Abstract
MAIN-LIKE1 (MAIL1) is a ubiquitously expressed nuclear protein, which has a crucial function during root development. We have recently described loss of function mutants for MAIL1, in which the organization and function of the primary root meristem is lost soon after germination. Moreover cell differentiation is impaired resulting in primary root growth arrest soon after emergence. Here we show that mail1 mutants form several anchor roots from the hypocotyl to root junction. These anchor roots show similar defects in the organization of the stem cell niche as the primary root. In contrast, differentiation processes are not impaired and thus anchor roots seem to be able to compensate for the loss of primary root function. Our data show that MAIL1 is essential for specification of cell fate in the primary root but not in anchor roots.
Keywords: arabidopsis, anchor root, differentiation, primary root, root development
We have recently identified a so far uncharacterized protein subfamily in Arabidopsis thaliana that contains 4 members which we named MAIN, MAIL1, MAIL2 and MAIL3 (MAINTENANCE OF MERISTEMS-LIKE). The MAIN protein family is characterized by a conserved aminotransferase-like, plant mobile domain. We have described loss-of-function mutants for MAIN (main-1 and main-2) and MAIL1 (mail1) which both show strong developmental defects during primary root development. Our detailed studies of the mail1 mutant phenotype revealed that the organization of the stem cell niche in the RAM was lost soon after germination and stem cells and differentiating cells were unable to acquire or maintain their specific cell fate. At 6 d after germination the mail1 primary root arrested growth as a short root-like structure that contained mainly dead or undifferentiated cells. Soon after primary root growth arrest, mail1 seedlings developed several anchor roots from the hypocotyl-root junction (Fig. 1A and B).1 It has been shown previously that in WT plants the formation of anchor roots can be induced by excising the tip of the primary root. This indicates that anchor root formation might represent a generic mechanism to compensate for the loss of primary root growth potential.2 Hence, the formation of anchor roots in the mail1 mutant might compensate for the progressive loss of primary root function. Here we analyzed the structure and function of anchor roots in the mail1 mutant in detail. WT plants produce normally 1 or 2 very short anchor roots from pericycle cells of the hypocotyl, in a similar way as lateral roots are formed from pericycle cells of the primary root.3 Mail1 mutants produced callus-like structures at the hypocotyl-root junction and between 5 to 10 anchor roots from these cells. (Fig. 1C and D).
Figure 1.
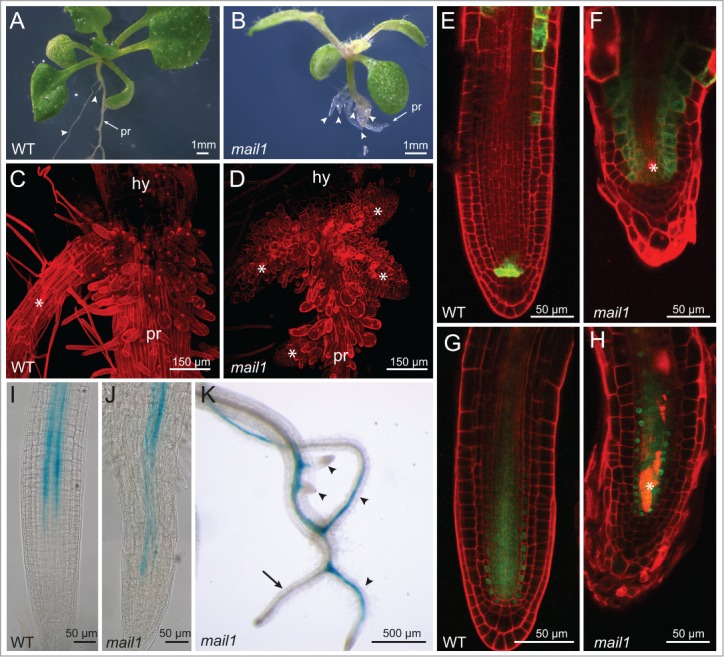
Microscopic studies of anchor root formation in mail1 seedlings. Eighteen-day-old WT seedling (A) and 18-day-old mail1 seedling (B), pr = primary root, arrowheads point to anchor roots. Confocal image of hypocotyl-root junction of a WT seedling (C) and of a mail1 seedling (D), hy = hypocotyl; asterix marks anchor roots. Confocal image of an anchor root of a WT (E) and of a mail1 seedling (F) expressing ProWOX5::erGFP (green) and stained with PtdIns (red), asterix indicates dead cells. Confocal image of an anchor root of a WT (G) and of a mail1 mutant (H) expressing ProSHR::SHR-GFP and stained with PI (red). Expression pattern of ProAPL::GUS in the primary root of a 3-day-old WT seedling (I) and of a 3-day-old mail1 seedling (J). (K) ProAPL::GUS expression in a 12-day-old mail1 seedling. Arrow indicates primary root, arrowheads point to anchor roots.
Confocal imaging of propidium iodide (PI) -stained root tips revealed that the RAM of anchor roots exhibited similar developmental defects as the RAM of the primary root: quiescent center cells (QC) and stem cells showed abnormal cell division patterns and the organization of the stem cell niche became progressively lost. Analysis of marker lines for genes essential for root stem cell niche maintenance and activity revealed that both WOX5 (WUSCHEL-RELATED HOMEOBOX) and SHR (SHORTROOT) were mis-expressed in anchor roots in a similar way as in the primary root. The ProWOX5::erGFP marker showed GFP fluorescence in additional cells above the presumptive QC (Fig. 1E and F). Furthermore, cellular expression of ProSHR::SHR-GFP was disturbed in anchor roots and showed a similar expression pattern as described previously for the primary root (Fig. 1G and H).4,5
A characteristic feature of the mail1 phenotype was the accumulation of dead cells within the division zone of the primary root tip. Cell death was also observed in the division zone of anchor roots (Fig. 1F and H), but the number of dead cells was clearly reduced.
We have previously shown that in the mail1 primary root cell differentiation was severely disturbed, since specification of different cell types did not occur. Here, we followed the expression of the transcription factor APL (ALTERED PHLOEM DEVELOPMENT), which is specifically required for the differentiation of phloem cells using ProAPL::GUS marker lines.6 ProAPL::GUS is expressed throughout vascular strands in WT plants and shows a continuous expression pattern in protophloem and metaphloem sieve element cells and in companion cells of the primary root.6 In contrast, in the mail1 primary root Pro::APL-GUS displayed a non-continuous expression pattern as shown here for 3-day-old seedlings (Fig. 1I and J). In 12-day-old mail1 seedlings, ProAPL::GUS expression disappeared in the primary root, but it showed a continuous expression pattern in anchor roots (Fig. 1K).
These observations show that in mail1 mutants the cellular organization of the stem cell niche is disturbed in anchor roots in a similar way as in primary roots. However, cell differentiation is less affected in anchor roots. Therefore, MAIL1 seems to have an essential function in maintaining cell fate in the cells of the primary root deriving from the RAM while it is less important for cell fate maintenance in anchor roots, which arise from cells of the SAM.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ikä Helariutta (University of Helsinki, Finland) for ProAPL::GUS seeds.
References
- 1. Uhlken C, Horvath B, Stadler R, Sauer N, Weingartner M. MAIN-LIKE1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J 2014; 78:107-20; PMID:24635680; http://dx.doi.org/ 10.1111/tpj.12455 [DOI] [PubMed] [Google Scholar]
- 2. Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 2011; 1 55:384-98; http://dx.doi.org/ 10.1104/pp.110.165126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development 1993; 119:71-84; PMID:8275865 [DOI] [PubMed] [Google Scholar]
- 4. Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 1993; 119:57-70; PMID:8275864 [DOI] [PubMed] [Google Scholar]
- 5. van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 1995; 378:62-5; PMID:7477287; http://dx.doi.org/ 10.1038/378062a0 [DOI] [PubMed] [Google Scholar]
- 6. Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature 2003; 426:181-6; PMID:14614507; http://dx.doi.org/ 10.1038/nature02100 [DOI] [PubMed] [Google Scholar]


