Abstract
Human leukocyte antigen II (HLA-II) plays an important role in host immune responses to cancer cells. Changes in gene methylation may result in aberrant expression of HLA-II, serving a key role in the pathogenesis of Kazakh esophageal squamous cell carcinoma (ESCC). We analyzed the expression level of HLA-II (HLA-DP, -DQ, and -DR) by immunohistochemistry, as well as the methylation status of HLA-DRB1 and HLA-DQB1 by MassARRAY spectrometry in Xinjiang Kazakh ESCC. Expression of HLA-II in ESCC was significantly higher than that in cancer adjacent normal (ACN) samples (P < 0.05). Decreased HLA-II expression was closely associated with later clinical stages of ESCC (P < 0.05). Hypomethylation of HLA-DRB1 and hypermethylation of HLA-DQB1 was significantly correlated with occurrence of Kazakh ESCC (P < 0.01), and mainly manifested as hypomethylation of CpG9, CpG10-11, and CpG16 in HLA-DRB1 and hypermethylation of CpG6-7 and CpG16-17 in HLA-DQB1 (P < 0.01). Moreover, hypomethylation of HLA-DQB1 CpG6-7 correlated with poor differentiation in ESCCs, whereas hypermethylation of HLA-DRB1 CpG16 and hypomethylation of HLA-DQB1 CpG16-17 were significantly associated with later stages of ESCC (P < 0.05). A significant inverse association between HLA-DRB1 CpG9 methylation and HLA-II expression was found in ESCC (P < 0.05). These findings suggest aberrant HLA-DRB1 and HLA-DQB1 methylation contributes to the aberrant expression of HLA-II. These molecular changes may influence the immune response to specific tumor epitopes, promoting the occurrence and progression of Kazakh ESCC.
Keywords: Esophageal squamous cell carcinoma, DNA methylation, HLA, Kazakh, Massarray
Abbreviations:
- HLA-II
Human leukocyte antigen II
- EC
Esophageal carcinoma
- ESCC
esophageal squamous cell carcinoma
- IHC
Immunohistochemistry
- ACN
cancer adjacent normal
- NE
normal esophageal tissues
- CpG
CG dinucleotides
- HPV
human papillomavirus
- CIITA
class II transactivator
- MHC
major histocompatibility complex
Introduction
Esophageal carcinoma (EC) is one of most common malignant tumors in the world. The incidence rate varies in different physiographic regions, nations, and races. China has a high incidence of EC with a high mortality rate for these patients.1 The Kazakh national (ethnic) minority living in Xinjiang (northwest of China) has been reported to be one of the ethnicities with the highest incidence of EC.2 The occurrence of EC is a complex process involving multiple factors, stages, and interactions.3 In our previous study, we observed a human leukocyte antigen II (HLA-II) allele polymorphism that may influence the immune response to human papillomavirus (HPV)-encoded epitopes and the risk of Kazakhs esophageal squamous cell carcinoma (ESCC).4
HLA class II molecules are encoded mainly by DP, DQ, and DR genes, expressed in immune cells, and responsible for presenting antigenic peptides to CD4+ T-cells to trigger the expansion and differentiation of these T-cells and induce an array of antigen-specific immune responses.5 Considering the key functions of these molecules, it is not surprising that certain HLA-II locus alleles, such as HLA-DRB1 and HLA-DQB1, which are known to play important roles in controlling the immune response, are closely associated with diseases.6,7 Proper expression of HLA-II proteins is critical for the normal function of HLA-II genes, while aberrant expression of HLA-II may result in an insufficient immune response or autoimmunity to some disease, especially in the case of tumors.8,9 In our previous study, we found that HLA-II (HLA-DP, DQ, and DR) exhibited aberrant expression in Kazakh ESCCs,10 but the mechanism underlying this association remains unclear. Previously, class II transactivator (CIITA), which is a major transactivator of major histocompatibility complex (MHC) class II, was shown to be a global regulator of HLA-II genes expression11,12; this finding has been confirmed in many tumors.13,14 Epigenetic alterations, such as DNA methylation of the HLA-II gene, are more likely involved in aberrant expression of HLA-II and cancer pathogenesis, but few studies have investigated such a relationship. Therefore, we used quantitative DNA methylation analysis (MassARRAY spectrometry) to examine whether methylation of HLA-II loci HLA-DRB1 and HLA-DQB1 is associated with aberrant expression of the HLA-II gene, and whether this is associated with the occurrence and progression of Kazakh ESCC.
Results
HLA-II expression in Kazakh ESCC and cancer adjacent normal tissue: correlation between HLA-II expression and ESCC clinicopathological parameters
Immunohistochemical staining for HLA-II revealed diffuse staining of the membranes and cytoplasm of tumor cells (Fig. 1). HLA-II protein was observed in 24 (36.4%) out of 66 ESCC tissue samples, while HLA-II was only observed in 1 (5.0%) out of 20 cancer adjacent normal (can) tissue samples. Furthermore, expression of HLA-II in ESCC was higher than that in ACN (P < 0.05). To assess the role of HLA-II expression in ESCC, we examined possible correlations between HLA-II expression and clinicopathological parameters, including age, gender, histological grade, depth of invasion, nodal status, and clinical stages. Interestingly, we found that although expression of HLA-II was higher in ESCC than in ACN, it followed the progression of ESCC (including deeper invasion, nodal metastasis, and later clinical stage). Expression of HLA-II decreased in later clinical stage (P < 0.05; Table 2), suggesting that expression of HLA-II was correlated with aggressive clinicopathological characteristics.
Figure 1.
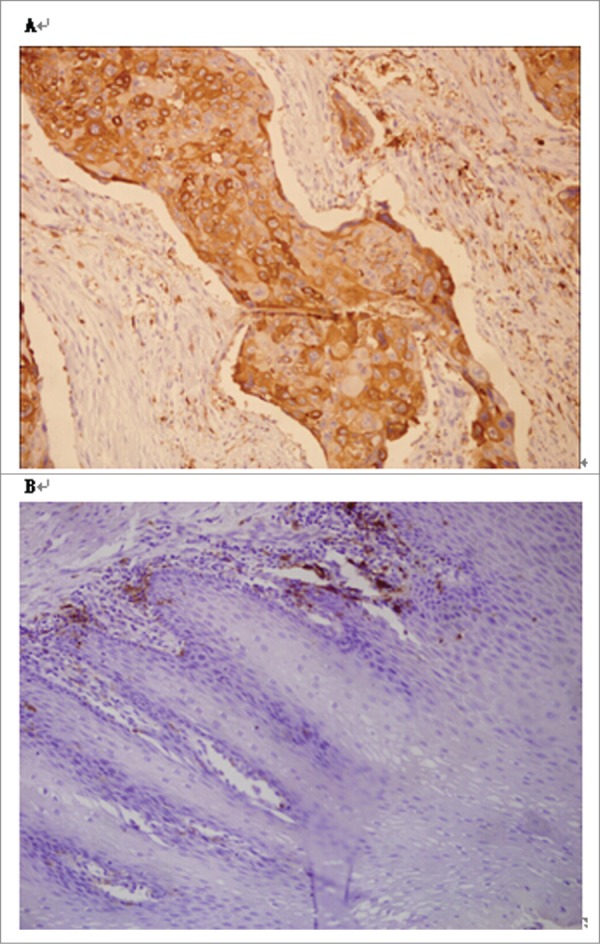
Immunohistochemical staining of esophageal squamous cell carcinoma. (A) Esophageal squamous cell carcinoma tissue. Note strong staining of HLA-II in squamous epithelial cancer cells (original magnification × 200). (B) Adjacent normal esophageal tissue. Note the absent staining of HLA-II in squamous epithelial cells (original magnification × 200).
Table 2.
Correlation between clinicopathologic data and HLA-II expression in the Kazakh ESCC
| Expression of HLA-II |
|||||
|---|---|---|---|---|---|
| Clinicopathologic Parameter | No. of Cases | Positive n(%) | Negative n(%) | X2 | P |
| Age (y) | |||||
| ≤ Median (53y) | 21 | 13(43.3%) | 17(56.7%) | ||
| > Median | 45 | 11(30.6%) | 25(69.4%) | 1.155 | 0.314 |
| Gender | |||||
| M | 38 | 22(57.9%) | 16(42.1%) | ||
| F | 28 | 20(71.4%) | 8(28.6%) | 1.276 | 0.308 |
| Histologic grade | |||||
| Well | 24 | 17(70.8%) | 7(29.2%) | ||
| Moderate + poor | 42 | 25(59.5%) | 17(40.5% | 0.844 | 0.431 |
| Depth of invasion | |||||
| TI-T2 | 44 | 30(68.2%) | 14(31.8%) | ||
| T3-T4 | 22 | 9(40.9%) | 13(59.1%) | 4.513 | 0.062 |
| Nodal status | |||||
| PN− | 29 | 20(69.0%) | 9(31.0%) | ||
| pN+ | 37 | 17(45.9%) | 20(54.1%) | 3.497 | 0.082 |
| TNM stage | |||||
| I-II | 34 | 23(67.6%) | 11(32.4%) | ||
| III-IV | 32 | 13(40.6%) | 19(59.4%) | 4.855 | 0.047* |
P < 0.05.
Analysis of the methylation status of the HLA-II gene promoter region
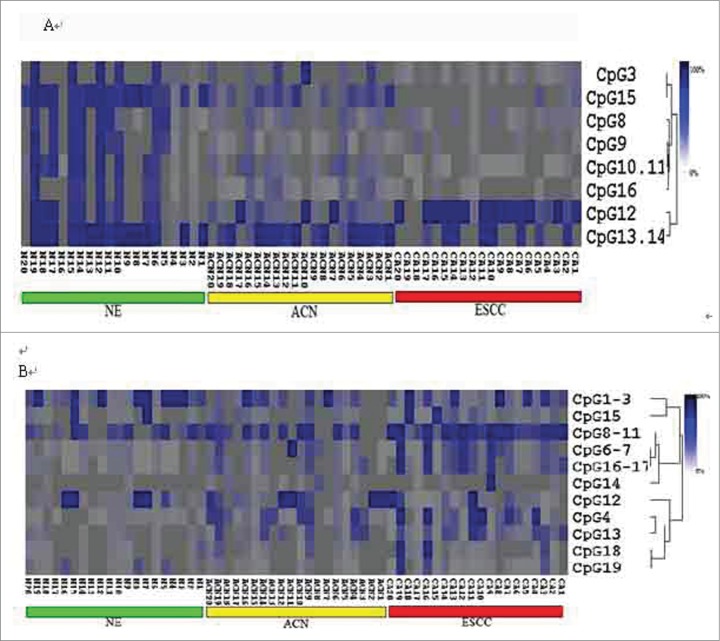
MassARRAy spectrometry was used to detect the methylation status of the promoter region of HLA-DRB1 and HLA-DQB1. The 311-bp region of the HLA-DRB1 promoter contains 16 CpG units; the 284-bp region of the HLA-DQB1 promoter contains 19 CpG units. Of these CpG units, only 10 in the HLA-DRB1 promoter and 18 in the HLA-DQB1 promoter could be analyzed, and the coverage rates were 63% and 95%, respectively. Methylation PCR products were cut into small pieces by the RNase enzyme. In fact, not all the pieces of the PCR products could be analyzed, as some fragments were too big or too small (>7000 Da or < 1500 Da) or the partial methylation mass spectra overlapped. Using hierarchical cluster analysis to examine the CpG units methylation status, as shown in Figure 2, CpG methylation levels of the samples could be identified by color, with dark blue indicating 100% methylation rate of the CpG unit and decreasing color indicating lower methylation rates.
Figure 2.
Hierarchical cluster analysis of HLA-DRB1 and -DQB1 CpG unit methylation status. (A) 2-way hierarchical cluster analysis of the methylation levels of HLA-DRB1 in esophageal squamous cell carcinoma (ESCC), cancer adjacent normal tissues (ACN), and normal esophageal tissues (NE). (B) 2-way hierarchical cluster analysis of the methylation levels of HLA-DQB1 in ESCC, ACN, and NE tissues. The colors indicate the percent of methylation in each CpG.
Methylation levels of HLA-DRB1 and HLA-DQB1 in Kazakh ESCC, ACN, and normal esophageal tissue samples
The overall average methylation level of HLA-DRB1 was significantly higher in normal esophageal tissues (NE) than in Kazakh ESCC (P < 0.01). Using MassARRAY spectrometry, we narrowed our analysis to identify the methylation status of each CpG unit in the gene sequence. We found that, except for CpG12, the methylation levels of other CpG units were higher in NE and ANC samples than in ESCC samples. Using Kruskal-Wallis H analysis, we found that the hypomethylation of 3 HLA-DRB1 CpG units (CpG9, CpG10-11, and CpG16) was significantly associated with ESCC (P < 0.01). On the contrary, the overall average methylation level of HLA-DQB1 was significantly higher in Kazakh ESCC than in NE (P < 0.01), and the methylation levels of each CpG unit were also higher in Kazakh ESCC than in the control groups. Using Kruskal-Wallis H analysis, we found that the hypermethylation of HLA-DQB1 CpG6-7 and CpG16-17 was significantly associated with Kazakh ESCC (P < 0.01; Fig. 3).
Figure 3.
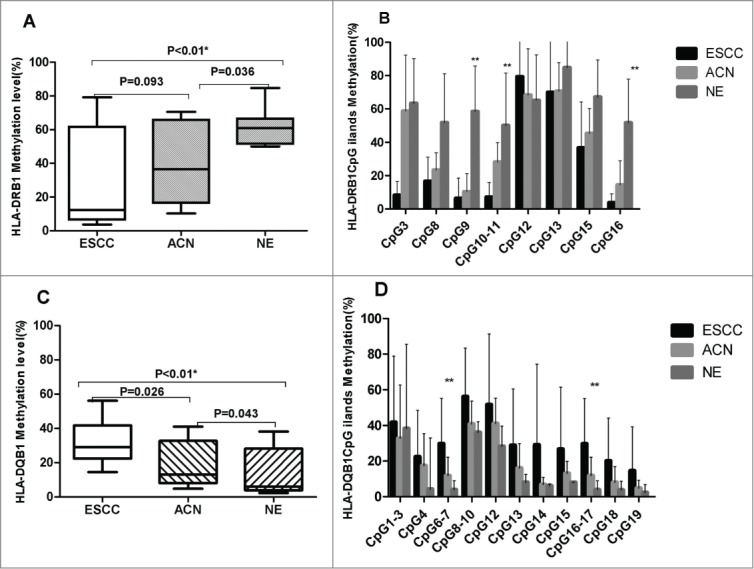
HLA-DRB1 and HLA-DQB1 methylation levels in Kazakh ESCC, ACN, and NE samples. (A) Overall methylation level of HLA-DRB1 in Kazakh ESCC, ACN, and NE samples. The overall methylation level of NE is significantly higher than that of ESCC and ACN (both P < 0.05). (B) Methylation levels of different CpG units of HLA-DRB1 contrast in Kazakh ESCC, ACN, and NE samples. 3 CpG (CpG9, 10-11, and 16) units exhibited significantly decreased methylation levels in ESCC compared to NE samples (all P < 0.05). Methylation levels of 2 CpG units (10-11 and 16) are significantly lower in ESCC than in ACN samples (all P < 0.05). The methylation levels of 2 CpG units (9 and 10-11) are significantly decreased in ESCC compared to ACN levels (all P < 0.05). (C) Overall methylation levels of HLA-DQB1 in Kazakh ESCC, ACN, and NE samples. The overall methylation level in ESCC is significantly higher than that in ACN and NE (all P < 0.05). (D) Methylation levels of different CpG units of HLA-DQB1 in Kazakh ESCC, ACN, and NE samples. Methylation levels of 2 CpG units (CpG6-7 and 16-17) are significantly increased in ESCC compared to that in NE samples (all P < 0.05).
Correlation between methylation status of HLA-DRB1 and HLA-DQB1 and clinical pathological parameters in ESCC
To assess the correlation between methylation status of HLA-DRB1 and HLA-DQB1 and the ESCC clinical pathological parameters, we chose specific CpG units, including HLA-DRB1 CpG9, CpG10-11, and CpG16 and HLA-DQB1 CpG6-7 and CpG16-17, as methylation of these units is significantly different between ESCC and control samples. HLA-DQB1 CpG6-7 exhibited higher methylation levels in well-differentiated ESCCs than in poorly differentiated ESCCs (P < 0.05), suggesting that hypomethylation of HLA-DQB1 CpG6-7 correlated with poor differentiation in ESCCs. The methylation level of HLA-DRB1 CpG16 in stage III-IV ESCC was higher than that in stages I-II, while the methylation level of HLA-DQB1 CpG16-17 in stage III-IV ESCC was lower than that in stage I-II ESCC (P < 0.05), Suggesting that hypermethylation of HLA-DRB1 CpG16 and hypomethylation of HLA-DQB1 CpG16-17 were significantly associated with later stages of ESCC. No significant differences in methylation status were observed for other clinical parameters (Table 3).
Table 3.
Methylation status of specific HLA-DRB1 and HLA-DQB1 CpG units according to different clinicopathologic parameters in Kazakh ESCCs
| Specific HLA-DRB1 CpG site (% Methylaton) |
Specific HLA-DQB1 CpG site (% Methylaton) |
|||||
|---|---|---|---|---|---|---|
| Clinicopathologic Parameter | No. of Cases | CpG9 | CpG10-11 | CpG16 | CpG6-7 | CpG16-17 |
| Age (y) | ||||||
| ≤ Median (53 y) | 30 | 4.36 | 9.35 | 17.30 | 26.86 | 28.14 |
| > Median | 36 | 9.25 | 8.10 | 5.00 | 31.45 | 31.91 |
| Gender | ||||||
| M | 38 | 4.40 | 10.36 | 4.38 | 21.11 | 28.00 |
| F | 28 | 6.44 | 7.31 | 1.13 | 33.22 | 32.89 |
| Histologic grade | ||||||
| Well | 24 | 8.57 | 12.17 | 15.00 | 47.50* | 47.00 |
| Moderate + poor | 42 | 3.45 | 6.27 | 14.00 | 18.27 | 19.91 |
| Depth of invasion | ||||||
| TI-T2 | 44 | 6.57 | 9.29 | 3.33 | 26.89 | 30.56 |
| T3-T4 | 22 | 1.50 | 7.50 | 1.00 | 32.44 | 30.33 |
| Nodal status | ||||||
| PN− | 29 | 3.18 | 7.94 | 1.00 | 31.88 | 32.75 |
| pN+ | 37 | 9.00 | 10.27 | 5.67 | 12.05 | 12.00 |
| TNM stage | ||||||
| I-II | 34 | 13.6 | 9.94 | 1.36* | 37.82 | 40.09* |
| III-IV | 32 | 2.00 | 7.36 | 12.50 | 16.86 | 15.29 |
P < 0.05.
Correlation between promoter methylation and expression of HLA-DP, DQ, and DR in ESCCs
To evaluate whether HLA-DRB1 and HLA-DQB1 promoter methylation correlated with aberrant expression of HLA-DP, DQ, and DR, we analyzed the methylation status of 5 specific CpG units (HLA-DRB1 CpG9, CpG10-11, and CpG16 and HLA-DQB1 CpG6-7 and CpG16-17). Among ESCCs that did not express HLA-II protein, the methylation rate of HLA-DRB1 CpG 9 (8%) was significantly higher than that in HLA-II-positive ESCC samples (0.3%), suggesting a significant inverse association between HLA-DRB1 CpG9 methylation and HLA-II expression in Kazakh ESCC (P < 0.05). No obvious difference was noted for the other CpG units (Table 4).
Table 4.
Correlation between HLA-II (DRB1 and DQB1) methylation and the expression of HLA-II (HLA-DR, DP, and DQ) in ESCCs
| Specific HLA-DRB1 CpG site (% Methylaton) |
Specific HLA-DQB1 CpG site (% Methylaton) |
|||||
|---|---|---|---|---|---|---|
| HLA-DP, DQ, and DR expression | No. of Cases | CpG9 | CpG10-11 | CpG16 | CpG6-7 | CpG16-17 |
| Positive | 24 | 0.3* | 8.4 | 1.5 | 28 | 29.67 |
| Negative | 42 | 8 | 9.18 | 3.17 | 30.5 | 30.83 |
P < 0.05.
Discussion
HLA-II is an important immune system protein that plays a key role in tumor antigen presentation, immune response, and immune surveillance.8 In recent years, several studies reported that aberrant expression of HLA-II is closely related to the pathogenesis of tumors.15-17 Aberrant expression of HLA-II results in dysfunctional presentation of exogenous antigen to CD4+ T-cells, allowing some tumor cells to escape immune surveillance. In this study, we found that HLA-II (DP, DQ, and DR) exhibited abnormally high expression in Kazakh ESCC. These results are similar to those in Sumiyoshi's report,18 which revealed that aberrant HLA-II expression may be involved in the occurrence of Kazakh ESCC. Examination of the correlations between HLA-II expression and clinicopathological parameters showed that decreased expression of HLA-II is related to some aggressive clinicopathological characteristics, such as depth of invasion and nodal metastasis, although these findings were not statistically significant. The decreased expression of HLA-II was more significant in later clinical stages than in early clinical stages, suggesting that lower expression of HLA-II may lead to a reduction in tumor antigen presentation, immune response, and immune surveillance, in close relationship with the development of ESCC.
Although Kazakh ESCC exhibit aberrant expression of HLA-II, the underlying mechanism is still unclear. We speculated that methylation of the promoter region of HLA-DRB1 and HLA-DQB1 is associated with aberrant expression of the HLA-II gene, leading to the development of Kazakh ESCC. Thus, we examined the methylation status of HLA-DRB1 and HLA-DQB1 using MassARRAY spectrometry, which is a high-throughput tool for quantitative analysis of DNA methylation and provides mass spectrum information that can be used to determine the methylation status of the gene and also to assess the methylation level of each CpG unit.19
This analysis revealed that the overall average methylation level of HLA-DRB1 was clearly reduced in Kazakh ESCC compared to NE samples. Furthermore, we found that the methylation levels of 3 CpG units (CpG9, CpG10-11, and CpG 16) were most dramatically decreased. HLA-DQB1, however, presented different methylation status in Kazakh ESCC from HLA-DRB1. The average methylation levels of HLA-DQB1 were increased in cancerous tissues, and CpG6-7 and CpG16-17 showed significantly higher methylation in ESCC compared with control tissue samples (P < 0.05). We propose that the 2 differing methylation statuses in Kazakh ESCC samples may result from the complex structure of the HLA-II gene, in which the different regions of the gene sequence yield different structures and functions.
In this study, we showed that methylation of HLA-II and aberrant expression of HLA-II were closely related to the occurrence of Kazakh ESCC. We analyzed 5 CpG units of HLA-II (DRB1 CpG9, 10-11, and 16 and DQB1 CpG6-7 and 16-17) and discovered a significant inverse relationship between HLA-DRB1 methylation and HLA-DP, DQ, and DR protein expression, especially for the HLA-DRB1 CpG9 unit. This results suggests that hypomethylation of HLA-DRB1 is one of the reasons for aberrant high expression of HLA-II in Kazakh ESCC. Previously, others have shown that the epigenetics of class II transactivator (CIITA) may be important for regulation of the expression of HLA-II,20 suggesting that HLA-II expression is regulated by multiple factors.
In addition, we evaluated the relationship between HLA-DRB1 and HLA-DQB1 methylation status and some clinical parameters. Hypermethylation of HLA-DRB1 CpG16 was significantly correlated with characteristics of tumors in later clinical stages, and this relationship may be related to the decreased expression of HLA-II in later clinical stages of ESCC. This finding suggests that hypermethylation of HLA-DRB1 CpG16 may be one of crucial causes of dysfunction of HLA-II and may promote the development of Kazakh ESCCs. On the contrary, the methylation level of HLA-DQB1 CpG6-7 was significantly lower in poorly differentiated ESCC, and the methylation level of HLA-DQB1 CpG16-17 was lower in later stages of tumors, suggesting that hypomethylation of HLA-DQB1 CpG6-7 and CpG16-17 may predict more aggressive of Kazakh ESCCs. In Wang et al. serous epithelial ovarian carcinoma study,21 the authors also found hypomethylation of CpGs located in HLA-DQB1, and increased CD8+ T-tumor cells infiltration, further suggesting that hypomethylation of HLA-DQB1 may promote the progression of some epithelial malignant tumors. Interestingly, this study also revealed that the methylation level of HLA-DRB1 was decreased from NE to ESCC, while, with progression of ESCC, the methylation level of HLA-DRB1 increased, a trend that is contrary to HLA-II expression. Before carcinogenesis, HLA-DRB1 methylation levels gradually decreased, increasing the expression of HLA-II. This increase may enhance the activity of antigen presentation and increase the ability of the immune system to protect against cancer and promote body immune surveillance. After carcinogenesis with the increased level of HLA-DRB1 methylation, HLA-II gene expression decreases and the activity of antigen presentation is reduced, possibly inhibiting the body's immune response against tumor and thereby promoting the progression of ESCC.
In conclusion, the results presented here illustrate aberrant HLA-DRB1 and HLA-DQB1 promoter methylation, which acts via a complex process leading to HLA-II loss and decreased immunosurveillance functions against cancer cells. These characteristics may be one of the reasons for aberrant protein expression, which promotes the occurrence and progression of Kazakh ESCC. The methylation of certain CpG units was significantly different between Kazakh ESCC and control groups and in the different clinical stages, suggesting that this methylation status may serve as a candidate biomarker for Kazakh ESCC.
Materials and Methods
Study population
A total of 66 Kazakan specimens were collected between 2000 and 2007. The patients were 34-79 years old (38 men and 28 women) and had been diagnosed with ESCC. These patients did not receive radiotherapy or chemotherapy before surgery. Twenty specimens were collected from cancer adjacent normal (ACN) tissues, and 20 specimens were collected from normal Kazakan esophageal tissues (NE) as controls. The NE group participants were 30 to 73 years old (12 men and 8 women). All individuals were recruited from the Yili Friendship Hospital in Xinjiang, China. Each participant provided written informed consent, and the study was approved by the participating hospital.
All ESCC specimens obtained after the surgery were embedded in paraffin, subsequently sectioned into 5-μm slices, and subjected to conventional H&E staining. The diagnosis of ESCCs were confirmed by 2 pathologists according to WHO histological tumor classification criteria22: 25 cases of well-differentiated ESCC and 41 cases of poorly differentiated ESCC. ACN specimens, which were sampled from more than 5 cm away from the cancer region, were confirmed to be free of cancer tissue. NE tissues were gastroscopy biopsy tissues that were preserved in paraffin and diagnosed by 2 pathologists as mild esophagitis.
Immunohistochemical analysis
The paraffin-embedded tissue samples were cut into 4-μm thick sections and mounted on polylysine-coated slides. The samples were dewaxed in xylene and rehydrated using a graded series of ethanol solutions. After deparaffinization, endogenous peroxidase activity was blocked by incubation in a 3% peroxide-methanol solution at room temperature for 10 minutes, and then antigen retrieval was performed at 100°C (in an autoclave) for 7 minutes in a 10 nmol/L sodium citrate buffer (pH 6.0). Afterwards, sections were incubated with goat serum to block nonspecific adsorption at room temperature for 10 minutes. Sections were incubated with the primary anti–HLA-II (HLA-DP, DQ, DR) monoclonal antibody (1:100; DAKO; Glostrup, Denmark) overnight at 4°C. Next, a thorough washing with phosphate-buffered saline (PBS) was performed. Subsequently, binding of the primary antibody was visualized using the EnVision kit (DAKO; Glostrup, Denmark) in accordance with the manufacturer's instructions. Finally, sections were faintly counterstained with hematoxylin and mounted with glycerol gelatin. A positive control (HLA-II-positive sample) and a negative control (PBS) were included in these experiments. All sections were analyzed under a light microscope by 2 experienced pathologists. Results were scored as positive or negative by the percentage and intensity of positive cells. The percentage of positive cells was scored as 0 in the absence of staining, 1 for less than 25% stained cells, 2 for 25%–50% stained cells, and 3 for more than 50% stained cells .The intensity of staining was scored as 0, 1, 2, or 3 in reference to absent, weak, clear, or strong expression. The staining results were divided into 2 categories based on the sum of both scores; 0-2 was considered negative (−), while scores of 3-6 were considered positive (+).
DNA preparation
Briefly, each formalin-fixed and paraffin-embedded sample was cut into 5-μm thick sections, and 10-15 slides were placed into a new high pressure EP-tube. Next, genomic DNA was isolated from paraffin-embedded tissues using a phenol-chloroform method23 and dissolved in sterile double-distilled water for 12-24 h. Samples were stored at -80°C until further analysis.
Bisulfite treatment with EZ DNA methylation kit
To perform bisulfite conversion of the target sequence, the EZ DNA Methylation Kit was used according to the instruction manual.24 During the bisulfite treatment chemical mixture conversion reaction, PCR was performed as follows: 95°C for 30 s, 50°C for 15 minutes, repeating these 2 steps for 20 cycles.
Primer design and PCR tagging for EpiTYPER assay
HLA-DRB1 and HLA DQB1 CpG units were identified via the UCSC website (http:genome.ucsc.edu). First “Table Browser” software was used to obtain DNA sequence located in the promoter region and that was also rich of CpG islands by “CpG island set”. Next, the sequence was used to design the gene primer using the EpiDesigner software (http:// epidesigner.com), and upstream and downstream primers were chosen in accordance with the recommended size for PCR amplicons (200–600 base pairs). For each reverse primer, an additional T7 promoter tag was added for in vivo transcription, and a 10-mer tag was added to the forward primer to adjust for the melting temperature differences. HLA-DRB1 and HLA DQB1 CpG specificity primers are shown in Table 1. The cycle program was as follows: 95°C for 4 minutes, 95°C for 20 s, 56°C for 30 s, 95°C for 1 minute (repeating the 3 steps for 45 cycles), and 72°C for 3 minutes. After the dephosphorylation of unincorporated dNTPs by shrimp alkaline phosphatase(SEQUENOM), transcription and digestion were performed simultaneously at 37°C for 3 h by RNase A and T7 polymerase. The cleavage reactants were purified with CLEAN resin (Sequenom; San Diego, CA) and dispensed onto silicon chips preloaded with matrix (Spectro-CHIPS, Sequenom). Mass spectra were collected using a MassARRAY mass spectrometer (Bruker-Sequenom) and analyzed using proprietary peak picking and signal-to-noise calculations (Sequenom Epityper v1.0.5). In MassARRAY analysis, initially, quality control (QC) was performed for each CpG unit. The non-applicable reading and its corresponding site were eliminated in calculation.
Table 1.
The primers used to amplify the HLA-DRB1 and DQB1 translational start codon for Massarray DNA methylation analysis
| HLA gene | Forward primer | Reverse primer | Amplified fragment length |
|---|---|---|---|
| HLA-DRB1 | 5′-aggaagagagttttagaataggttggaggtaggg-3′ | 5′-cagtaatacgactcactatagggagaaggcttcccattaaaaaaataacactcaaa-3′ | 311 bp |
| HLA-DQB1 | 5′-aggaagagagttagaaggatattttggagaggaaa-3′ | 5′-cagtaatacgactcactatagggagaaggctcctatccccctactctaccctaaat-3′ | 284 bp |
10mer tag “aggaagagag” and T7 promoter tag “cagtaatacgactcactatagggagaaggctc” was added for forward primer and reverse primer separately to adjust for the melting temperature difference.
Statistical analysis
The SPSS version 13.0 software was employed for all statistical analyses. Correlations between the HLA-DP, DQ, and DR staining were calculated using the Pearson χ2 test. Kruskal-Wallis H test was used to assess the overall methylation levels and different CpG unit methylation levels among ESCC, can, and NE groups. Wilcoxon test was used to compare differences in CpG unit methylation levels between clinicopathological parameters (including age, gender, histologic grade, depth of invasion, nodal status, and tumor-node-metastasis stages), which were divided into 2 classifications. 2-way hierarchical cluster was used to analyze the methylation levels of HLA-DRB1 and DQB1 in case and control groups. P-values were calculated by the Epi-Info program,. P-values lower than 0.05 were considered statistically significant in Pearson χ2 and Wilcoxon test. P values lower than 0.017 (0.05/3) were considered statistically significant in Kruskal-Wallis H test (multiple comparison among ESCC, can, and NE).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Li Yi Gu and Chunming Ge for assistance with tissue collection and Shungang Li for assistance with statistical analysis.
Funding
This work was supported by Ministry of Science and Technology of China (Nos. 2012AA02A503, 2009BAI82B03 and 2010DFB34100), the National Natural Science Foundation(No.81160301, No.81260301, and No.81460363), and the Shihezi University Science and Technology Research Foundation (No. 2012ZRKXYQ-YD32).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a Cancer J Clin 2005; 55(2): 74-108; PMID:15761078 [DOI] [PubMed] [Google Scholar]
- 2.Y Z. The distribution of esophageal cancer in Xinjiang. J Xinjiang Med Univ (China) 1988; 11: 139-144 [Google Scholar]
- 3.Cheung WY, Liu G. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am 2009; 38(1): 75-91; viii PMID:19327568; http://dx.doi.org/ 10.1016/j.gtc.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Li L, Pang L, Chen Y, Yang L, Liu C, Zhao J, Chang B, Qi Y, Liang W, Li F. HLA-DRB1*1501 and HLA-DQB1*0301 alleles are positively associated with HPV16 infection-related Kazakh esophageal squamous cell carcinoma in Xinjiang China. Cancer Immunol, Immunother : CII 2012; 61(11): 2135-2141; PMID:22588649; http://dx.doi.org/ 10.1007/s00262-012-1281-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mota F, Rayment N, Chong S, Singer A, Chain B. The antigen-presenting environment in normal and human papillomavirus (HPV)-related premalignant cervical epithelium. Clin Exp Immunol 1999; 116(1):33-40; PMID:10209502; PMCID:1905217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollaee A, Ghaffarpor M, Ghlichnia HA, Ghaffari SH, Zamani M. The influence of the HLA-DRB1 and HLA-DQB1 allele heterogeneity on disease risk and severity in Iranian patients with multiple sclerosis. Int J Immunogenet 2012; 39(5): 414-422; PMID:22404765; http://dx.doi.org/ 10.1111/j.1744-313X.2012.01104.x [DOI] [PubMed] [Google Scholar]
- 7.Madeleine MM, Johnson LG, Smith AG, Hansen JA, Nisperos BB, Li S, Zhao LP, Daling JR, Schwartz SM, Galloway DA. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res 2008; 68(9):3532-3539; PMID:18451182; PMCID:2662593; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baccar A, Ferchichi I, Troudi W, Marrakchi R, Ben Hmida N, Jebini S, Mrad K, Ben Romdhane K, Benammar Elgaaied A. CD99 and HLA-II immunostaining in breast cancer tissue and their correlation with lymph node metastasis. Disease Markers 2013; 34(5):363-371; PMID:23481630; http://dx.doi.org/ 10.3233/DMA-130982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordanova ES, Philippo K, Giphart MJ, Schuuring E, Kluin PM. Mutations in the HLA class II genes leading to loss of expression of HLA-DR and HLA-DQ in diffuse large B-cell lymphoma. Immunogenetics 2003; 55(4):203-209; PMID:12756506; http://dx.doi.org/ 10.1007/s00251-003-0563-z [DOI] [PubMed] [Google Scholar]
- 10.Hu Jianming, Chen YZ, Liu Chuxia, Wenting Li, Yin liang, Kang Xueling, Wang Weiwei, Liu Dong, Liang Weihua, Jiang Jinfang, Li Hongan, Li Feng. Expression and Significance Of Human Leukocyte Antigen HLA-DP,DQ,DR in Xinjiang Kazakh Esophageal Carcinoma. J Shihezi University(China) 2009; 6:731-734 [Google Scholar]
- 11.Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med 1995; 181(2):765-767; PMID:7836928 PMCID:2191893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell 2002; 109 Suppl:S21-33; PMID:11983150 [DOI] [PubMed] [Google Scholar]
- 13.Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, Johnson NA, Zhao Y, Telenius A, Neriah SB, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011; 471 (7338):377-381; PMID:21368758; http://dx.doi.org/ 10.1038/nature09754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie XW, Mei MH, Liao WJ, Qian LH, Yu X, Fei R, Qin LL, Zhang HH, Peng JR, Shen DH, et al. Expression of CIITA-related MHCII molecules in tumors linked to prognosis in hepatocellular carcinoma. Int JOncol 2009; 34(3):681-688; PMID:19212673 [DOI] [PubMed] [Google Scholar]
- 15.Zhou JH, Ye F, Chen HZ, Zhou CY, Lu WG, Xie X. Altered expression of cellular membrane molecules of HLA-DR, HLA-G and CD99 in cervical intraepithelial neoplasias and invasive squamous cell carcinoma. Life Sci 2006; 78(22):2643-2649; PMID:16434060; http://dx.doi.org/ 10.1016/j.lfs.2005.10.039 [DOI] [PubMed] [Google Scholar]
- 16.Glew SS, Duggan-Keen M, Cabrera T, Stern PL. HLA class II antigen expression in human papillomavirus-associated cervical cancer. Cancer Res 1992; 52(14): 4009-4016; PMID:1377602 [PubMed] [Google Scholar]
- 17.Jo YS, Lee JC, Li S, Choi YS, Bai YS, Kim YJ, Lee IS, Rha SY, Ro HK, Kim JM, et al. Significance of the expression of major histocompatibility complex class II antigen, HLA-DR and -DQ, with recurrence of papillary thyroid cancer. International journal of cancer Journal international du cancer 2008; 122(4): 785-790; PMID:17957790; http://dx.doi.org/ 10.1002/ijc.23167 [DOI] [PubMed] [Google Scholar]
- 18.Sumiyoshi K, Kuwano H, Watanabe M, Kitamura M, Toh Y, Sugimachi K. HLA-DR antigen expression in squamous epithelial dysplasia and squamous cell carcinoma of the esophagus: an immunohistochemical study. Oncol Rep 1999; 6(2):301-306; PMID:10022993 [DOI] [PubMed] [Google Scholar]
- 19.Radpour R, Haghighi MM, Fan AX, Torbati PM, Hahn S, Holzgreve W, Zhong XY. High-throughput hacking of the methylation patterns in breast cancer by in vitro transcription and thymidine-specific cleavage mass array on MALDI-TOF silico-chip. Mol Cancer Res : MCR 2008; 6(11):1702-1709; PMID:19010818; http://dx.doi.org/ 10.1158/1541-7786.MCR-08-0262 [DOI] [PubMed] [Google Scholar]
- 20.De Lerma Barbaro A, De Ambrosis A, Banelli B, Li Pira G, Aresu O, Romani M, Ferrini S, Accolla RS. Methylation of CIITA promoter IV causes loss of HLA-II inducibility by IFN-gamma in promyelocytic cells. Int Immunol 2008; 20(11):1457-1466; PMID:18829986; PMCID:2572003; http://dx.doi.org/ 10.1093/intimm/dxn103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Cicek MS, Charbonneau B, Kalli KR, Armasu SM, Larson MC, Konecny GE, Winterhoff B, Fan JB, Bibikova M, Chien J, Shridhar V, Block MS, Hartmann LC, Visscher DW, Cunningham JM, Knutson KL, Fridley BL, Goode EL. Tumor hypomethylation at 6p21.3 associates with longer time to recurrence of high-grade serous epithelial ovarian cancer. Cancer Res 2014; 74(11):3084-3091; http://dx.doi.org/ 10.1158/0008-5472 CAN-13-3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley RHLAA. WHO Classification Tumours of the Digestive System. IARC Press Lyon; 2000; 9-11 [Google Scholar]
- 23.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y, Liu D, Lim H, Taylor CR. DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J Histochem Cytochem 2002; 50(8):1005-1011; PMID:12133903 [DOI] [PubMed] [Google Scholar]
- 24.Thompson RF, Suzuki M, Lau KW, Greally JM. A pipeline for the quantitative analysis of CG dinucleotide methylation using mass spectrometry. Bioinformatics 2009; 25(17):2164-2170; PMID:19561019; PMCID:2800352; http://dx.doi.org/ 10.1093/bioinformatics/btp382 [DOI] [PMC free article] [PubMed] [Google Scholar]