Abstract
Plant growth is controlled by intrinsic developmental programmes and environmental cues. Jasmonate (JA) has important roles in both processes, by regulating cell division and differentiation, as well as in defense responses and senescence. We report an increase in rice plant height caused by overexpression of a gene encoding a cytochrome P450 enzyme, CYP94C2b, which promoted deactivation of JA-Ile. The height increase occurred through enhanced elongation of internodes in the absence of concomitant cell elongation, unlike previous findings with coi1 knock-down plants. Thus, modulating JA metabolism can increase the number of elongated cells in an internode. Based on these and previous findings, we discuss the difference in the effects of CYP94C2b overexpression vs. coi1 knock-down.
Keywords: cell division, cell elongation, cytochrome P450, internode, jasmonate, plant height
Abbreviations
- bHLH
basic helix-loop-helix
- COI1
CORONATINE INSENSITIVE1
- CYP
cytochrome P450
- JA
Jasmonate
- JAZ
JASMONATE ZIM DOMAIN
- RSS3
RICE SALT SENSITIVE3
- SCF
Skp1-cullin-F-box
The plant hormone jasmonate (JA) and its conjugates have important roles in defense responses, growth control and leaf senescence.1-3 JA has also been suggested to be involved in plant response and adaptation to abiotic stresses.4 In line with these studies, we recently showed that repressive regulation of JA signaling is important for growth and viability of rice under stressful conditions.5,6
In plant cells, JA is converted into bioactive JA-Ile, which is recognized by binding to SCFCOI1, together with JAZ (jasmonate ZIM domain) proteins.7,8 This recognition causes ubiquitination and subsequent degradation of the JAZ factors that repress activity of basic helix–loop–helix (bHLH) transcription factors, such as MYC factors, leading to de-repression of JA-responsive genes.9-12 A nuclear factor called RICE SALT SENSITIVE 3 (RSS3) that interacts with both class C bHLH and JAZ factors is also involved in the repression of JA-responsive gene expression in rice. This repression is disrupted in the root tips of rss3 mutants, in which root elongation is severely inhibited under saline conditions. Thus, repression of JA signaling is important for root elongation, which is elaborately controlled according to growth conditions.5
In Arabidopsis, JA-Ile is deactivated by its conversion into 12-OH-JA-Ile and 12-COOH-JA-Ile, catalyzed by 2 cytochrome P450 enzymes, CYP94B3 and CYP94C1.13-15 Kurotani et al.6 reported that CYP94C2b, a rice homolog of CYP94C1, also promotes deactivation of JA-Ile. CYP94C2b overexpression causes reduced levels of JA-Ile after wounding and alleviates responses to wounding or exogenous JA, in the context of gene expression and inhibition of shoot and root growth.6 Notably, increased CYP94C2b expression enhances viability of rice plants under saline conditions, although extremely high levels of CYP94C2b expression have detrimental effects on plant survival. These findings imply that repression of exaggerated JA responses is important for plant survival under harsh environmental conditions.
Using the same transgenic lines as those tested for salt tolerance,6 we investigated whether enhanced CYP94C2b expression affects growth of rice under non-stress conditions. We compared the length of the second leaf sheath, as it is reliable to evaluate shoot elongation of rice seedlings. As shown in Figure 1A, increased CYP94C2b expression resulted in enhanced elongation of the leaf sheath. Interestingly, in contrast to the effect under saline conditions, extremely high levels of CYP94C2b expression under non-stress conditions did not negatively affect shoot growth (Fig. 1B). Thus, different mechanisms downstream of JA signaling may operate in the shoot growth under non-stress conditions and viability under salinity stress conditions. In Arabidopsis, one study observed that wounding induces growth stunting accompanied by reduction in leaf area and cell number, but not in cell size, thereby suggesting that JA inhibits cell division and thereby plant growth.16 This wound-induced stunting is not observed in mutants of JA synthesis and signaling, such as aos, opr3, coi1, myc2 and jaz3. Moreover, the expression of a cyclinB1;2::GUS reporter gene is reduced in shoot meristem and young leaves after treatment with JA.16 In the case of lines overexpressing CYP94C2b, however, expression levels of the cell cycle marker genes, PCNA and CycB2;1, are not increased,6 suggesting that the enhanced shoot elongation by deactivation of JA-Ile is not associated with activation of cell division at the seedling stage.
Figure 1.
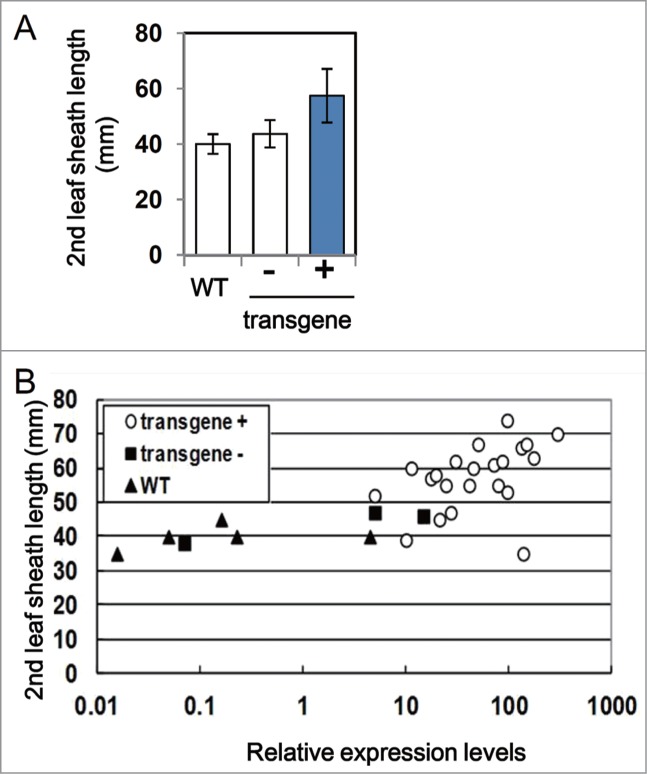
Shoot elongation in the wild-type (WT) and CYP94C2b-overexpressing lines at seedling stage under non-stress conditions. (A) Length of the second leaf sheath in 1-week-old rice seedlings with (+) and without (−) the CYP94C2b transgene, segregated from the same line, was compared to that in non-transgenic WT plants. Average and standard deviation in WT and siblings of the transgenic lines are indicated. n ≥ 3. (B) Plot of shoot length and relative expression levels of CYP94C2b in each plant. Expression levels of CYP94C2b were analyzed by quantitative reverse transcription PCR and normalized to those of 25S rRNA as reported in ref. 6.
In the greenhouse, plants overexpressing CYP94C2b showed increased height in comparison with non-transgenic wild-type (WT) plants (Fig. 2A). In rice, a major determinant of plant height is the internode elongation that occurs during vegetative-to-reproductive phase transition. Enhancement of internode elongation by CYP94C2b overexpression was observed with the 4 uppermost internodes and was most apparent in the second internode from the node beneath the panicle (Fig. 2B). The enhanced growth in shoots and internodes observed in the plants overexpressing CYP94C2b implies a repressive role for JA in rice growth under non-stress conditions. Superficially, this seems consistent with the observation of Yang et al.17 that suppression of COI1 function by RNA interference causes enhanced growth in both rice seedlings and mature plants.
Figure 2.

Increase in culm and internode length in lines overexpressing CYP94C2b. (A) Representative culm lengths of the wild-type (WT) and the CYP94C2b-overexpressing line (FE047) at seed maturation stage after flowering. Internodes are numbered from top to bottom as described.19 The longest culm in each plant was used for this and the following comparison. Bar, 10 cm. (B) Length of the first to fourth internodes in plants (+) overexpressing CYP94C2b or in WT plants (−). Internode positions are numbered as in (A). Mean and standard deviation for respective internodes are presented. n = 5 (no transgene); n = 18 (with transgene). Asterisks indicate significant differences in internode length between the plants with and without the CYP94C2b transgene (**p < 0.01; *p < 0.05, Student's t-test). On average, internode length was increased 1.3-fold by CYP94C2b overexpression (first internode, 1.2-fold increase; second, 1.5-fold; third, 1.2-fold; fourth, 1.3-fold). (C and D) Length of cells in the uppermost internode just below the panicle of WT lines and the FE047 line overexpressing CYP94C2b. (C) Longitudinal tissue sections were prepared by hand sectioning with a razor blade, stained with 1% safranin and observed under a dark-field microscope. Representative images for WT and FE047 are shown. Bar, 50 μm. (D) Cell length at the indicated part of the uppermost internode. Top, at 1 cm below the upper node. Middle, mid-region of the internode. Bottom, 1 cm above the lower node. Length of cells located in the third layer from the medullary cavity was measured. For each part, the average length of 50–120 cells within the 1-cm height region was scored. In each graph bar, the average of scores obtained with 4 plants is presented. Error bars indicate standard deviations.
In rice, internode elongation is caused by cell division in the intercalary meristem, followed by cell elongation in the elongation zone.18,19 In the coi1 knock-down rice plants, longitudinal internode elongation is associated with increased cell elongation.17 It has been well known that gibberellins is involved in internode elongation in rice.17,20,21 In addition, coi1 knock-down plants appear to exhibit hallmarks phenotypes of GA hypersensitive mutants.17 Therefore, a possible crosstalk might exist between JA and GA signaling pathways to control internode growth. Consistent with this, overexpression of GA-deactivating enzyme (EUI1) also suppresses the effect of coi1 knock-down in the increase in internode elongation that is associated with enhanced cell elongation.17 In the case of deepwater rice, however, GA induces growth in internodes by promoting both cell division and cell elongation.20 Noticeably, in the lines overexpressing CYP94C2b, we found no significant changes in the extent of cell elongation in the middle and bottom regions of the internode and the cells in the top region showed even reduced elongation (Fig. 2C and D). Thus, the enhanced internode elongation observed in the lines overexpressing CYP94C2b cannot be explained by enhanced cell elongation. Our results instead indicate that the number of cells, including those undergoing elongation, is increased by CYP94C2b overexpression. This can be achieved by enhanced cell division in the intercalary meristem and/or delayed arrest or termination of the meristem. It is also possible that CYP94C2b overexpression increases the ratio of cells that undergo elongation, which is often regulated in coordination with cell division control. Whatever the explanation, our results raise a question as to what mechanisms are responsible for the difference between the modes of internode growth observed in the coi1 knock-down plants and CYP94C2b overexpressing plants.
A possible explanation for the difference is that production of 12-OH-JA-Ile and 12-COOH-JA-Ile is facilitated in the CYP94C2b-overexpressing but not coi1-knocked down rice, resulting in different effects on cell number and cell size in internodes. In Arabidopsis, it has recently been reported that CYP94B3 and CYP94C1 overexpression increases the accumulation of 12-O-glucosyl-JA and 12-HSO4-JA.22 Such changes of the levels of jasmonate derivatives might also occur in CYP94C2b-overexpressing rice and affect physiological states. Another possibility is that the dynamical pattern of JA signals is differentially affected by the 2 types of modulation. CYP94C2b is likely involved in negative feedback regulation of JA signaling, as in the case of CYP94B3 and CYP94C1.6 Accordingly, enhanced CYP94C2b expression may affect mainly the duration of the JA signal. In contrast, COI1 functions as a receptor for bioactive JA-Ile. Thus, COI1 dysfunction should result in reduced JA signaling amplitude. Conceivably, such dynamical properties of signals are important for controlling gene regulation and cell differentiation, as has been suggested in mammals.23 In our case, prolonged JA signals might be involved in controlling the number of elongated cells, whereas increased amplitude of JA signals would be required to regulate cell elongation. It could be argued that overexpression of enzymes that deactivate JA-Ile also causes reduced amplitude of JA signals. However, this largely depends on the strength of affinity to JA-Ile of CYP94C2b as compared with that of COI1. Interestingly, similar to our observation, overexpression of a JAZ/TIFFY gene, OsTIFY11b, has also been shown to cause enhanced leaf elongation in mature rice plants, but this is not associated with increased cell elongation.24 All of these studies, along with the observation that JAZ factors are involved in repressive regulation of JA signaling, may reflect that downstream factors of JA signaling are differentially activated, depending on the dynamical pattern of JA signals. It would be interesting to investigate how such regulation could be achieved.
Funding
This work was supported in part by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, AMR0002), and JSPS KAKENHI [grant No. 24580493].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Ahmed Ismail and Kenji Hayashi for helpful discussion, Drs. Hiroaki Ichikawa and Makoto Hakata for useful comments and providing seeds of the FOX line FE047, Mami Sugiura, Chiho Kanayama, Rena Endo, Yayoi Komeda and Yoshiko Nagai for technical assistance.
References
- 1.Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol Biol 1997; 48:355–81; PMID:15012267; http://dx.doi.org/ 10.1146/annurev.arplant.48.1.355 [DOI] [PubMed] [Google Scholar]
- 2.Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 2007; 100:681–97; PMID:17513307; http://dx.doi.org/ 10.1093/aob/mcm079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shyu C, Brutnell TP. Growth-defence balance in grass biomass production: the role of jasmonates. J Exp Bot 2015. [Epub ahead of print]; PMID:25711704; doi: 10.1093/jxb/erv011 [DOI] [PubMed] [Google Scholar]
- 4.Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 2015; 20:219–29; PMID:25731753; http://dx.doi.org/ 10.1016/j.tplants.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 5.Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, Ando T, Sugimoto K, Mitsuda N, Katoh E, et al.. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013; 25:1709–25; PMID:23715469; http://dx.doi.org/ 10.1105/tpc.113.112052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurotani K, Hayashi K, Hatanaka S, Toda Y, Ogawa D, Ichikawa H, Ishimaru Y, Tashita R, Suzuki T, Ueda M, et al.. Elevated Levels of CYP94 Family Gene Expression Alleviate the Jasmonate Response and Enhance Salt Tolerance in Rice. Plant Cell Physiol 2015; I56:779-89; PMID:25637374; http://dx.doi.org/ 10.1093/pcp/pcv006 [DOI] [PubMed] [Google Scholar]
- 7.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 2008; 105:7100–5; PMID:18458331; http://dx.doi.org/ 10.1073/pnas.0802332105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al.. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010; 468:400–5; PMID:20927106; http://dx.doi.org/ 10.1038/nature09430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al.. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666–71; PMID:17637675; http://dx.doi.org/ 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- 10.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007; 448:661–5; PMID:17637677; http://dx.doi.org/ 10.1038/nature05960 [DOI] [PubMed] [Google Scholar]
- 11.Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-Domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 2011; 4:279–88; PMID: 21242320; http://dx.doi.org/ 10.1093/mp/ssq073 [DOI] [PubMed] [Google Scholar]
- 12.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al.. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011; 23:701–15; PMID:21335373; http://dx.doi.org/ 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitaoka N, Matsubara T, Sato M, Takahashi K, Wakuta S, Kawaide H, Matsui H, Nabeta K, Matsuura H. Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol 2011; 52:1757–65; PMID:21849397; http://dx.doi.org/ 10.1093/pcp/pcr110 [DOI] [PubMed] [Google Scholar]
- 14.Koo AJ, Cooke TF, Howe GA. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA 2011; 108:9298–303; PMID:21576464; http://dx.doi.org/ 10.1073/pnas.1103542108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, et al.. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoy-L-isoleucine for catabolic turnover. J Biol Chem 2012; 287:6296–306; PMID:22215670; http://dx.doi.org/ 10.1074/jbc.M111.316364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Turner JG. Wound-Induced Endogenous Jasmonates stunt plant growth by inhibiting mitosis. PLoS One 2008; 3:e3699; PMID:19002244; http://dx.doi.org/ 10.1371/journal.pone.0003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al.. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 2012; 109:E1192–200; PMID:22529386; http://dx.doi.org/ 10.1073/pnas.120161610917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshikawa K. Stem in The Growing Rice Plant, Tokyo, Nobunkyo, 1989. p. 123–48. [Google Scholar]
- 19.Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000; 12:1591–606; PMID:11006334; http://dx.doi.org/http://dx.doi.org/ 10.1105/tpc.12.9.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauter M, Mekhedov SL, Kende H. Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J 1995; 7:623–32; PMID:7742859 [DOI] [PubMed] [Google Scholar]
- 21.Ayano M, Kani T, Kojima M, Sakakibara H, Kitaoka T, Kuroha T, Angeles-Shim RB, Kitano H, Nagai K, Ashikari M. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ 2014; 37:2313–24; PMID:24891164; http://dx.doi.org/ 10.1111/pce.12377.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubert Y, Widemann E, Miesch L, Pinot F, Heitz T. CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J Exp Bot 2015; [Epub ahead of print]; http://dx.doi.org/ 10.1093/jxb/erv190.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science 2012; 336:1440–4; PMID:22700930; http://dx.doi.org/ 10.1126/science.1218351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakata M, Kuroda M, Ohsumi A, Hirose T, Nakamura H, Muramatsu M, Ichikawa H, Yamakawa H. Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci Biotechnol Biochem 2012;76:2129–34; PMID:23132589; http://dx.doi.org/ 10.1271/bbb.12054524 [DOI] [PubMed] [Google Scholar]


