Abstract
pH-dependent antibodies are engineered to release their target at a slightly acidic pH, a property making them suitable for clinical as well as biotechnological applications. Such antibodies were previously obtained by histidine scanning of pre-existing antibodies, a labor-intensive strategy resulting in antibodies that displayed residual binding to their target at pH 6.0. We report here the de novo isolation of pH-dependent antibodies selected by phage display from libraries enriched in histidines. Strongly pH-dependent clones with various affinity profiles against CXCL10 were isolated by this method. Our best candidate has nanomolar affinity for CXCL10 at pH 7.2, but no residual binding was detected at pH 6.0. We therefore propose that this new process is an efficient strategy to generate pH-dependent antibodies.
Keywords: antibody recycling, chemokine, histidine, monoclonal antibody, pH-dependency, phage display, phage libraries
Abbreviations
- BLI
bio-layer interferometry
- CDR
complementary determining region
- CDRH
CDR of the heavy chain
- CDRL
CDR of the light chain
- ELISA
enzyme-linked immunosorbent assay
- GPCR
G protein-coupled receptor
- KB
kinetic buffer
- mAb
monoclonal antibody
- PBS
phosphate buffered saline
- scFv
single-chain variable fragment
- SPR
surface plasmon resonance
Multiple approaches are currently available for the generation of monoclonal antibodies (mAbs) against an antigen, ranging from standard hybridoma fusions to a wide spectrum of in vitro selection and evolution technologies.1-3 Interactions occurring at antibody-antigen interfaces have been extensively studied and the structural, thermodynamic and kinetic principles guiding these molecular recognitions are well understood.4 The exquisite specificity and high affinity of mAbs make them highly sought after as drugs to efficiently bind and potentially neutralize a biological target to achieve therapeutic benefits. Thus, they represent a rapidly growing class of drugs, with some currently ranking as bestselling biologics and most used in cancer or autoimmune indications.5 Furthermore, high affinity mAbs are invaluable reagents for research applications, such as protein purification or the development of highly specific biosensors or immunoassays such as enzyme-linked immunosorbent assay (ELISA).6 These techniques take advantage of the high specificity of the interaction between a mAb and its target to allow the detection of a defined molecule even in complex biological samples.7 The specificity of mAbs has, for instance, been exploited to develop kits allowing a rapid identification of pathogenic viruses or bacteria for human and cattle diseases.8 In addition, antibodies are powerful purification tools and antibody-coupled resins are used to purify not only single proteins, but also enzymes, drugs, proteins complexes and lipids by immunoaffinity chromatography.9-13 Finally, antibodies can be used to create biosensors monitoring the interaction between the antibody and its antigen, allowing the specific detection of virtually any protein in biological samples but also of contaminants in food or of pollutants in the environment.14-16
High affinity mAbs are thus powerful and versatile reagents used in diverse applications. However, high affinity antibody-antigen interactions are difficult to reverse. Harsh conditions, such as extreme pH or high salt concentrations, are required to dissociate these complexes and such conditions often lead to denaturation, aggregation and precipitation of proteins.17 This situation occurs, for example, when regenerating surfaces ends the process. The end result is that antibody-based reagents can only be used a limited number of times.
High affinity binding can also lead to unwanted effects during therapeutic intervention. For instance, mAbs directed against soluble targets such as cytokines, can induce significant target accumulation due to the formation of stable antibody-antigen complexes that are recycled via the interaction with the neonatal Fc receptor (FcRn).18-22 FcRn is responsible for the long serum half-life of IgGs that bind this receptor in the slightly acidic environment of the early endosome and are then returned to the extra-cellular compartment.23 Interestingly, if the antibody-antigen interaction is stable under these conditions, the bound antigen is recycled with the antibody and persists in circulation.
For these reasons, the development of antibodies capable of specific and high affinity binding to an antigen under physiological conditions, but that release the target upon mild modification of the environment, could be of great utility both for research and therapeutic applications. An attractive strategy is to generate pH-dependent mAbs capable of binding an antigen at a neutral pH of 7–7.4 and releasing the antigen under slightly more acidic conditions (pH 5.5–6.0, instead of pH 1–3 as it is the case with traditional mAbs). Such antibodies would enable the dissociation of immune complexes without affecting the structure and stability of the antibody or target protein, therefore allowing the regeneration and re-use of antibody-based matrices with minimal loss of performance between cycles. In addition, pH-dependent antibodies capable of neutralizing their target in the extracellular environment and releasing the bound antigen upon internalization in the endosome hold the promise of superior therapeutic potential.24 Due to the recycling of free antibodies, instead of immune complexes, pH-dependent mAbs are expected to display an extended half-life, allowing the reduction of the dose or dosing frequency. This approach would also be particularly suited to the targeting of abundant proteins. Several examples of pH-dependent mAbs directed against soluble and membrane proteins, such as IL6, IL6R and PCSK9, have been previously described.25-27 These studies exploited the properties of histidine residues, which have a logarithmic acid dissociation constant (pKa) around 6.0, and are therefore neutral at pH 7.2 but become positively charged at a pH below 6.0.28 These additional positive charges can destabilize the interaction between 2 proteins, a principle that was initially demonstrated with granulocyte colony-stimulating factor (G-CSF) mutants containing histidine residues, which dissociated from their receptor in the endosome.29 In the context of mAbs, histidines are typically introduced at the interface between the antibody and its target, more specifically in the complementary-determining regions (CDRs). Although the results obtained so far with pH-dependent mAbs are promising, studies published to date were based on the modification of pre-existing mAbs to confer pH-dependent antigen binding.25-27,30,31 These studies involved extensive histidine scanning mutagenesis of residues in the CDRs and characterization of the resulting individual mutants. As the original therapeutic mAb was selected for specific properties (high affinity, neutralization potency), its sequence and CDR conformation may not be optimal to further incorporate pH-dependent properties. Supporting this hypothesis, pH-dependent mAbs engineered using non-pH dependent mAbs as a starting point retained a significant level of target binding at pH 6.0.25,26 To circumvent these limitations and generate mAbs with enhanced properties, we aimed at isolating pH-dependent antibodies de novo from phage display antibody libraries enriched in histidine content and applying specific selection strategies favoring the amplification of phage displaying pH-dependent single-chain variable fragments (scFv). As a proof of concept for this approach, we report the isolation of highly pH-dependent mAbs binding the chemokine CXCL10 with nanomolar affinities and capable of neutralizing biological activity at pH 7.4, while showing no binding at pH 6.0.
We first built a synthetic scFv library to be used in conjunction with phage display technology that was enriched in histidine residues in the CDRH3 of the VH domain, a region where histidines are found at low frequency in natural repertoires.32,33 The library was based on a single VH (VH3–23) and 7 VL germlines (including both kappa and lambda families). With the aim to retain sufficient diversity to obtain antibodies with good affinities against various targets while introducing a histidine content sufficient to confer strong pH-dependent properties, YAT and NHT codons were alternated in CDRH3 of 8 to 15 amino acids (Fig. 1A). The YAT codon encodes either for histidine or tyrosine, thus allowing for a significant enrichment in histidines, and also tyrosines, that mediate different types of interactions between proteins. Tyrosine has indeed been successfully used in antibody libraries based on a minimalistic design, incorporating tyrosine and serine residues for CDR diversification.34 The NHT codon encodes for 12 different amino acids, excluding cysteine and stop codons. Synthetic diversity unbiased for histidine using NNS codons was introduced in the CDRL3 of the VL domain, while CDRL1 and CDRL2 as well as CDRH1 and CDRH2 maintained germline sequences. Following electroporation, the final size of the library was determined to be 1.3 × 109 transformants. Sequencing of 10 randomly picked clones confirmed that sequences were in frame, diverse and contained between 1 and 4 histidines in their CDRH3.
Figure 1.

De novo isolation of pH-dependent antibodies. (A). A phage display library enriched in histidine in the CDRH3 was constructed by introducing degenerated oligonucleotides in an acceptor library. NHT and YAT codons, encoding for the indicated amino acids, were alternated at the indicated positions. Inserts encoding for 8 to 15 amino acids were used. (B). Scheme of selections against hCXCL10. Phage particles were added on beads coated with hCXCL10 at pH 7.4. Following seven washes, the pH was lowered to 5.4 and the supernatant of the mixture was recovered and amplified before being used for the subsequent round of selection.
To favor the isolation of pH-dependent antibodies from this library, 3 rounds of selection against human CXCL10 were performed under different pH conditions. The binding of phage to hCXCL10 was performed at pH 7.4 while the elution was performed at pH 5.4 to ensure that all histidines were protonated (Fig. 1B). Under these conditions, the phage displaying at their surface pH-dependent scFv should be enriched while those maintaining binding under the mild elution condition (i.e. pH 5.4) should remain bound to the target and lost in the process. The output of the third round of selection was screened using a pH-dependent ELISA to identify scFv binding to hCXCL10 at pH 7.4, but losing 50% or more of their binding following an elution step at pH 5.4. A large proportion (38%) of scFvs binding to hCXCL10 met these criteria and were considered as positive clones, while 11% were pH-independent binders of hCXCL10. All scFvs were also tested for binding at pH 7.4 to streptavidin, human interleukin-6 receptor (hIL6R) and human CCL5 to ensure specificity and did not show any unspecific binding (data not shown).
Sequencing of phagemids isolated from positive clones revealed 8 unique sequences containing one or 2 histidines in the CDRH3 (Table 1). Interestingly, all clones contained a histidine residue at position IMGT 107 of the CDRH3 and the VLs used were based on the IGLV6-57 germline, but contained CDRL3 of different lengths and sequences.35 The VH and VL were reformatted into human IgG1, expressed, purified and tested in a dose-dependent manner using an ELISA at pH 7.4 or pH 5.4, which confirmed their greater capacity to bind hCXCL10 at neutral pH (Figure S1). All mAbs were able to neutralize the biological activity of hCXCL10 in vitro in chemotaxis assays with IC50 ranging between 3 and 231 nM (Table 2). The sequences reformatted as IgGs of clones C7, H2, G11 and F11 were also tested for their capacity to bind in a pH-dependent manner and neutralize mouse CXCL10 (mCXCL10) (Table 2).
Table 1.
Positive pH-dependent clones selected by phage display. Sequences of positive clones displaying pH-dependent binding to hCXCL10 by ELISA. The frequency of each sequence as well as the CDRH3 and L3 sequences are presented in comparison to the C7 clone. (−) : same amino acid as C7; (.) : no amino acid at this position due to a shorter CDRL3
| Clone | CDRH3 | CDRL3 | Frequency | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C7 | A | R | H | D | Y | F | Y | L | Y | D | Y | A | F | D | Y | Q | S | W | D | M | G | L | Q | . | V | V | 2% |
| H2 | - | - | - | - | - | - | - | V | - | - | - | Y | - | - | - | - | - | - | - | G | L | E | W | R | L | - | 2% |
| G6 | - | - | - | - | - | A | - | L | - | - | - | F | - | - | - | - | - | - | - | F | - | W | . | . | - | - | 5% |
| F11 | - | - | - | - | - | D | - | L | - | - | - | S | - | - | - | - | - | - | - | Q | W | S | V | G | A | - | 2% |
| A2 | - | - | - | - | - | P | - | Y | - | - | - | Y | - | - | - | - | - | - | - | S | S | K | L | A | S | - | 14% |
| E2 | - | - | - | - | - | D | - | A | - | - | - | Y | - | - | - | - | - | - | - | V | R | R | M | . | A | - | 5% |
| B11 | - | - | - | - | - | A | - | F | H | - | - | F | - | - | - | - | - | Y | - | A | R | R | G | K | P | - | 7% |
| G11 | - | - | - | - | - | P | - | L | H | - | - | F | - | - | - | - | - | - | - | P | M | G | G | . | A | - | 5% |
| E10 | - | - | - | - | - | D | - | F | H | - | - | Y | - | - | - | - | - | - | - | G | R | S | . | . | P | - | 10% |
Table 2.
Binding and inhibitory properties of pH-dependent antibodies targeting CXCL10. The binding (EC50) of each clone for human or murine CXCL10 was measured by ELISA. The pH of the binding and of the elution phases are indicated at the top of each column (pH binding/pH elution). For each antibody, the pH-dependency is quantified by the pH 7.4/6.0 to pH 7.4/7.4 ratio. Inhibitory properties were investigated by the inhibition of the migration of L1.2/CXCR3 transfectants to 1 nM hCXCL10 or 5 nM mCXCL10. n.d.: not determined
| Clone |
human CXCL10 |
murine CXCL10 |
||||||
| EC50 pH 7.4/7.4 [nM] |
EC50 pH 7.4/6.0 [nM] |
pH-dependency (ratio EC50) |
IC50 chemotaxis [nM] |
EC50 pH 7.4/7.4 [nM] |
EC50 pH 7.4/6.0 [nM] |
pH-dependency (ratio EC50) |
IC50 chemotaxis [nM] |
|
| C7 | 0.05 | 0.13 | 2.6 | 8 | 0.67 | 7.06 | 10.5 | 295 |
| H2 | 0.12 | 0.21 | 1.8 | 11 | 0.32 | 1.21 | 3.8 | 517 |
| G6 | 0.06 | 0.10 | 1.8 | 21 | n.d. | n.d. | n.d. | n.d. |
| F11 | 1.05 | 3.41 | 3.2 | 3 | 2.83 | 6.38 | 2.2 | 279 |
| A2 | 0.16 | 0.51 | 3.3 | 62 | n.d. | n.d. | n.d. | n.d. |
| E2 | 2.09 | 11.30 | 5.4 | 79 | n.d. | n.d. | n.d. | n.d. |
| G11 | 0.12 | 1.47 | 11.8 | 104 | 0.76 | 1.49 | 2.0 | 679 |
| E10 | 6.15 | 25.52 | 4.2 | 232 | n.d. | n.d. | n.d. | n.d. |
As C7 showed similar binding properties to human and mouse CXCL10 and was also able to inhibit chemotaxis induced by the chemokines of both species (Fig. 2A), it was selected for further characterization and engineering. First, the binding of C7 to hCXCL10 was characterized by surface plasmon resonance (SPR; Fig. 2B). Three conditions were used: i) association and dissociation at pH 7.4; ii) association at pH 7.4 and dissociation at pH 6.0; iii) association and dissociation at pH 6.0. A high affinity anti-hCXCL10 mAb, CF1, was used as a pH-independent control. By coating biotinylated hCXCL10 on a streptavidin chip and using the IgGs as analytes, we confirmed the pH-dependency of C7. As expected, the complex formed between hCXCL10 and C7 dissociated faster at pH 6.0 than at pH 7.4. Moreover, the binding of C7 to hCXCL10 was reduced when the association was performed at pH 6.0 (Fig. 2B). In comparison, pH variations did not affect the binding of CF1 to hCXCL10. By performing kinetic analyses, we determined the affinity constant of C7 for hCXCL10 at pH 7.4 (KD 4.0 nM) and at pH 6.0 (KD 9.4 nM), highlighting a 2.4-fold lower affinity at pH 6.0. This decrease in affinity was due to a faster dissociation of the complex (kd(pH 7.4) = 5.74×10−4 [s−1] versus kd(pH 6.0) = 1.51 × 10−3 [s−1]), while the association constants were similar independent of the pH (ka(pH 7.4) = 1.44 × 105 [M−1s−1] vs. ka(pH 6.0) = 1.62 × 105 [M−1s-1]).
Figure 2.
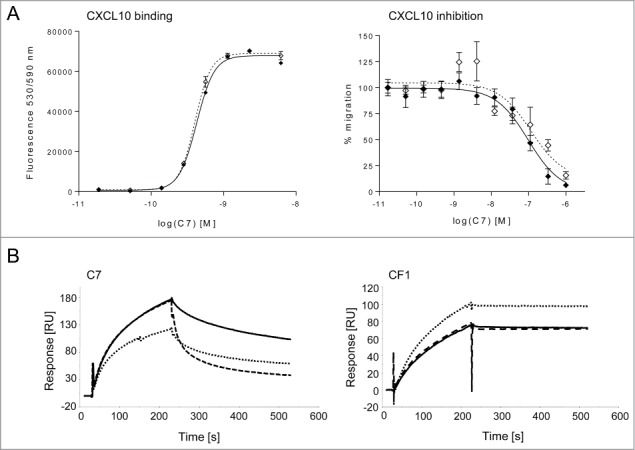
Characterization of C7 IgG. (A). Cross-reactivity of C7. C7 binds to murine (⋄) and human (⧫) CXCL10 with similar affinities by ELISA (left panel) and inhibits both murine and human CXCL10-mediated chemotaxis of L1.2 cells transfected with hCXCR3 (right panel). The ELISA as well as the chemotaxis assay were performed at pH 7.4. Chemotaxis data are presented as a percentage of cell migration, with 100 % being the migration observed with either 5 nM hCXCL10 or 5 nM mCXCL10. In both graphs, data are presented as the mean of triplicates +/− SEM and are representative of at least 2 independent experiments. (B). Binding of C7 to hCXCL10 by SPR. The biotinylated chemokine was captured on a streptavidin chip and the antibody was used as analyte. Following association at pH 7.4, the complex between C7 and hCXCL10 dissociate faster at pH 6.0 (- -) than at pH 7.4 (—). The signal obtained following association and dissociation at pH 6.0 (.....) was lower than at pH 7.4, suggesting that the affinity of C7 for hCXCL10 is reduced at slightly acidic pH. In contrast, the binding of the reference antibody CF1 to hCXCL10 was not affected by variations in pH.
We next investigated whether C7 could be simultaneously improved to increase its pH-dependency and CXCL10 neutralization potential. Lead optimization libraries were constructed by introducing diversity in the CDRs 1, 2 and 3 of the VH or the VL of C7 while the other variable domain was kept unmodified (Fig. S2). In CDRs 1 and 2, diversity was introduced at positions known to be involved in antigen-antibody interactions 36,37 whereas other positions were maintained as germline. In total, 23 positions were diversified in the CDRL1, L2, H1 and H2 using alternated NAT (encoding D, N, Y and H) and NNT (encoding 15 amino acids, including histidine) codons. In the CDRH3 and L3, residues were mutated in groups of 4, using YAT and NNT codons. Following transformation, we determined the final size of these libraries to be 1.3 × 108 and 2.5 × 108 transformants for the VH and the VL libraries respectively.
As we were more interested in obtaining antibodies against mCXCL10 than hCXCL10, selections against the former were performed as described in Figure 1B with the exception that the elution step in the third round was either reduced to 15 minutes or performed at pH 6.0 in order to preferentially recover phage that would dissociate quickly under these conditions. In addition, the concentration of mCXCL10 was decreased to 10 nM during the 3 rounds of selection in order to select for scFvs with higher affinity. The output of the second and third rounds were screened in a homogenous binding assay using fluorometric microvolume assay technology (FMAT) performed at pH 7.4 and pH 6.0. Clones were considered positive if binding was observed at pH 7.4, but not at pH 6.0 nor with the irrelevant protein hIL6R (Fig. 3A). No positive clone was retrieved from the library containing a diversified VL. By sequencing positive clones obtained from the library containing a diversified VH, 5 different sequences were identified that contained between one and 4 additional histidines in the CDRs of the VH compared to the parental C7 sequence (Fig. 3B). All positive candidates from the output of the third round of selection corresponded to the sequences of 1A4, indicating that this clone dominated the selection process when the stringency was increased. The sequences were also enriched in aspartate, tyrosine and asparagine residues.
Figure 3.
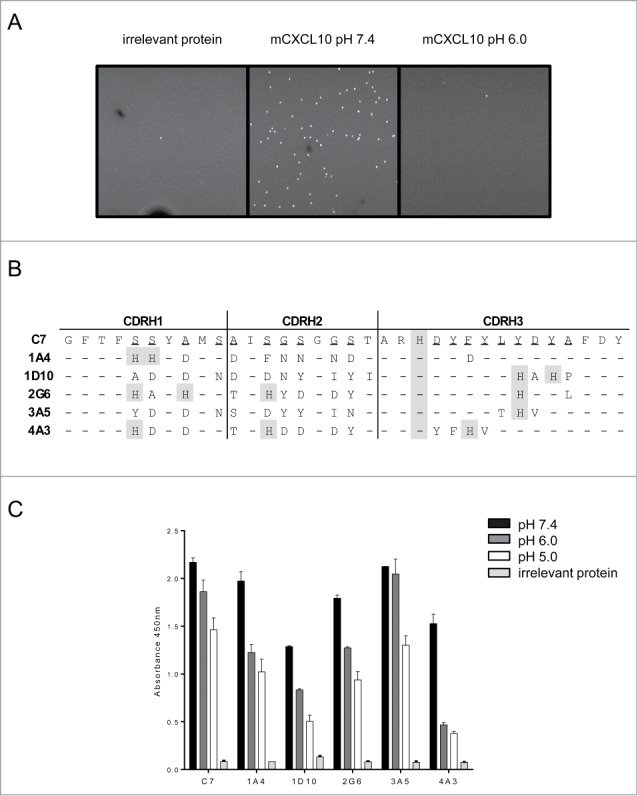
Optimization of C7. (A). pH-dependent screening of selection outputs by FMAT. Streptavidin beads coated with mCXCL10 were incubated either at pH 6.0 or 7.4 with scFv isolated from periplasmic extracts. Candidates were considered as positive if a binding was observed at pH 7.4 but not at pH 6.0 nor with an irrelevant protein (hIL6R). (B). CDRH sequences of 5 positive candidates, presented in comparison to the C7 clone. Randomized positions are underlined and histidines are shaded in gray. (−) : same amino acid as C7. (C). Sequences reformatted onto the hIgG1 backbone of hits described in B were tested for pH-dependency by ELISA. Biotinylated mCXCL10 was captured into streptavidin wells and following binding of the clones to the chemokine, an elution step was performed at the indicated pH and the binding remaining after one hour was assessed.
The five candidates were reformatted into human IgG1 and further characterized by in vitro assays. By ELISA, we demonstrated that all IgGs were able to bind mCXCL10 at pH 7.4 and the signals obtained were reduced following incubation at pH 6.0 or 5.0 (Fig. 3C). In this format, the binding of C7 to mCXCL10 was reduced by 30% following elution at pH 5.0, whereas 1A4, 1D10 and 4A3 lost 50–70% of their binding under the same conditions, indicating a stronger pH-dependency. 1A4, 1D10 and 4A3 were tested in a chemotaxis assay for their capacity to inhibit migration of cells in response to 5 nM mCXCL10 (Fig. 4A). While 1D10 and 4A3 exhibited an activity similar to that of C7 (IC50 295 nM), the candidate 1A4 was significantly more potent, with an IC50 of 1.6 nM, corresponding to a 180-fold improvement of potency compared to C7. Similarly, when performing dose-response ELISA, 1A4 displayed the strongest pH-dependency with a 167-fold higher apparent affinity for mCXCL10 at pH 7.4 than at pH 6.0, whereas the original C7 clone displayed a 24-fold difference between its apparent affinity at pH 7.4 and at pH 6.0 (Fig. 4B).
Figure 4.
Characterization of candidates isolated from C7 optimization selections. (A). Inhibition of 5 nM mCXCL10-mediated chemotaxis of L1.2/CXCR3 cells with C7 (black), 1A4 (red), 1D10 (green) and 4A3 (blue). Data are presented as a percentage of cell migration, with 100 % being the migration observed with 5 nM mCXCL10 and as the mean of 3 measurements ± SEM. They are representative of 2 independent experiments. (B). Dose-response curves demonstrating the binding of C7 (open diamonds) and 1A4 (filled circles) to mCXCL10 in a pH-dependent ELISA. The chemokine was captured on the plate via a biotin tag and the binding of the antibodies to their target was followed by an elution step at pH 7.4 or 6.0. The EC50 of 1A4 shifts from 0.19 nM to 56.5 nM when the pH of the elution step is lowered to 6.0, while the EC50 of C7 increases from 1.4 nM to 34.4 nM. (C). Binding of mCXCL10 to C7 (left) or 1A4 (right) coated on Protein A sensors by BLI. A comparison was performed between dissociation at pH 7.2 (red) and at pH 6.0 (blue) following association at pH 7.2. The experiment was also performed with conditions at pH 6.0 (gray). (D). Binding of C7 (left) or 1A4 (right) to biotinylated mCXCL10 coated on streptavidin biosensors. The color code is similar to that used in C.
These data indicate that, as 1A4 has improved characteristics, C7 could be simultaneously engineered for increased pH dependency and CXCL10 neutralization potential. We further characterized the binding kinetics of C7 and 1A4 under different pH conditions using bio-layer interferometry (BLI). We performed these assays in 2 different formats, either immobilizing the antibody or the chemokine at the extremity of the biosensor (Fig. 4C and D).
First, antibodies were captured on Protein A biosensors and transferred to buffer at pH 7.2 that contained chemokine to monitor association. The dissociation was then followed either at pH 7.2 or 6.0, and 1A4 exhibited a sharp dissociation at pH 6.0 in contrast to C7 (Fig. 4C). In addition, when both association and dissociation were performed at pH 6.0, C7 was able to capture significant levels of mCXCL10 whereas the signal obtained with 1A4 remained close to baseline, thus confirming the ELISA data. Surprisingly, the dissociation at pH 7.2 was slow for both IgGs, probably due to an avidity effect caused by multiple interactions between antibodies and chemokine. Indeed, chemokines are known to form tetramers as well as higher order oligomers, 38 and thus can provide a matrix for multiple antibodies to bind, leading to very slow dissociation kinetics as schematically represented in Fig. 4C (right panel). This avid interaction may also be the reason that the signal does not return to baseline after the sharp drop during the dissociation phase at pH 6.0. Such SPR profiles have been reported for several analytes and are most likely explained by biologically irrelevant artifacts due to the format used during the experiment.39 In agreement with the hypothesis that these interactions may mask the different kinetic properties of 1A4 and C7, when the chemokine was immobilized and the antibody used as analyte the difference in affinity between 1A4 and C7 was more pronounced when association and dissociation were performed at pH 7.2 (Fig. 4D). In this format, both IgGs showed a fast dissociation at pH 6.0 and the binding of C7 to mCXCL10 at pH 6.0 was also confirmed. It is worth noting that following the dissociation phase, a residual binding was also observed with this format, most likely due to bivalent interactions of mAbs with their target. We nevertheless performed kinetic analysis for both antibodies using the latter format because it is less prone to avidity effects. At pH 7.2, 1A4 showed a greater affinity than C7 for mCXCL10, with a KD of 1 nM, whereas the affinity constant of C7 was determined to be 11.2 nM (Table 3). At pH 6.0, 1A4 did not bind to mCXCL10, whereas binding was observed for C7, with a dissociation rate 4-fold faster than at pH 7.2, a pH-dependency similar to that initially observed with the human chemokine. In order to quantify the pH-dependency of 1A4, we performed kinetic studies with the association phase at pH 7.2 and the dissociation at pH 6.0. Using this format, a 21-fold difference in dissociation rate between pH 7.2 and 6.0 was observed for the binding of 1A4 to mCXCL10 compared to a 4-fold difference for C7 (Table 3). These results were confirmed by the determination of the affinity of C7 and 1A4 antigen-binding fragments (Fab) for biotinylated mCXCL10 loaded onto streptavidin biosensors. Using these fragments, we confirmed that 1A4 binding to mCXCL10 was of higher affinity and of higher pH-dependency than that of C7, even in a monovalent format (Table 3 and Figure S3). Overall, our data demonstrate that 1A4 is a potent neutralizer of CXCL10 and binding is highly dependent on pH as 1A4 shows nanomolar affinity at physiological pH, but is unable to bind CXCL10 at a pH of 6.0.
Table 3.
Binding kinetics of the pH-dependent anti-mCXCL10 clones C7 and 1A4. The affinity of C7 and 1A4 (IgG and Fab formats) for mCXCL10 was determined by BLI. The pH of the association and of the dissociation phases are indicated at the top of each column (pH association/pH dissociation). The pH 6.0 to pH 7.2 kd ratio of each antibody is shown
| pH 7.2/pH7.2 |
pH 7.2/pH6.0 |
Ratio kd2/kd1 | ||||
|---|---|---|---|---|---|---|
| ka *104 |
kd1 *10–4 |
KD |
kd2 *10–4 |
|||
| Clone | [M-1s-1] | [s-1] | [nM] | [s-1] | ||
| C7 | IgG | 43.6 | 48.6 | 11.2 | 186 | 4 |
| 1A4 | IgG | 80.4 | 8.3 | 1.0 | 180 | 22 |
| C7 | Fab | 1.7 | 7.1 | 41.6 | 88 | 12 |
| 1A4 | Fab | 2.7 | 3.2 | 11.7 | 478 | 151 |
As discussed above, mAbs are widely used as research, diagnostic, preparative and therapeutic reagents,6 and now form a rapidly growing class of drugs. In all cases, the high affinity of these highly selective molecules confers the disadvantage of forming complexes that are so stable that they require harsh conditions to dissociate the antibody from its target. This usually results in denaturation of the antibody, rendering it unsuitable for repeated use in applications such as diagnostics kits and preparative reagents. The development of pH-dependent antibodies will thus be valuable because they can be used repeatedly, with an obvious cost per sample improvement. Moreover, as already mentioned, such antibodies would also be useful in therapeutic settings as the pH-dependency of antibodies was reported to improve the half-life of mAbs in vivo by enhancing the recycling of free antibody (for a recent review, see ref. 24). Furthermore, a recent publication highlighted the use of pH-dependent mAb as so termed “sweeping antibodies:” the introduction of mutations to increase their affinity for FcRn resulted in antibodies that were able to selectively eliminate their target from plasma.30 The antigen concentration in circulation was reduced up to 1000-fold compared to standard antibodies, supporting the hypothesis that the use of pH-dependent mAbs could improve therapeutic efficacy.
Previous approaches aimed at conferring pH-dependency to a pre-existing antibody by introducing histidines in the antigen contact region, a strategy that is labor intensive because it requires: i) isolation and characterization of a non-pH-dependent antibody by immunization or phage display; ii) introduction of histidines in the CDRs of this candidate to produce an extensive range of single mutants; iii) characterization of these single mutants; and iv) combination of the individual mutations isolated to obtain an antibody with appropriate pH-dependent binding characteristics. This strategy may be risky as it is probable that not all existing antibodies exhibit a sequence or conformation suitable for transformation into a pH-dependent mAb. In this study, we substituted this process by the de novo isolation of pH-dependent antibodies using libraries enriched in histidine residues combined with phage display selections including an elution step at slightly acidic pH for the preferential amplification of pH-dependent clones. Histidine-enriched libraries are universal and could be used to isolate mAbs directed against theoretically any biological target, therefore allowing a faster process. Furthermore, we could simultaneously improve 2 characteristics of a selected candidate as we obtained variants that are more potent neutralizers and display greater pH-dependency. The resulting IgG, 1A4, had nanomolar affinity for CXCL10 at pH 7.2, while completely detaching from its target in less than 30 seconds when the pH was set to 6.0. Moreover, this antibody had no detectable binding at pH 6.0, which is in contrast to most previously described antibodies that retained some level of binding at pH 6.0.25,26 The only antibody published so far that did not bind its target at pH 6.0 was the result of mutagenesis of an initial antibody showing naturally reduced binding at the lower pH, suggesting that greater pH-dependency can potentially be achieved within a favorable combining site.27 It is worth noting that our results were obtained starting with a relatively small histidine-enriched library (1.3 × 109 transformants). The construction of larger libraries (e.g. > 1010 individual clones) should increase the chances of success of isolating strongly pH-dependent binders and therefore reduce the need to perform additional lead optimization. We therefore anticipate that the de novo isolation of pH-dependent antibodies will represent an efficient route, relying on the upfront selection of optimal conformations or sequences rather than via the re-engineering of an antibody combining site that was not initially selected for pH-dependent binding properties.
Material and Methods
Material, methods and associated references are available online as supplementary material.
Disclosure of Potential Conflicts of Interest
The authors are current or former employees of NovImmune SA.
Acknowledgments
We thank G. Magistrelli for help in protein expression and purification, P. Malinge for advice in SPR and BLI experiments, N. Bosson, S. Calloud, N. Costes, M. Galby-Charreton and M. Guerrier for their help with library construction and phage display selections.
Funding
The research leading to these results has received funding from the European Union Seventh Framework Programme [FP7–2007–2013] under grant agreement n° HEALTH-F4–2011–281608 (TIMER).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Yagami H, Kato H, Tsumoto K, Tomita M. Monoclonal antibodies based on hybridoma technology. Pharm Pat Anal 2013; 2:249-63; http://dx.doi.org/ 10.4155/ppa.13.2 [DOI] [PubMed] [Google Scholar]
- 2. Hairul Bahara NH, Tye GJ, Choong YS, Ong EB, Ismail A, Lim TS. Phage display antibodies for diagnostic applications. Biologicals 2013; 41:209-16; PMID:23647952; http://dx.doi.org/ 10.1016/j.biologicals.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 3. Vincent KJ, Zurini M. Current strategies in antibody engineering: Fc engineering and pH-dependent antigen binding, bispecific antibodies and antibody drug conjugates. Biotechnol J 2012; 7:1444-50; PMID:23125076; http://dx.doi.org/ 10.1002/biot.201200250 [DOI] [PubMed] [Google Scholar]
- 4. Sela-Culang I, Kunik V, Ofran Y. The structural basis of antibody-antigen recognition. Front Immunol 2013; 4:302; PMID:24115948; http://dx.doi.org/ 10.3389/fimmu.2013.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aggarwal RS. What's fueling the biotech engine-2012 to 2013. Nat Biotechnol 2014; 32:32-39; PMID:24406926; http://dx.doi.org/ 10.1038/nbt.2794 [DOI] [PubMed] [Google Scholar]
- 6. Campbell AM. ed. Monoclonal antibody technology: the production and characterization of rodent and human hybridomas. New York: Elsevier Science Ltd; 1984. [Google Scholar]
- 7. Zeng X, Shen Z, Mernaugh R. Recombinant antibodies and their use in biosensors. Anal Bioanal Chem 2012; 402:3027-38; http://dx.doi.org/ 10.1007/s00216-011-5569-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deb R, Chakraborty S, Veeregowda B, Veram A, Tiwari R, Dhama K. Monoclonal antibody and its use in the diagnosis of livestock diseases. Adv Biosci Biotechnol 2013; 50-62; http://dx.doi.org/ 10.4236/abb.2013.44A008 [DOI] [Google Scholar]
- 9. Moser AC, Hage DS. Immunoaffinity chromatography: an introduction to applications and recent developments. Bioanalysis 2010; 2:769-90; PMID:20640220; http://dx.doi.org/ 10.4155/bio.10.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tomomori-Sato C, Sato S, Conaway RC, Conaway JW. Immunoaffinity purification of protein complexes from Mammalian cells. Methods Mol Biol 2013; 977:273-87; PMID:23436370; http://dx.doi.org/ 10.1007/978-1-62703-284-1_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davoli E, Fanelli R, Bagnati R. Purification and analysis of drug residues in urine samples by on-line immunoaffinity chromatography/high-performance liquid chromatography/continuous-flow fast atom bombardment mass spectrometry. Anal.Chem. 1993; 65:2679-85. [DOI] [PubMed] [Google Scholar]
- 12. Jorge I, Burillo E, Mesa R, Baila-Rueda L, Moreno M, Trevisan-Herraz M, Silla-Castro JC, Camafeita E, Ortega-Munoz M, Bonzon-Kulichenko E, et al. The human HDL proteome displays high inter-individual variability and is altered dynamically in response to angioplasty-induced atheroma plaque rupture. J Proteomics 2014; 106:61-73; PMID:24747125; http://dx.doi.org/ 10.1016/j.jprot.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 13. Ehle H, Horn A. Immunoaffinity chromatography of enzymes. Bioseparation 1990; 1:97-110; PMID:1368167 [PubMed] [Google Scholar]
- 14. Luppa PB, Sokoll LJ, Chan DW. Immunosensors–principles and applications to clinical chemistry. Clin Chim Acta 2001; 314:1-26. [DOI] [PubMed] [Google Scholar]
- 15. Gonzalez-Martinez MA, Puchades R, Maquieira A. Optical immunosensors for environmental monitoring: how far have we come? Anal Bioanal Chem 2007; 387:205-18; PMID:17072601; http://dx.doi.org/ 10.1007/s00216-006-0849-8 [DOI] [PubMed] [Google Scholar]
- 16. Ricci F, Volpe G, Micheli L, Palleschi G. A review on novel developments and applications of immunosensors in food analysis. Anal Chim Acta 2007; 605:111-29; PMID:18036374; http://dx.doi.org/ 10.1016/j.aca.2007.10.046 [DOI] [PubMed] [Google Scholar]
- 17. Goding JW. ed. Monoclonal antibodies: principles and practice. London: Academic Press; 1986. [Google Scholar]
- 18. Chakraborty A, Van LM, Skerjanec A, Floch D, Klein UR, Krammer G, Sunkara G, Howard D. Pharmacokinetic and pharmacodynamic properties of canakinumab in patients with gouty arthritis. J Clin Pharmacol 2013; 53:1240-51; PMID:24122883; http://dx.doi.org/ 10.1002/jcph.162 [DOI] [PubMed] [Google Scholar]
- 19. Fetterly GJ, Aras U, Meholick PD, Takimoto C, Seetharam S, McIntosh T, de Bono JS, Sandhu SK, Tolcher A, Davis HM, et al. Utilizing pharmacokinetics/pharmacodynamics modeling to simultaneously examine free CCL2, total CCL2 and carlumab (CNTO 888) concentration time data. J Clin Pharmacol 2013; 53:1020-7; PMID:23878055; http://dx.doi.org/ 10.1002/jcph.140 [DOI] [PubMed] [Google Scholar]
- 20. Haringman JJ, Gerlag DM, Smeets TJ, Baeten D, van den Bosch F, Bresnihan B, Breedveld FC, Dinant HJ, Legay F, Gram H, et al. A randomized controlled trial with an anti-CCL2 (anti-monocyte chemotactic protein 1) monoclonal antibody in patients with rheumatoid arthritis. Arthritis Rheum. 2006; 54:2387-92; PMID:16869001; http://dx.doi.org/ 10.1002/art.21975 [DOI] [PubMed] [Google Scholar]
- 21. Siemers ER, Friedrich S, Dean RA, Gonzales CR, Farlow MR, Paul SM, Demattos RB. Safety and changes in plasma and cerebrospinal fluid amyloid beta after a single administration of an amyloid beta monoclonal antibody in subjects with Alzheimer disease. Clin Neuropharmacol 2010; 33:67-73; PMID:20375655; http://dx.doi.org/ 10.1097/WNF.0b013e3181cb577a [DOI] [PubMed] [Google Scholar]
- 22. Xiao JJ, Krzyzanski W, Wang YM, Li H, Rose MJ, Ma M, Wu Y, Hinkle B, Perez-Ruixo JJ. Pharmacokinetics of anti-hepcidin monoclonal antibody Ab 12B9m and hepcidin in cynomolgus monkeys. AAPS J 2010; 12:646-57; PMID:20737261; http://dx.doi.org/ 10.1208/s12248-010-9222-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giragossian C, Clark T, Piche-Nicholas N, Bowman CJ. Neonatal Fc receptor and its role in the absorption, distribution, metabolism and excretion of immunoglobulin G-based biotherapeutics. Curr Drug Metab 2013; 14:764-90; PMID:23952252; http://dx.doi.org/ 10.2174/13892002113149990099 [DOI] [PubMed] [Google Scholar]
- 24. Igawa T, Mimoto F, Hattori K. pH-dependent antigen-binding antibodies as a novel therapeutic modality. Biochim Biophys Acta 2014; http://dx.doi.org/ 10.1016/j.bbapap.2014.08.003; PMID:25125373 [DOI] [PubMed] [Google Scholar]
- 25. Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, Moriyama C, Watanabe T, Takubo R, Doi Y, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol 2010; 28:1203-7; PMID:20953198; http://dx.doi.org/ 10.1038/nbt.1691 [DOI] [PubMed] [Google Scholar]
- 26. Chaparro-Riggers J, Liang H, DeVay RM, Bai L, Sutton JE, Chen W, Geng T, Lindquist K, Casas MG, Boustany LM, et al. Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J Biol Chem 2012; 287:11090-7; PMID:22294692; http://dx.doi.org/ 10.1074/jbc.M111.319764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Devanaboyina SC, Lynch SM, Ober RJ, Ram S, Kim D, Puig-Canto A, Breen S, Kasturirangan S, Fowler S, Peng L, et al. The effect of pH dependence of antibody-antigen interactions on subcellular trafficking dynamics. MAbs. 2013; 5:851-9; PMID:24492341; http://dx.doi.org/ 10.4161/mabs.26389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stryer L, Berg JM, Tymoczko JL. ed. Biochemistry. New York: W.H. Freeman; 2002. [Google Scholar]
- 29. Sarkar CA, Lowenhaupt K, Horan T, Boone TC, Tidor B, Lauffenburger DA. Rational cytokine design for increased lifetime and enhanced potency using pH-activated "histidine switching". Nat Biotechnol 2002; 20:908-13; PMID:12161759; http://dx.doi.org/ 10.1038/nbt725 [DOI] [PubMed] [Google Scholar]
- 30. Igawa T, Maeda A, Haraya K, Tachibana T, Iwayanagi Y, Mimoto F, Higuchi Y, Ishii S, Tamba S, Hironiwa N, et al. Engineered monoclonal antibody with novel antigen-sweeping activity in vivo. PLoS One 2013; 8:e63236; PMID:23667591; http://dx.doi.org/ 10.1371/journal.pone.0063236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murtaugh ML, Fanning SW, Sharma TM, Terry AM, Horn JR. A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches. Protein Sci. 2011; 20:1619-31; PMID:21766385; http://dx.doi.org/ 10.1002/pro.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J.Mol.Biol. 2003; 334:733-49; http://dx.doi.org/ 10.1016/j.jmb.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 33. Venet S. Human antibody libraries capturing purpose oriented CDRH3 repertoires ; dissertation. Lausanne (Switzerland): Ecole Polytechnique Fédérale de Lausanne; 2013; 97-98. [Google Scholar]
- 34. Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J Mol Biol 2005; 348:1153-62; PMID:15854651; http://dx.doi.org/ 10.1016/j.jmb.2005.03.041 [DOI] [PubMed] [Google Scholar]
- 35. Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 2003; 27:55-77; PMID:12477501; http://dx.doi.org/ 10.1016/S0145-305X(02)00039-3 [DOI] [PubMed] [Google Scholar]
- 36. Honegger A, Pluckthun A. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol 2001; 309:657-70; PMID:11397087; http://dx.doi.org/ 10.1006/jmbi.2001.4662 [DOI] [PubMed] [Google Scholar]
- 37. Ignatovich O, Jespers L, Tomlinson IM, de Wildt RM. Creation of the large and highly functional synthetic repertoire of human VH and Vkappa domain antibodies. Methods Mol Biol 2012; 911:39-63; PMID:22886245; http://dx.doi.org/ 10.1007/978-1-61779-968-6_4 [DOI] [PubMed] [Google Scholar]
- 38. Salanga CL, Handel TM. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: the role of structural dynamics in function. Exp Cell Res 2011; 317:590-601; PMID:21223963; http://dx.doi.org/ 10.1016/j.yexcr.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rich RL, Myszka DG. Grading the commercial optical biosensor literature-Class of 2008: 'The Mighty Binders'. J Mol Recognit 2010; 23:1-64; PMID:20017116; http://dx.doi.org/ 10.1002/jmr.1004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



