Abstract
Plants survive adversity by modulating their growth in response to changing environmental signals. The phytohormone Gibberellic acid (GA) plays a central role in regulating these adaptive responses by stimulating the degradation of growth repressing DELLA proteins which accumulate during stress. The current model for GA signaling describes how this hormone binds to its receptor GID1 so promoting association of GID1 with DELLA, which then undergoes ubiquitin-mediated proteasomal degradation. Recent data revealed that conjugation of DELLAs to the Small Ubiquitin-like Modifier (SUMO) protein enables plants to modulate its abundance during environmental stress. This is achieved by SUMOylated DELLAs sequestering GID1 via its SUMO interacting motif (SIM) allowing non-SUMOylated DELLAs to accumulate leading to growth restraint under stress and potential yield loss. We demonstrate that GID1 proteins across the major cereal crops contain a functional SIM able to bind SUMO1. Site directed mutagenesis and yeast 2 hybrid experiments reveal that it is possible to disrupt the SIM-SUMO interaction motif without affecting the GA dependent DELLA–GID1 interaction and thereby uncoupling SUMO–mediated inhibition from DELLA degradation. Arabidopsis plants overexpressing a SIM mutant allele of GID1 perform better at relieving DELLA restraint than wild–type GID1. This evidence suggests that manipulating the SIM motif in the GA receptor may provide a possible route to developing stress tolerant crops plants.
DELLA proteins are the central repressors of molecular pathways governed by the growth promoting phytohormone GAs.1-4 Current evidence indicates that a key strategy employed by plants to survive adverse conditions is to restrain growth and development by increasing the abundance of DELLAs.5,6 GAs play a key role regulating these adaptive responses by stimulating the degradation of DELLA proteins. The integrative role of DELLAs is heavily reliant on the plant's ability to control DELLA protein levels. Until recently the only mechanism for regulating DELLA protein abundance was through modulating the levels of GA to trigger ubiquitin-mediated proteasomal degradation. Several other ubiquitin-like proteins have been described in plants including the Small Ubiquitin-like Modifier (SUMO) that can act to stabilize the proteins with which it is conjugated.7 In some cases SUMO proteases remove this protein tag to destabilize the de-conjugated protein.7
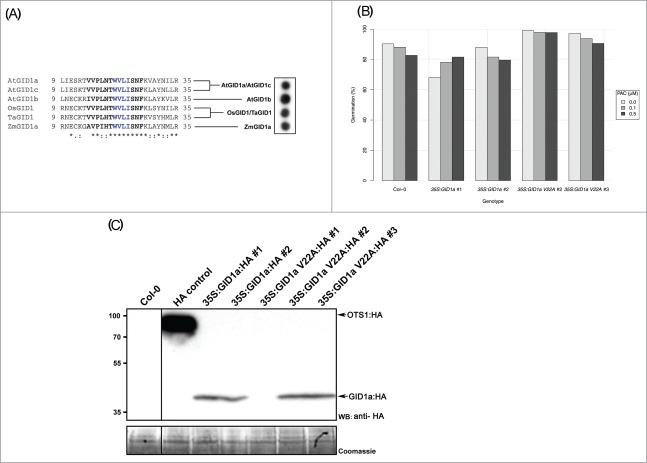
In Conti et al. (2014),8 we reported a new mechanism for modulating DELLA protein abundance independent of GA. We demonstrated that a proportion of DELLAs are conjugated to SUMO. DELLAs are more abundant in Arabidopsis plants deficient in SUMO protease activity overly tolerant to salt 1 and 2 (ots1 ots2 double mutants), despite no changes in GAs accumulation or DELLA genes transcript levels. DELLA accumulation during salt stress conditions is accompanied by a rapid increase in SUMO conjugated DELLA and this effect is enhanced in ots1 ots2.8 Increasing OTS SUMO protease activity results in strongly reduced DELLA accumulation and consequently, increased growth in a GAs deficient background. Similarly overexpressing GID1 can reverse growth restraint mediated by high DELLA levels in ots1 ots2. We identified a SUMO interacting motif (SIM) in GID1 and demonstrate that SUMO conjugated DELLA binds to this motif in a GA-independent manner.8 The consequent sequestration of GID1 by SUMO–conjugated DELLAs leads to an accumulation of non-SUMOylated DELLAs and subsequent growth restraint during stress. A key aspect of our study was the identification of a SIM in GID1a. We also observed that a core SIM motif is highly conserved among the GID1 GA receptors across the major cereal crop species (Fig. 1A). However we noted that the amino acid sequence surrounding the core SIM motif is variable which could affect SIM activity. We generated peptides that match the extended SIM sites (bold and blue, Fig. 1A) and demonstrated by Far Western experiments that all these motifs are functional SUMO binding peptides using Arabidopsis SUMO1 protein as a prey. This remarkable conservation of the SIM site in GID1 from divergent plant species is consistent with this mechanism playing a critical role in DELLA signaling.
Figure 1.
(A) Far Western interaction assays of GID1 SIMs from various plant species. The figure shows the amino acid alignment of the SIM regions in the GID1 protein among the indicated plant species. Sequences in bold were synthesized as peptides and tested for interaction with SUMO1 by Far Western blotting. Sequences in blue indicate core region in the SIM motifs in the various GID1 proteins. At; Arabidopsis; Os, Oryza Sativa; Ta, Triticum aestivum; Zm, Zea mays. (B) Germination assay of Wildtype (Col-0), HA:GID1a and HA:GID1aV22A overexpressing lines under various concentrations of PAC. The figure shows the percentage of seeds with an emerged cotyledon 2 days after stratified seeds were exposed to light. 35S:GID1aV22A lines show a statistically significantly higher germination rate than the 35S:GID1a and the Col-0 lines (P < 0.02). n ∼ 200 seeds /treatment/line. (C) Western blot showing levels of HA epitope tagged GID1a and GID1aV22A proteins in the transgenic Arabidopsis lines. Two transgenic lines for each construct show similar protein expression levels as detected by anti-HA antibodies (WB: anti-HA). A transgenic line expressing HA tagged OTS1 was used a HA epitope control for the Western blots. The lower panel shows the same blot that had been Coomassie stained to indicate equal protein loading in each lane.
Our previous data allowed us to postulate that a relatively small pool of SUMOylated DELLA could stabilize the larger pool of unmodified DELLA by titrating out GID1a protein via the SIM motif. This was confirmed in ots1 ots2 lines overexpressing GID1a which overcame growth restriction in salt and sensitivity to the GA-biosynthesis inhibitor paclobutrazol (PAC) due to increased DELLA levels. Furthermore overexpressing a variant GID1a with its SIM site disrupted (GID1aV22A) further enhanced ots1 ots2 growth under both control and salt conditions compared to the overexpression of wild–type GID1a. We now show that even in wild type genetic background (Col-0) over expressing GID1aV22A was more efficient in reducing sensitivity to PAC during germination compared to overexpressing wild type GID1 alone (Fig. 1B). Immunoblotting data indicated that the levels of GID1 and GID1aV22A proteins were comparable suggesting that the observed phenotype is not due to variability in protein levels (Fig. 1C).
Plants over-expressing GID1aV22A were better at rescuing the salt stress induced inhibition of growth and relieving sensitivity to PAC than overexpressing GID1a. We attributed this effect to reduced interaction of the GID1aV22A with the SUMO groups of SUMOylated DELLA proteins, which act to sequester the GID1a receptor molecule and prevent it from directing the degradation of DELLA proteins. However any alteration on the GA receptor could have unwanted side effects such as increased? DELLA or GA binding and the observed phenotypes could be thus due to alternative mechanisms. We therefore wanted to confirm that GID1aV22A protein was still able to bind to DELLA proteins in the presence of GAs and therefore still able to direct DELLA degradation.
Yeast two hybrid experiments using the DELLA protein RGA as prey and GID1a, GID1aV22A and GID1aV22S as bait shows RGA can bind to the mutagenized versions GID1a and this interaction retains its dependence on presence of GA3 (Fig. 2). However, even though yeast-2-hybrid experiments are semi quantitative at best, there appears to be a trend in the level of interaction with the native GID1a protein showing the strongest interaction, followed by the GID1aV22A mutant which was used in our previous study, which shows weak yeast growth under 5 μM of the inhibitor 3-Amino-1,2,4-triazole. Finally the GID1aV22S mutant shows the weakest interaction of all, with very little or no yeast growth under the 5 μM 3-AT. The yeast-2-hybrid experiment was repeated a further 2 times with different yeast clones and the results showed the same trend in interaction strength.
Figure 2.
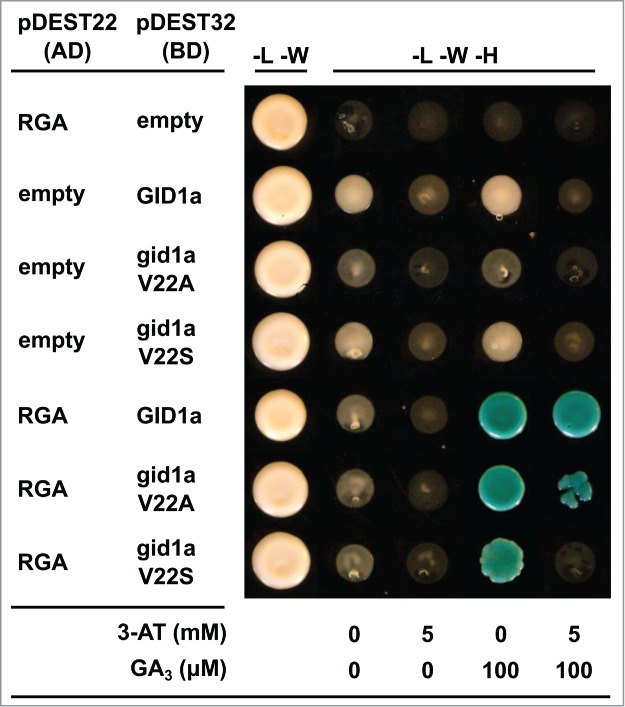
Yeast 2 hybrid assays showing the interaction between the RGA DELLA protein with GID1 or GID1aV22A or GID1aV22S SIM mutant proteins. The letters -L -W and -H correspond to media lacking Leucine, Tryptophan and Histidine respectively.
It has been established that GA–bound GID1 enhances ubiquitination and proteasomal degradation of DELLAs by promoting DELLA binding to the F-Box E3 ubiquitin ligase, SLY1. In this context GID1 plays a crucial link between the hormone and ubiquitin mediated degradation of DELLA repressor proteins. The results reported here and in Conti et al. (2014)8 support the conclusion that there is an alternative mechanism for DELLA proteins to recognize the GA receptor GID1 via the SIM-SUMO interaction. Our study also establishes that SUMOylation can be recruited to block GID1 receptor access to the DELLA motif in a novel manner. Ueguchi-Tanaka et al. (2007)9 showed that a triple alanine substation in the SIM region of the rice GID1 (20T - 22A) abolished the binding of the rice DELLA protein SLR but not the binding of GA3. Interestingly the SIM motif region in GID1 overlaps with the N-terminal ‘lid’ region that covers the GA docking pocket within the receptor. It is therefore tempting to speculate that binding of SUMOylated proteins to GID1 may also interfere with GA access to GID1 and consequently its binding DELLA proteins. Our yeast 2 hybrid results indicate that a single amino substitution in GID1aV22A does not abolish interaction with DELLA proteins nor GAs and supports the model that this protein mutant can direct the degradation of DELLA proteins but is released from inhibition by SUMO. The evidence suggests that the GID1aV22A protein is resistant to inhibition by SUMOylated DELLA and could target DELLA proteins for degradation even under low GA levels (i.e under stress conditions). The identification of these critical residues within the novel SIM motif in GID1 can potentially lead to fine tuning plant responses to salt or other stresses and could be used to generate stress resistant varieties of crop plants by manipulating specific amino acids in the GA receptor.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Funding
L.C. was supported by a research fellowship from the Biological and Biotechnological Research Council (BBSRC), the University of Milan and the Leverhulme Trust. S.N. was supported by a studentship from the BBSRC.
References
- 1. Peng J, Carol P, Richards D, King K, Cowling R, Murphy G, Harberd N. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 1997; 11:3194-205; PMID:9389651; http://dx.doi.org/ 10.1101/gad.11.23.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peng J, Richards D, Hartley N, Murphy G, Devos K, Flintham J, Beales J, Fish L, Worland A, Pelica F, et al. . Green revolution genes encode mutant gibberellin response modulators. Nature 1999; 400:256; PMID:10421366; http://dx.doi.org/ 10.1038/22307 [DOI] [PubMed] [Google Scholar]
- 3. Silverstone AL, Mak PY, Martínez EC, Sun TP. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 1997; 146:1087-99; PMID:9215910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001; 13:999-1010; PMID:11340177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006; 311:91-4; PMID:16400150; http://dx.doi.org/ 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- 6. Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 2008; 18:656-60; PMID:18450450; http://dx.doi.org/ 10.1016/j.cub.2008.04.034 [DOI] [PubMed] [Google Scholar]
- 7. Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 2009; 106:5418-23; PMID:19276109; http://dx.doi.org/ 10.1073/pnas.0811088106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, et al. . Small Ubiquitin-like modifier protein, SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 2014. 28:102-10; PMID:24434138; http://dx.doi.org/ 10.1016/j.devcel.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 9. Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I. et al. . Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007; 19:2140-55; PMID:17644730; http://dx.doi.org/ 10.1105/tpc.106.043729 [DOI] [PMC free article] [PubMed] [Google Scholar]