti U, Rettig J. Primed vesicles can be distinguished from docked vesicles by analyzing their mobility. J Neurosci. 2007;27:1386–1395. doi: 10.1523/JNEUROSCI.4714-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heuser JE, Reese TS, Landis DM. Functional changes in frog neuromuscular junctions studied with freeze-fracture. J Neurocytol. 1974;3:109–131. doi: 10.1007/BF01111936. [DOI] [PubMed] [Google Scholar]
- 37.Munson M. Synaptic-vesicle fusion: a need for speed. Nat Struct Mol Biol. 2015;22:509–511. doi: 10.1038/nsmb.3056. [DOI] [PubMed] [Google Scholar]
- 38.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 39.Kummel D, Krishnakumar SS, Radoff DT, Li F, Giraudo CG, Pincet F, Rothman JE, Reinisch KM. Complexin cross-links prefusion SNAREs into a zigzag array. Nat Struct Mol Biol. 2011;18:927–933. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brewer KD, Bacaj T, Cavalli A, Camilloni C, Swarbrick JD, Liu J, Zhou A, Zhou P, Barlow N, Xu J, et al. Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution. Nat Struct Mol Biol. 2015;22:555–564. doi: 10.1038/nsmb.3035. Advanced NMR techniques were used to elucidate the binding site of the neuronal SNARE regulatory factor synaptotagmin. Based on their in vitro structural and functional work in neurons, the authors propose a model where Ca2+-binding to synaptotagmin alters synaptotagmin’s binding to SNAREs to drive full SNARE complex assembly for membrane fusion
- 41.Krishnakumar SS, Radoff DT, Kummel D, Giraudo CG, Li F, Khandan L, Baguley SW, Coleman J, Reinisch KM, Pincet F, et al. A conformational switch in complexin is required for synaptotagmin to trigger synaptic fusion. Nat Struct Mol Biol. 2011;18:934–940. doi: 10.1038/nsmb.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trimbuch T, Xu J, Flaherty D, Tomchick DR, Rizo J, Rosenmund C. Re-examining how complexin inhibits neurotransmitter release. Elife. 2014;3:e02391. doi: 10.7554/eLife.02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ryu JK, Min D, Rah SH, Kim SJ, Park Y, Kim H, Hyeon C, Kim HM, Jahn R, Yoon TY. Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science. 2015;347:1485–1489. doi: 10.1126/science.aaa5267. Single molecule fluorescence experiments revealed that α-SNAP/NSF-mediated SNARE complex disassembly occurs via a single step and utilizes only a single round of ATP hydrolysis. This study speaks to the energetic and kinetic mechanism of SNARE disassembly by α-SNAP and NSF.
- 44. Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. Ultra-high resolution cryoEM structures of α-SNAP/NSF in different nucleotide states indicated large conformational changes, which suggests a mechanism for α-SNAP/NSF to induce shearing and rotation of the SNARE complex for SNARE disassembly.
- 45.Shah N, Colbert KN, Enos MD, Herschlag D, Weis WI. Three alphaSNAP and 10 ATP molecules are used in SNARE complex disassembly by N-ethylmaleimide-sensitive factor (NSF) J Biol Chem. 2015;290:2175–2188. doi: 10.1074/jbc.M114.620849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H. Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. EMBO J. 1995;14:2317–2325. doi: 10.1002/j.1460-2075.1995.tb07226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Littleton JT, Barnard RJ, Titus SA, Slind J, Chapman ER, Ganetzky B. SNARE-complex disassembly by NSF follows synaptic-vesicle fusion. Proc Natl Acad Sci U S A. 2001;98:12233–12238. doi: 10.1073/pnas.221450198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. Elife. 2014;3:e02272. doi: 10.7554/eLife.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zick M, Orr A, Schwartz ML, Merz AJ, Wickner WT. Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc Natl Acad Sci U S A. 2015;112:E2290–E2297. doi: 10.1073/pnas.1506409112. This study shows that the yeast α-SNAP homolog Sec17, can also promote SNARE complex assembly. This work could have interesting mechanistic implications in fast neuronal membrane fusion if an analogous function is indeed discovered for α-SNAP.