Abstract
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is a common birth defect of complex etiology. Several genes have been implicated in the etiology of NSCL/P, although only a few have been replicated across datasets. ARHGAP29 was suggested as a candidate gene for NSCL/P as it is located in close proximity to ABCA4 (1p22), a gene previously identified in a GWAS of NSCL/P. Rare, potentially damaging, coding variants in ARHGAP29 were found in NSCL/P cases, and its expression was detected during murine craniofacial development. In this study, we investigated whether variations in ARHGAP29 were associated with NSCL/P in our family based dataset. Five SNPs flanking and within ARHGAP29 were genotyped in our NSCL/P datasets consisting of simplex and multiplex families of non Hispanic white (NHW, primarily European) and Hispanic ethnicities. Results showed strong association of three ARHGAP29 SNPs with NSCL/P in the NHW families. Two intronic SNPs (rs1541098 and rs3789688) showed strong association with NSCL/P in all NHW families (P=0.0005 and P=0.0002, respectively), and simplex NHW families (P=0.003 for both SNPs). A SNP in the 3’ UTR (rs1576593) also showed strong association with NSCL/P in all NHW families (P=0.002), and the multiplex subset (P=0.002). ARHGAP29 SNP haplotypes were also associated with NSCL/P. Evidence of gene-gene interaction was found between ARHGAP29 and additional cleft susceptibility genes. This study further supports ARHGAP29 as a candidate gene for human NSCL/P in families of Caucasian descent.
Keywords: cleft lip/palate, ARHGAP29, association, haplotype, gene-gene interaction
INTRODUCTION
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is a common birth defect, notable for its significant lifelong morbidity and complex etiology. It affects 135,000 newborns worldwide with wide variability related to geographic origin and socioeconomic status. In general, Native American and Asian populations present the highest frequencies, sometimes at 1/500 or higher, followed by Caucasian, and African-derived populations showing the lowest frequencies at approximately 1/2500 births (Mossey et al., 2009).
The complex etiology of NSCL/P reflects multiple genetic and environmental factors acting individually or in concert. A variety of research approaches including candidate gene, genome-wide linkage, and genome-wide association studies (GWAS), have been used to identify the etiologic genes contributing to NSCL/P. While numerous genes and specific polymorphic variants have been suggested to confer an increased risk of NSCL/P, only a few have been replicated across datasets (Dixon et al., 2011). Of note, the association of IRF6 gene variants with NSCL/P has been confirmed in multiple populations (Dixon et al., 2011) and have been suggested to explain about ~12% of the genetic contribution to NSCL/P (Zucchero et al., 2004). Additionally, a gene desert region on chromosome 8q24 was consistently associated with NSCL/P in different GWA studies (Beaty et al., 2010; Birnbaum et al., 2009; Grant et al., 2009; Mangold et al., 2010), and independently replicated in various populations (Bagordakis et al., 2013; Beaty et al., 2013; Blanton et al., 2010; Brito et al., 2012; Lennon et al., 2012; Mostowska et al., 2010; Nikopensius et al., 2009; Rojas-Martinez et al., 2010; Velazquez-Aragon et al., 2012; Wang et al., 2012).
ARHGAP29 was suggested as a candidate gene for NSCL/P because it is located in close proximity to ABCA4 (1p22), a gene identified in a previous GWAS of NSCL/P (Leslie et al., 2012). Despite identifying a number of missense mutations in ABCA4 in individuals with NSCL/P, ABCA4 is unlikely to be the etiologic gene for clefting because expression is restricted to the retina (Beaty et al., 2010). Hence, it has been hypothesized that a neighboring gene, ARHGAP29, may be the true etiologic gene or locus. Rare, potentially damaging, coding variants in ARHGAP29 were found in NSCL/P cases, and its expression was detected during craniofacial development in mice (Leslie et al., 2012). ARHGAP29 encodes a Rho GTPase activating protein (GAP) 29 and is involved in essential cellular functions that are critical for craniofacial development (Mossey et al., 2009), and may be a downstream effector of Tgfb and Wnt (Kardassis et al., 2009; Schlessinger et al., 2009) signaling pathways, which are also involved in craniofacial development. This suggests that perturbation of ARHGAP29 may play a role in NSCL/P. In this study, we investigated whether variations in ARHGAP29 were associated with NSCL/P in families of Hispanic and non Hispanic white (NHW) ethnicities.
MATERIALS AND METHODS
Study Population
Our study population consisted of simplex and multiplex families of NHW (primarily European) (n=507) and Hispanic (primarily Mexican) (n=314) ethnicities. Details of the study families are presented in Table I. Families were ascertained through probands, and additional relatives were recruited. Individuals presenting with syndromic clefts, cleft palate only, or unknown cleft types were excluded. This study was approved by the University of Texas Health Science Center Committee for Protection of Human Subjects.
Table I.
Description of NSCL/P families
| Ethnic group | Mean pedigree size (min-max) |
Pedigrees | Unaffecteds typed |
Affecteds typed |
Total individuals |
|---|---|---|---|---|---|
| Hispanic | 2.8 (2–11) | 314 | 327 | 564 | 891 |
| NHW | 3.1 (2–32) | 507 | 575 | 998 | 1573 |
NHW = non Hispanic white.
Genotyping
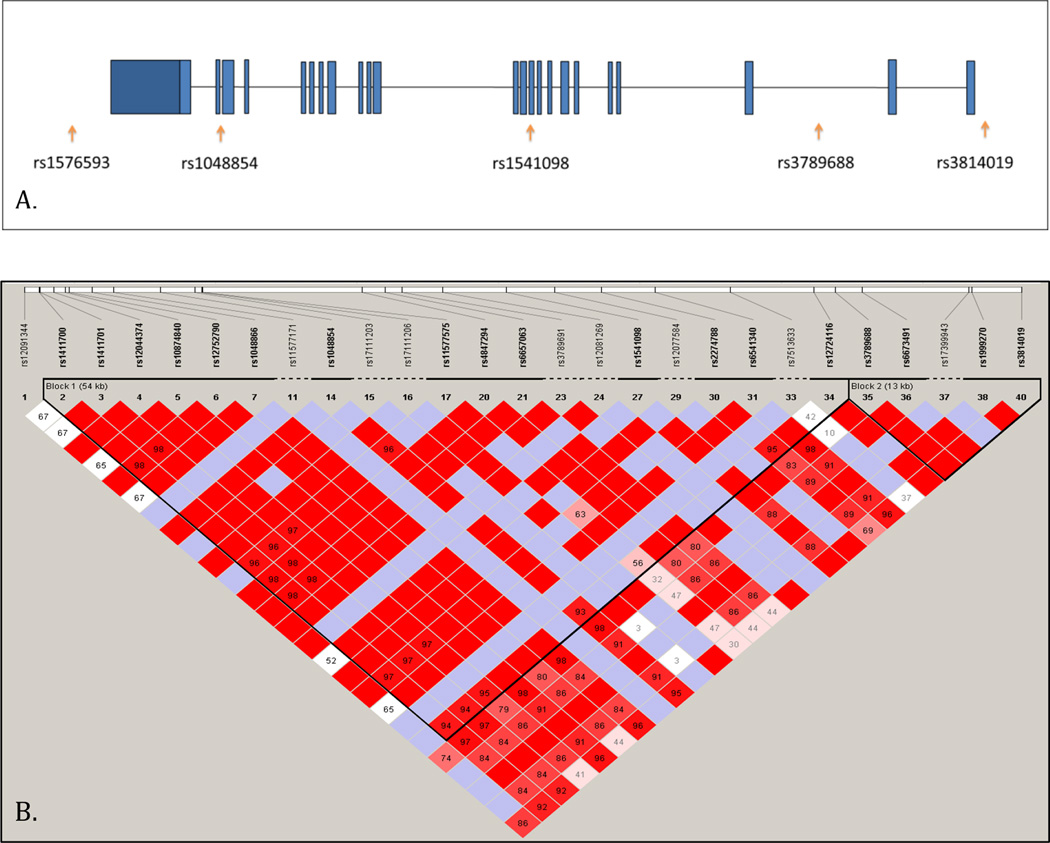
Five flanking and intragenic ARHGAP29 SNPs were genotyped in our NSCL/P datasets (Table II). SNPs were selected as tag-SNPs, considering heterozygosity values, gene structure, and the linkage disequilibrium block surrounding each gene, as previously described (Carlson et al., 2004) (Figure 1). Genotypes were generated using Taqman chemistry (Ranade et al., 2001) on an automatic sequence-detection instrument (ABI Prism 7900HT, Applied Biosystems, Foster City, CA).
Table II.
ARHGAP29 SNPs genotyped
| SNP Id. | Base position* | Function | Alleles** |
|---|---|---|---|
| rs3814019 | 94704396 | 5' near gene | A/G |
| rs3789688 | 94691240 | Intron | A/G |
| rs1541098 | 94667970 | Intron | T/C |
| rs1048854 | 94643531 | Coding synonymous | A/G |
| rs1576593 | 94631751 | 3' UTR | T/C |
According to NCBI dbSNP Build 137.
Ancestral allele listed first.
Figure 1.
Schematic representation of the ARHGAP29 gene structure and linkage disequilibrium plot. A. Gene structure of ARHGAP29 with colored bars representing exons, horizontal lines representing introns. Arrows denote location of genotyped SNPs. B. Linkage disequilibrium plot with boxes representing the marker pair relationship plotted between two markers. The color of the boxes (intensity of the color) is based on the raw score for that marker pair. Squares are colored darker if the D’ value is high (i.e., LD is strong), lighter shades indicate less LD. Empty dark squares mean D’= 1 (i.e., complete LD between two SNPs).
Statistical Analysis
Family-based single SNP association tests were performed using CAPL (Chung et al., 2011). Analysis were performed for all families stratified by ethnicity and then stratified by ethnicity and family history of NSCL/P. Pairwise haplotype analysis was performed using APL (Chung et al., 2006). Bonferroni correction was used to adjust for multiple testing and P-values ≤0.01 were considered statistically significant for the single SNP analyses.
Gene-Gene Interaction Analysis
We examined ARHGAP29 SNP interactions with genes recently suggested to be involved with NSCL/P in humans and/or mice: IRF6 (Zucchero et al., 2004), P63 (Leoyklang et al., 2006), PBX1 and PBX2 (Ferretti et al., 2011), WNT3 and WNT9B (Juriloff and Harris, 2008). Transmission of all possible intergenic 2-SNP pairs was examined with APL (Chung et al., 2010; Chung et al., 2011). Genotype data for twenty-three SNPs in these genes were used for the gene-gene interaction analyses (Supplementary Material).
In Silico Prediction of SNP Function
AliBaba2.1 and PATCH (www.gene-regulation.com), SNP function prediction methods, were used to determine whether the genotyped ARHGAP29 SNPs harbored transcription factor binding sites.
RESULTS
Overall, we found strong association with individual ARHGAP29 SNPs and haplotypes in our NHW dataset. One SNP in the 3’UTR (rs1576593) and two intronic SNPs (rs1541098 and rs3789688) showed evidence for association (p=0.002, p=0.0005, and p=0.0002, respectively) with NSCL/P in the NHW dataset. When stratified by family history, only the association with rs1576593 remained significant in the multiplex families (p=0.002), whereas rs1541098 and rs3789688 remained significant in the simplex families (p=0.003 for both) (Table III). No associations were found for ARHGAP29 SNPs and NSCL/P in Hispanics (Table III).
Table III.
Association results by ethnicity
| Families | SNP | P-value* | |
|---|---|---|---|
| NHW | Hispanic | ||
| All | rs1576593 | 0.002 (T) | 0.13 |
| rs1541098 | 0.0005 (T) | 0.96 | |
| rs3789688 | 0.0002 (A) | 0.79 | |
| Multiplex | rs1576593 | 0.002 (T) | 0.03 (C) |
| Simplex | rs1541098 | 0.003 (T) | 0.58 |
| rs3789688 | 0.003 (A) | 0.86 | |
APL test, p<0.01 indicates statistical significance; Letters in parenthesis indicate overtransmitted allele.
NHW = non Hispanic white
Haplotype analyses detected modest evidence for association of ARHGAP29 SNP alleles with NSCL/P in the NHW dataset. In the pooled dataset, associations were detected for several haplotypes: rs1576593 and rs3789688 (p=0.003), rs1541098 and rs3814019 (p=0.006), and rs1541098 and rs3789688 (p=0.008) (Table IV). There was evidence for altered transmission of the rs1541098 and rs3789688 haplotypes in the multiplex families (p=0.003), whereas in simplex families there was evidence for altered transmission of several haplotypes: rs1576593 and rs1048854 (p=0.004), rs1048854 and rs3789688 (p=0.009), and rs1048854 and rs1541098 (p=0.009).
Table IV.
NHW haplotype results
| Families | ARHGAP29 | Haplotypes with Greatest Altered Transmission |
Global p-value* | |
|---|---|---|---|---|
| SNP1 | SNP2 | |||
| ALL | rs1541098 | rs3814019 | C -G↓ | 0.008 |
| rs1541098 | rs3789688 | T - G↑ C - A↓ |
0.006 | |
| rs1576593 | rs3789688 | T - A↓ | 0.003 | |
| MULTIPLEX | rs1541098 | rs3789688 | T - G↑ | 0.003 |
| SIMPLEX | rs1048854 | rs3789688 | A - A↓ G - G↑ |
0.009 |
| rs1048854 | rs1541098 | G - T↑ A - C↓ |
0.009 | |
| rs1576593 | rs1048854 | T - A↓ C - G↑ |
0.004 | |
APL test, all p<0.01 included.
Gene-gene interaction analyses for ARHGAP29 and additional cleft susceptibility genes showed evidence for statistical interaction in the pooled NHW families. Significant interactions were observed between ARHGAP29 rs1576593 and PBX1 rs6426870 (p=0.0002) in simplex families, as well as between ARHGAP29 rs3789688 and P63 rs4575879 (p=0.00001), PBX2 rs204993 (p=0.0004), WNT3 rs7216231 (p=0.0001), and WNT9B rs12602434 and rs1530364 (p=0.0001 and p=0.0004, respectively) in all NHW families (Table V).
Table V.
NHW gene-gene interaction results
| Gene/SNP | ARHGAP29 SNP | ||||||
|---|---|---|---|---|---|---|---|
| rs3789688 | rs1541098 | rs1576593 | |||||
| ALL | Simplex | ALL | Simplex | ALL | Simplex | ||
| PBX1 | rs6426870 | --- - | 0.003 | ---- | 0.003 | ---- | 0.0002 |
| P63 | rs4575879 | 0.00001 | 0.001 | 0.001 | 0.003 | 0.003 | ---- |
| PBX2 | rs204993 | 0.0004 | 0.0045 | ---- | 0.007 | 0.003 | ---- |
| WNT3 | rs7216231 | 0.0001 | 0.002 | 0.004 | 0.008 | 0.007 | ---- |
| WNT9B | rs12602434 | 0.0001 | ---- | 0.006 | ---- | ---- | ---- |
| WNT9B | rs1530364 | 0.0004 | 0.004 | 0.001 | ---- | 0.007 | ---- |
Only interactions with at least one P-value ≤ 0.005 are reported.
NHW = non Hispanic white
The results of the in silico analyses showed differential binding partners for each allele in rs3814019 and in rs1576593 located in the 5’ and 3’ UTR of ARHGAP29, respectively (Table VI). While the ancestral allele of rs3814019 was predicted to bind to the transcription factors GATA-3, CPC1, and p40X, the alternate allele did not harbor biding sites for these transcription factors. Rather, it was predicted to bind to ELF-1, ISGF-3, NF-Kb, and RelA. For rs1576593, the ancestral allele T, associated with NSCL/P, was predicted to bind to a micro-RNA (mIR-1194), whereas analyses for the alternate allele using both prediction programs failed to identify a transcription factor-binding site.
DISCUSSION
Recent evidence suggests a role for ARHGAP29 in NSCL/P based on craniofacial expression during murine development and identification of several variants in ARHGAP29, that collectively were overrepresented in cases with NSCL/P from Filipino and US populations, compared with unaffected controls (Leslie et al., 2012). In this study, we replicate the association of ARHGAP29 gene variants with NSCL/P in our large family-based NHW dataset, further supporting the suggestion that ARHGAP29 may be a cleft susceptibility gene. A SNP in the 3’UTR (rs1576593) and two intronic SNPs (rs1541098 and rs3789688) showed strong association with NSCL/P in all NHW families. Notably, the association with rs1576593 was stronger in multiplex families, whereas the association with rs1541098 and rs3789688 was stronger in simplex families. Although these latter two SNPs are located in different LD blocks (Figure 1), they are in strong LD with each other and may be transmitting the same information. No individual associations were observed for SNPs rs3814019 and rs1048854, nonetheless altered transmission of haplotypes including these SNPs were also associated with NSCL/P in the NHW dataset. While rs3814019 is located in the 5’ UTR of the ARHGAP29 gene with potential regulatory effects on gene transcription, rs1048854 is a synonymous variant (Gln891Gln) in exon 21 with no known impact on the final protein.
Although intronic variants are unlikely to have effects on gene transcription and/or on the final protein structure, variants located in regulatory regions in many cases are known to have deleterious effects. Our in silico analysis of potential regulatory SNPs in the 5’ and 3’ UTR of ARHGAP29 (rs3814019 and rs1576593, respectively) showed distinct allele-specific binding partners. The ancestral allele A was predicted to bind to GATA-3 (GATA binding protein 3), a member of the GATA family of transcription factors, which acts as an enhancer-binding protein and regulator of T-cell development with an important role in endothelial cell biology (Song et al., 2009). In contrast, the alternate allele G was predicted to bind to transcription factors (ISGF3, NFKB, and RELA) that act both as enhancers and as repressors to regulate transcription of various genes. ISGF3 (interferon stimulated gene factor 3) comprises a gene complex, which in response to stimuli such as cytokines and growth factors, are phosphorylated and translocate to the cell nucleus to act as transcriptional activators (Bluyssen et al., 1996). Similarly, NFKB (nuclear factor kappa-B) and RELA (v-rel avian reticuloendotheliosis viral oncogene homolog A) belong to the NFKB family, a protein complex that controls DNA transcription and represent the endpoint of a series of signal transduction events that are initiated by a vast array of stimuli related to many biological processes such as inflammation, immunity, differentiation, cell growth, tumorigenesis and apoptosis. NFKB binds to the Rel-like domain-containing proteins (i.e., RELA/p65, RELB, REL) and form dimers that bind at kappa-B sites in the DNA of their target genes; and the different dimer combinations act as transcriptional activators or repressors, respectively (Rayet and Gelinas, 1999). For the 3’ UTR SNP rs1576593, the ancestral allele T, associated with NSCL/P, was predicted to present a microRNA (mIR-1194) binding site, whereas analyses for the alternate allele using both prediction programs failed to demonstrate a transcription factor-binding site. This suggests that rs1576593 may have a regulatory role in post-transcriptional regulation of ARHGAP29 gene expression. In mouse, mIR-1194 presents 169 conserved sites including a site for fibroblast growth factor 5 (FGF5) (Calabrese et al., 2007). FGF5 is a member of the fibroblast growth factor signaling pathway, which is involved in several aspects of craniofacial development, including formation of the lip and the palate (Ferguson, 1988). Many FGF ligands and receptors are expressed at specific stages and at precise locations during normal palatogenesis and an absolute requirement of some has been demonstrated by their (conditional) inactivation resulting in a cleft palate phenotype (Lee et al., 2001; Rice et al., 2004). In humans, FGF gene variants have also been associated with NSCL/P (Menezes et al., 2008; Nikopensius et al., 2011; Riley et al., 2007; Riley and Murray, 2007; Wang et al., 2013).
In addition to the allele and haplotype associations found for ARHGAP29 with NSCL/P, we detected evidence of statistical interaction between ARHGAP29 and other cleft susceptibility genes (IRF6, P63, PBX1, PBX2, WNT3, and WNT9B) identified in previous human and mouse studies (Ferretti et al., 2011; Juriloff and Harris, 2008; Leoyklang et al., 2006; Zucchero et al., 2004). In complex disorders such as NSCL/P, gene-gene interactions should be considered as additional thresholds for genetic predisposition (Cordell, 2009). While most genetic studies have used a single-locus analysis strategy, whereby each variant is tested individually for association with some complex phenotypes, an oft-cited reason for the lack of success in genetic studies of these disorders is the challenge of identifying interactions between loci. If a genetic factor operates primarily through a complex mechanism involving multiple other genes, the concern is that the effect will be missed if one examines it in isolation, without allowing for its potential interactions with other genetic factors (Cordell, 2009). Therefore testing for gene-gene interactions might increase the power to detect effects, and also statistical interactions between loci that are informative about the biological and biochemical pathways underpinning NSCL/P. Our results suggested possible allelic interactions between ARHGAP29 and WNT3, WNT9B, PBX1, PBX2, and P63 genes in the NHW dataset. WNT3 and WNT9B belong to the wingless-type MMTV integration site (Wnt) signaling pathway, which plays an important role in craniofacial development. Wnt signaling genes are conserved among species and are essential to the development of several processes, including face morphogenesis (Jiang et al., 2006). Wnt3 and Wnt9b are located on chromosome 11 within the clf1 locus, which is associated with the spontaneous development of cleft lip and/or cleft palate in A/WySn mouse strains (Juriloff et al., 2005, 2006). In addition, polymorphic variants in numerous WNT genes including WNT3 and WNT9B have been associated with NSCL/P in different populations (Chiquet et al., 2008; Fontoura et al., 2014; Menezes et al., 2010; Mostowska et al., 2012; Yao et al., 2011). Recent evidence has suggested that PBX1 and PBX2 genes are clefting genes in mice, possibly through interactions with Wnt9b and p63 (Ferretti et al., 2011). Inactivation of Pbx genes in the epithelia and mesenchyme of the craniofacial region, as observed in compound Pbx1−/−; Pbx2+/− (Pbx1/2) mutants, resulted in animals with broader face, mandibular hypoplasia, cleft lip, and cleft palate. Wnt3 and Wnt9b were markedly downregulated contributing to perturbed p63 expression in the midface of Pbx compound mutants. Further, a Pbx-dependent Wnt-p63-Irf6 regulatory module was suggested, which when disrupted, led to localized suppression of apoptotic programs and CL/P (Ferretti et al., 2011). Taken together, these observations highlight the complex etiology of NSCL/P and point towards the need for considering genetic interactions in studies of NSCL/P. Of note, no evidence of interaction was found between ARHGAP29 and IRF6. IRF6 is suggested to be one of the largest contributors to the underlying genetics of human NSCL/P, contributing to as much as 12% of the genetic risk for clefting (Zucchero et al., 2004). This is an intriguing finding as Arhgap29 was reported to act downstream of Irf6, showing decreased expression in the palatal epithelium and skin of Irf6 deficient mice (Leslie et al., 2012).
In summary, we provide further evidence for the role of ARHGAP29 in NHW families with NSCL/P but not Hispanic families. Studies in additional families from other populations are needed to determine the role of this gene across populations. Importantly, we found evidence of genetic interactions involving ARHGAP29, PBX1, PBX2, P63, WNT3, and WNT9B indicating that variants in these genes and their interactions should also be evaluated across populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank the study families for their participation. We thank Maria Elena Serna, Rosa Martinez, and Dr. Syed Hashmi for recruiting patients and managing our patient database. This study was supported by NIH grants R01-DE11931 (to JTH and SHB) and R00-DE018954 (to AL).
Footnotes
The authors have no conflict of interest to declare.
REFERENCES
- Bagordakis E, Paranaiba LM, Brito LA, de Aquino SN, Messetti AC, Martelli-Junior H, et al. Polymorphisms at regions 1p22.1 (rs560426) and 8q24 (rs1530300) are risk markers for nonsyndromic cleft lip and/or palate in the Brazilian population. Am J Med Genet. 2013;161A:1177–1180. doi: 10.1002/ajmg.a.35830. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013;132:771–781. doi: 10.1007/s00439-013-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Blanton SH, Burt A, Stal S, Mulliken JB, Garcia E, Hecht JT. Family-based study shows heterogeneity of a susceptibility locus on chromosome 8q24 for nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol. 2010;88:256–259. doi: 10.1002/bdra.20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluyssen AR, Durbin JE, Levy DE. ISGF3 gamma p48, a specificity switch for interferon activated transcription factors. Cytokine Growth Factor Rev. 1996;7:11–17. doi: 10.1016/1359-6101(96)00005-6. [DOI] [PubMed] [Google Scholar]
- Brito LA, Paranaiba LM, Bassi CF, Masotti C, Malcher C, Schlesinger D, et al. Region 8q24 is a susceptibility locus for nonsyndromic oral clefting in Brazil. Birth Defects Res A Clin Mol Teratol. 2012;94:464–468. doi: 10.1002/bdra.23011. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Nat Acad Sci USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, Ma D, Stal S, Mulliken JB, et al. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Molec Genet. 2008;17:2212–2218. doi: 10.1093/hmg/ddn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61:189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Chung RH, Schmidt MA, Morris RW, Martin ER. CAPL: a novel association test using case-control and family data and accounting for population stratification. Genet Epidemiol. 2010;34:747–755. doi: 10.1002/gepi.20539. [DOI] [PubMed] [Google Scholar]
- Chung RH, Schmidt MA, Martin ER. CAPL: an efficient association software package using family and case-control data and accounting for population stratification. BMC Bioinformatics. 2011;12:201. doi: 10.1186/1471-2105-12-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MW. Palate development. Development. 1988;103(Suppl):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Develop Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura C, Silva RM, Granjeiro JM, Letra A. Association of WNT9B Gene Polymorphisms With Nonsyndromic Cleft Lip With or Without Cleft Palate in Brazilian Nuclear Families. Cleft Palate Craniofac J. 2014 Jan 17; doi: 10.1597/13-146. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediat. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Develop Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ. Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82:63–77. doi: 10.1002/bdra.20430. [DOI] [PubMed] [Google Scholar]
- Kardassis D, Murphy C, Fotsis T, Moustakas A, Stournaras C. Control of transforming growth factor beta signal transduction by small GTPases. FEBS J. 2009;276:2947–2965. doi: 10.1111/j.1742-4658.2009.07031.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Crisera CA, Erfani S, Maldonado TS, Lee JJ, Alkasab SL, et al. Immunolocalization of fibroblast growth factor receptors 1 and 2 in mouse palate development. Plast Reconst Surg. 2001;107:1776–1784. doi: 10.1097/00006534-200106000-00021. [DOI] [PubMed] [Google Scholar]
- Lennon CJ, Birkeland AC, Nunez JA, Su GH, Lanzano P, Guzman E, et al. Association of candidate genes with nonsyndromic clefts in Honduran and Colombian populations. Laryngoscope. 2012;122:2082–2087. doi: 10.1002/lary.23394. [DOI] [PubMed] [Google Scholar]
- Leoyklang P, Siriwan P, Shotelersuk V. A mutation of the p63 gene in non-syndromic cleft lip. J Med Genet. 2006;43:e28. doi: 10.1136/jmg.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, et al. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol. 2012;94:934–942. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Menezes R, Letra A, Ruff J, Granjeiro JM, Vieira AR. Studies of genes in the FGF signaling pathway and oral clefts with or without dental anomalies. Am J Med Genet A. 2008;146A:1614–1617. doi: 10.1002/ajmg.a.32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R, Letra A, Kim AH, Kuchler EC, Day A, Tannure PN, et al. Studies with Wnt genes and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol. 2010;88:995–1000. doi: 10.1002/bdra.20720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374:1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- Mostowska A, Hozyasz KK, Wojcicki P, Biedziak B, Paradowska P, Jagodzinski PP. Association between genetic variants of reported candidate genes or regions and risk of cleft lip with or without cleft palate in the polish population. Birth Defects Res A Clin Mol Teratol. 2010;88:538–545. doi: 10.1002/bdra.20687. [DOI] [PubMed] [Google Scholar]
- Mostowska A, Hozyasz KK, Biedziak B, Wojcicki P, Lianeri M, Jagodzinski PP. Genotype and haplotype analysis of WNT genes in non-syndromic cleft lip with or without cleft palate. Eur J Oral Sci. 2012;120:1–8. doi: 10.1111/j.1600-0722.2011.00938.x. [DOI] [PubMed] [Google Scholar]
- Nikopensius T, Ambrozaityte L, Ludwig KU, Birnbaum S, Jagomagi T, Saag M, et al. Replication of novel susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24 in Estonian and Lithuanian patients. Am J Med Genet. 2009;149A(11):2551–2553. doi: 10.1002/ajmg.a.33024. [DOI] [PubMed] [Google Scholar]
- Nikopensius T, Kempa I, Ambrozaityte L, Jagomagi T, Saag M, Matuleviciene A, et al. Variation in FGF1, FOXE1, and TIMP2 genes is associated with nonsyndromic cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2011;91:218–225. doi: 10.1002/bdra.20791. [DOI] [PubMed] [Google Scholar]
- Ranade K, Chang MS, Ting CT, Pei D, Hsiao CF, Olivier M, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, et al. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, et al. Impaired FGF signaling contributes to cleft lip and palate. Proc Nat Acad Sci USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Murray JC. Sequence evaluation of FGF and FGFR gene conserved non-coding elements in non-syndromic cleft lip and palate cases. Am J Med Genet. 2007;143A:3228–3234. doi: 10.1002/ajmg.a.31965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Martinez A, Reutter H, Chacon-Camacho O, Leon-Cachon RB, Munoz-Jimenez SG, Nowak S, et al. Genetic risk factors for nonsyndromic cleft lip with or without cleft palate in a Mesoamerican population: Evidence for IRF6 and variants at 8q24 and 10q25. Birth Defects Res A Clin Mol Teratol. 2010;88:535–537. doi: 10.1002/bdra.20689. [DOI] [PubMed] [Google Scholar]
- Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Develop. 2009;23:265–277. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- Song H, Suehiro J, Kanki Y, Kawai Y, Inoue K, Daida H, et al. Critical role for GATA3 in mediating Tie2 expression and function in large vessel endothelial cells. J Biol Chem. 2009;284:29109–29124. doi: 10.1074/jbc.M109.041145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Aragon JA, Alcantara-Ortigoza MA, Estandia-Ortega B, Reyna-Fabian ME, Cruz-Fuentes C, Villagomez S, et al. Association of interactions among the IRF6 gene, the 8q24 region, and maternal folic acid intake with non-syndromic cleft lip/palate in Mexican Mestizos. Am J Med Genet. 2012;158A(12):3207–3210. doi: 10.1002/ajmg.a.35641. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang T, Wu T, Hetmanski JB, Ruczinski I, Schwender H, et al. The FGF and FGFR Gene Family and Risk of Cleft Lip With or Without Cleft Palate. Cleft Palate Craniofac J. 2013;50:96–103. doi: 10.1597/11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Pan Y, Zhang Z, Wang L. Three polymorphisms in IRF6 and 8q24 are associated with nonsyndromic cleft lip with or without cleft palate: evidence from 20 studies. Am J Med Genet. 2012;158A:3080–3086. doi: 10.1002/ajmg.a.35634. [DOI] [PubMed] [Google Scholar]
- Yao T, Yang L, Li PQ, Wu H, Xie HB, Shen X, et al. Association of Wnt3A gene variants with non-syndromic cleft lip with or without cleft palate in Chinese population. Arch Oral Biol. 2011;56:73–78. doi: 10.1016/j.archoralbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. New Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.