Abstract
Liver regeneration has been well studied with hope of discovering strategies to improve liver disease outcomes. Nevertheless, the signals that initiate such regeneration remain incompletely defined, and translation of mechanism-based pro-regenerative interventions into new treatments for hepatic diseases has not yet been achieved. We previously reported the isoform-specific regulation and essential function of zinc-dependent histone deacetylases (Zn-HDACs) during mouse liver regeneration. Those data suggest that epigenetically regulated anti-proliferative genes are deacetylated and transcriptionally suppressed by Zn-HDAC activity or that pro-regenerative factors are acetylated and induced by such activity in response to partial hepatectomy (PH). To investigate these possibilities, we conducted genome-wide interrogation of the liver histone acetylome during early PH-induced liver regeneration in mice using acetyL-histone chromatin immunoprecipitation and next generation DNA sequencing. We also compared the findings of that study to those seen during the impaired regenerative response that occurs with Zn-HDAC inhibition. The results reveal an epigenetic signature of early liver regeneration that includes both hyperacetylation of pro-regenerative factors and deacetylation of anti-proliferative and pro-apoptotic genes. Our data also show that administration of an anti-regenerative regimen of the Zn-HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) not only disrupts gene-specific pro-regenerative changes in liver histone deacetylation but also reverses PH-induced effects on histone hyperacetylation. Taken together, these studies offer new insight into and suggest novel hypotheses about the epigenetic mechanisms that regulate liver regeneration.
Keywords: chromatin immunoprecipitation, histone acetylation, histone deacetylase, partial hepatectomy, suberoylanilide hydroxamic acid
Abbreviations
- Ac-H3K9
histone H3 acetylated on lysine 9
- CDKI
cyclin dependent kinase inhibitor
- ChIP-Seq
chromatin immunoprecipitation-next generation DNA sequencing
- GO
gene ontology
- PH
partial hepatectomy
- qRT-PCR
semi-quantitative real-time reverse-transcription polymerase-chain-reaction
- SAHA
suberoylanilide hydroxamic acid
- TSS
transcription start sites
- Zn-HDAC
zinc-dependent histone deacetylase
Introduction
Recovery from all liver injuries depends on the ability of the liver to regenerate. Such regeneration has been extensively studied with hope of discovering new therapeutic strategies that improve human liver disease outcomes. Mouse two-thirds partial hepatectomy (PH) has been the paradigm most commonly used to study the regulation of liver regeneration.1 Experiments using this model show that partial liver resection induces a characteristic hepatocellular proliferative response regulated by specific cytokines, growth and transcription factors and intracellular signaling events. This response restores normal hepatic mass and function, after which hepatocytes return to their pre-regenerative state of proliferative inactivity. Nevertheless, the earliest events that initiate hepatic regeneration remain incompletely defined, and translation of mechanism-based, pro-regenerative interventions into new treatments for liver diseases has not yet been achieved.
We recently reported the isoform-specific regulation and essential function of zinc-dependent histone deacetylases (Zn-HDACs) during mouse liver regeneration.2 Those studies showed that total Zn-HDAC activity increases and the global abundance of histone H3 acetylated on lysine residue 9 (Ac-H3K9, an epigenetic mark of transcriptional activation) coincidentally declines in regenerating liver. We also discovered that some Zn-HDACs (e.g., HDACs 1, 4, and 8) exhibit increased hepatic expression, others (HDACs 9 and 11) show decreased expression, and HDAC5 undergoes nuclear translocation in early regenerating liver. Finally, we evaluated Zn-HDAC regenerative function using suberoylanilide hydroxamic acid (SAHA), a broad inhibitor of these enzymes, and showed that SAHA suppresses PH-induced hepatocellular proliferation. Those data suggest a model of liver regeneration in which epigenetically-regulated anti-proliferative factors are deacetylated and transcriptionally suppressed by direct Zn-HDAC-dependent histone deacetylation in response to PH. Consistent with that idea, the cyclin dependent kinase inhibitor (CDKI) Cdkn2d (i.e., p19Ink4d) is concordantly hyperacetylated and transcriptionally-induced by SAHA in early regenerating liver.2 Alternatively, Zn-HDAC inhibition might also suppress regeneration by indirectly promoting the deacetylation and suppressing the expression of pro-regenerative factors; however, such regulation has not yet been described in any model of liver regeneration. Based on these considerations, we undertook genome-wide interrogation of the liver histone acetylome during early regeneration in the absence and presence of Zn-HDAC inhibition, using Ac-H3K9 chromatin immunoprecipitation combined with next-generation DNA sequencing (ChIP-Seq), to elucidate the epigenetic regulation of liver regeneration. The results of these analyses are reported here.
Results
Identification of PH-induced changes in liver histone acetylation
AcetyL-histone H3K9 ChIP-Seq was performed on liver tissue harvested 12 hours after PH or sham surgery from 3 replicate animals in each surgical group. The sequence data were first examined to characterize the abundance and distribution of acetylated sequences with respect to gene transcription start sites (TSS). This analysis showed accumulation of acetylated sequence in close proximity to TSS, with 76.6% of sequences immunoprecipitated from regenerating liver and 78.3% of those recovered from sham-operated liver within ±2000 base pairs (bp) of such sites (Supplementary Fig. 1A).
Next, the sequence data were analyzed to define and compare the heights of individual sequence peaks between experimental groups. The specific genes containing or proximate to sequence peaks identified as differentially abundant in regenerating vs. sham-operated liver were also determined. Gene sequences over-represented in DNA immunoprecipitated from regenerating (vs. sham-operated) liver correspond to loci that are hyperacetylated in response to PH, while under-represented sequences represent genomic sites that are deacetylated during regeneration. Using a (Benjamini-Hochberg) false discovery rate threshold of q < 0.1 (with which <10% of genes identified as differentially acetylated are expected to be false positives3), this analysis identified 454 gene sequences with decreased acetylation and 480 with increased acetylation in regenerating (vs. sham-operated) liver (Table 1 and Supplementary Table 1). These sequences corresponded to 392 and 410 unique genes, respectively (i.e., several genes are represented by multiple distinct sequences in this dataset). These data establish the methodology for and feasibility of using acetyL-histone ChIP-Seq to define genome-wide patterns of histone acetylation during liver regeneration.
Table 1.
Summary of differentially acetylated genes in liver 12 hr after PH vs. Sham Surgery (FDR<0.1)
| Total (unique) | GO terms enriched for differentially acetylated genes | Number of genes linked to3: | ||
|---|---|---|---|---|
| Effect of PH | genes1 | (FDR<0.1)2 | Cell proliferation4 | Cell Death4 |
| Decreased Acetylation | 454 (392) | cellular ketone metabolic process, regulation of cell death, lipid metabolic process, regulation of programmed cell death, sulfur amino acid metabolic process, organic acid metabolic process | 37 | 31 |
| Increased Acetylation | 480 (410) | gland development, cellular process, system development, anatomical structure development, organ development, multicellular organismal development, lactation, developmental process, cellular metabolic process, regulation of cell cycle, vasculature development | 36 | 8 |
1In PH vs. Sham replicates.
2Identified using DAVID bioinformatics database (see references 11–12) from GOTERM_BP_ALL, PANTHER_BP_ALL, KEGG_PATHWAY, PANTHER_PATHWAY on significantly de- or hyper-acetylated genes, respectively.
3Based on examination of all terms into which DAVID sorted differentially acetylated genes.
4See Table 2 for specific genes.
Functional classification of regenerative changes in liver histone acetylation
Next, to characterize functional patterns of change in liver histone acetylation during early regeneration, gene ontology (GO) and similar classification schema were used to categorize genes identified as differentially acetylated in liver after PH vs. sham surgery. First, we identified gene category terms that were significantly enriched (using q < 0.1) for genes that are significantly hyper- or de-acetylated by PH (also using q < 0.1). The results showed enrichment, among hyperacetylated genes, for those associated with ‘regulation of cell cycle’ (Table 1). Genes linked to development- and metabolism-related categories were also enriched in this group. Of note, the total numbers of hyper- and de-acetylated genes identified in early regenerating liver were comparable (Table 1 and Supplementary Table 1). Nevertheless, in contrast to the analysis of hyperacetylated genes, functional classification of the deacetylated genes did not show enrichment in any cell proliferation-associated term but did demonstrate these genes to be enriched in categories associated with regulation of cell death (Table 1). The deacetylated genes, like those that are hyperacetylated, were also enriched in metabolism-related categories.
Hyperacetylated genes
Based on the findings described above, functional annotation tools were subsequently used to more comprehensively identify all differentially-acetylated genes associated with any cell proliferation- or cell death-related gene category term (whether or not those gene categories were significantly enriched for such differentially acetylated genes). Using this approach, 36 of the 410 unique, hyperacetylated genes in regenerating liver were linked to terms associated with regulation of cell cycle and 8 were linked to cell death terms (Tables 1 and 2). Many of these hyperacetylated genes are known to be induced during, to positively regulate, or to interact with factors that promote liver regeneration. For example, Ccnd1 (i.e., cyclin d1), Cdkn1a (i.e., p21), Gadd45g, Lcn2, and Myc are induced in early regenerating liver 4-7; Ccnd1, Egfr, and Prlr promote liver regeneration 8-11; and Tcf7l2 (also known as Tcf4) binds to β-catenin, whose signaling is activated during and promotes regeneration.12
Table 2.
Cell proliferation & cell death-associated genes differentially acetylated 12 hr after PH vs. Sham Surgery
| Increased Acetylation by PH vs. Sham Surgery | Decreased Acetylation by PH vs. Sham Surgery | ||||
|---|---|---|---|---|---|
| Gene category terms1 | Cell Proliferation2 | Cell Death2 | Gene category terms1 | Cell Proliferation2 | Cell Death2 |
| Cell Proliferation | Adipor2 | Dap | Cell Proliferation | Acvr2b | Bcl6 |
| -Bladder cancer | Atf5 | Lgals9 | -Cell cycle control | Arid5b | Bik |
| -Oncogenesis | Bcr | Myc | -Cell proliferation and differentiation | Bcar1 | Birc5 |
| -Endometrial cancer | Calml3 | Mycl1 | -Cell cycle arrest | Bcl6 | Bmf |
| -Glioma | Ccnd1 | Nlrp12 | -Developmental growth | Cebpa | Bok |
| -Melanoma | Cda | Tnfrsf1a | -Growth | Coro1c | Cadm1 |
| -Prostate cancer | Cdkn1a | Tnfsf14 | -Negative regulation of cell proliferation | Csk | Clec2d |
| -Prostate gland growth | Cish | Tns1 | Cell death | Ddah1 | Dapk1 |
| -Regulation of cell cycle | Cxadr | -Apoptosis | Dusp6 | Dpm1 | |
| -Regulation of growth | Cyr61 | Cell Death | Erbb3 | Erbb3 | |
| -Regulation of mitotic cell cycle | Egfr | -Death | Ern1 | Ern1 | |
| Cell Death | Elf3 | -Induction of apoptosis | Fgfrl1 | Foxo1 | |
| -Induction of apoptosis | Epgn | -Induction of programmed cell death | Foxa3 | Foxo3 | |
| Ets2 | -Negative regulation of apoptosis | Foxo1 | Gas1 | ||
| Gadd45g | -Negative regulation of cell death | Foxo3 | Gclc | ||
| Gpam | -Negative regulation of neuron apoptosis | Fyn | Gclm | ||
| Hpgd | -Negative regulation of programmed cell death | Gas1 | H2-bl | ||
| Igf1 | -Positive regulation of apoptosis | Gtpbp4 | H2-K1 | ||
| Kdm2b | -Positive regulation of cell death | Hdac5 | H2-q6 | ||
| Lcn2 | -Positive regulation of cell killing | Il6ra | Itm2b | ||
| Myc | -Positive regulation of programmed cell death | Jmy | Jmy | ||
| Npm2 | -Programmed cell death | Lats2 | Khdc1a | ||
| Pik3r1 | -Regulation of apoptosis | Lst1 | Lst1 | ||
| Prlr | -Regulation of cell death | Mafb | Map3k5 | ||
| Prox1 | -Regulation of cell killing | Mkl2 | Msh2 | ||
| Psap | -Regulation of neuron apoptosis | Mreg | Notch2 | ||
| Rai1 | -Regulation of programmed cell death | Msh2 | Nr3c1 | ||
| Rarg | Ndfip1 | Pim1 | |||
| Sertad2 | Neo1 | Sgpl1 | |||
| Sik1 | Notch2 | Tmbim6 | |||
| Smad7 | Prl2c5 | Traf4 | |||
| Smc1a | Sesn2 | ||||
| Tcf7l2 | Smarca2 | ||||
| Tns1 | Sned1 | ||||
| Vav2 | Tbce | ||||
| Zfp655 | Tspan3 | ||||
| Ulk1 | |||||
1Identified using DAVID bioinformatics database as described in text (see references 11–12).
2Listed by “Official gene symbol” (genes in bold are over-connected in the interactome analysis in Figures 2A-B; 2 hyper- and 8 deacetylated genes shown in italics were sorted by DAVID to both cell proliferation- and cell death-related gene category terms).
Deacetylated genes
Similar examination of genes deacetylated in response to PH showed 37 of 392 associated with cell cycle- and 31 linked to cell death-related gene category terms (Tables 1 and 2). Unlike the hyperacetylated genes, this list included genes linked to ‘cell cycle arrest’ and ‘negative regulation of cell proliferation’ (Table 2). Thus, many more genes deacetylated by PH are associated with anti-proliferative and/or pro-apoptotic activity compared to those hyperacetylated by PH. Several of these deacetylated genes, including Cebpa, Foxo3, Gas1, and Lats2, are also known to be suppressed during early liver regeneration 4,13-15 and Cebpa was recently shown to be essential for termination of regeneration.16 Nevertheless, the regenerative regulation and function of many deacetylated genes in Table 2 is not yet well-characterized.
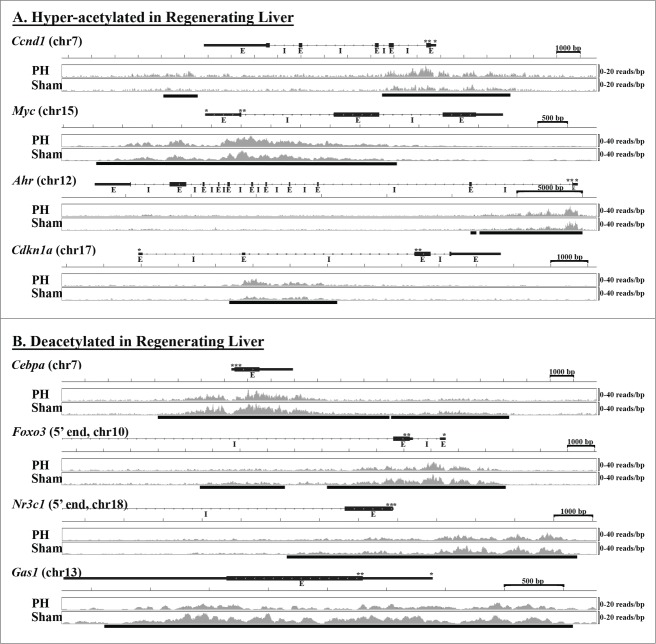
Together, these data define stereotypical, early regenerative changes in liver histone acetylation, and show that those patterns include both hyperacetylation of pro-regenerative and pro-proliferative genes and deacetylation of anti-proliferative and pro-apoptotic genes. Specific examples of histone acetylation patterns in regenerating and sham-operated liver for several differentially acetylated genes listed in Table 2 are shown in Figure 1.
Figure 1.
Patterns of Histone H3K9 Acetylation in Early Regenerating Liver. Examples of gene-specific patterns of histone acetylation in regenerating vs. sham-operated liver are shown for several specific genes identified as (A) hyperacetylated (Ccnd1, Myc, Ahr, Cdkn1a) or (B) deacetylated (Cebpa, Foxo3, Nr3c1, Gas1) 12 hours after PH. UCSC gene maps (transcription (*) and translation (**) start sites, exons (E), and introns (I) as designated) are aligned with abundance of immunoprecipitated sequence (sequence reads per bp with scale indicated to the right) integrated from livers of 3 replicates each after PH or sham surgery. The sequence abundance images were generated using the Integrative Genomics Viewer (IGV) genome browser.40,41 The bars below the sequence abundance data indicate specific sequence(s) identified as differentially acetylated.
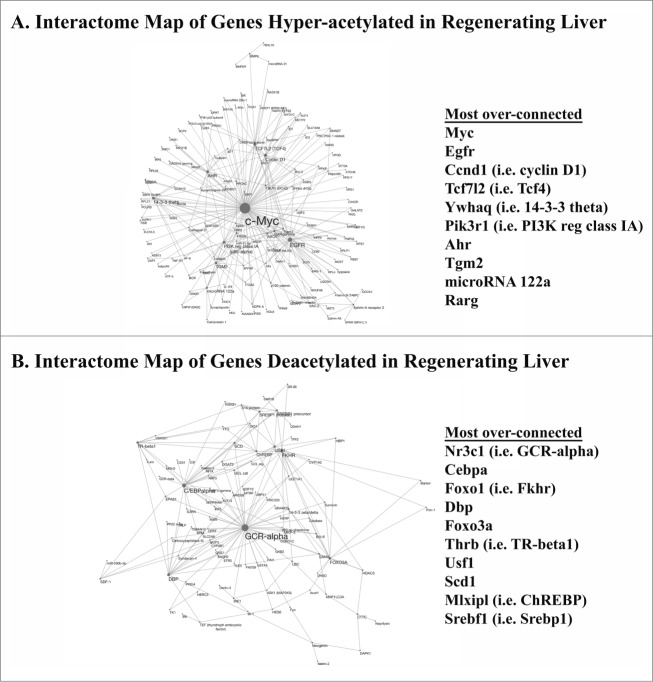
Next, we examined biological interactions between genes identified as differentially acetylated after PH. The goal of this interactome analysis was to assess whether differences in patterns of functional connectivity between hyper- vs. de-acetylated genes could be identified, and, if so, to determine the specific genes with the greatest levels of connectivity to other hyper- and de-acetylated genes. Such genetic “nodes” are candidate drivers of the biological functions associated with hyper- vs. de-acetylated genes (e.g., in this case regenerative function), and also might be useful for distinguishing regenerative patterns of connectivity between such genes. This analysis showed that several genes linked to cell proliferation- and cell death-associated gene category terms (Table 2) were also among the most highly connected nodes within genes hyper- or de-acetylated, respectively, by PH (Figs. 2A, B). For example, Myc, Egfr, Ccnd1, Tcf7l2, Pik3r1, and Rarg were identified as over-connected, hyperacetylated genes (Table 2 and Fig. 2A) and Nr3c1 (i.e., glucocorticoid receptor α), Cebpa, Foxo1, and Foxo3 were highly connected among deacetylated genes (Table 2 and Fig. 2B) in early regenerating liver. Several of these over-connected genes are also known to be transcriptional regulators, and many of the genes to which they are connected are targets of their regulation. Thus, these data show that epigenetic regulation of liver regeneration is characterized by coincident, concordant alterations in the histone acetylation of specific transcription factors and their targets.
Figure 2.
Interactome Plots of Differentially Acetylated Genes in Early Regenerating Liver. Patterns of biological interaction between (A) hyper- or (B) de-acetylated genes in regenerating vs. sham-operated liver are illustrated (with interactions identified using MetaCoreTM from GeneGo and images generated using Cytoscape version 3.1.1 (http://cytoscape.org)). Functional interactions are indicated by lines between designated genes, with increased connectivity represented by increased node size. The most over-connected genes in each set are listed.
The influence of SAHA on regenerative changes in liver histone acetylation
Anti-proliferative genes that are deacetylated (and transcriptionally suppressed) by Zn-HDACs during normal liver regeneration could mediate the anti-regenerative activity of Zn-HDAC inhibition, and discovery of such genes should elucidate the epigenetic mechanisms that control regeneration. We previously identified Cdkn2d (i.e., p19Ink4d) as one such candidate.2 Notably, the analysis here demonstrated that Cdkn2d is deacetylated after PH (with q = 0.11; Supplementary Table 1 and Supplementary Fig. 2). To identify other gene candidates whose SAHA-dependent reversal of PH-induced histone deacetylation might mediate the anti-regenerative effects of Zn-HDAC inhibition, ChIP-Seq was used to compare acetylation patterns in regenerating liver harvested 12 h after PH from replicates of SAHA- vs. vehicle-treated mice (n = 4 each). This analysis identified 1198 gene sequences with decreased acetylation and 1257 with increased acetylation (corresponding to 1162 and 1229 unique genes respectively) in SAHA- (vs. vehicle-) treated liver (for q<0.1; Table 3 and Supplementary Table 2). Surprisingly, the number of genes identified as hyperacetylated (the predicted direct effect of Zn-HDAC inhibition) was comparable to the total deacetylated (which is necessarily an indirect effect of such inhibition) by SAHA in regenerating liver (Table 3). As with the analysis of regenerating and sham-operated liver, the acetylated sequences identified in this experiment also clustered around TSS. In this case, 65.8% of acetylated sequences recovered from SAHA-treated mouse liver and 74.4% of such sequences from vehicle-treated controls were within ±2000 bp of those sites (Supplementary Fig. 1B). This experiment also demonstrated selective enrichment of genes hyper- (but not de-) acetylated by SAHA after PH in classification terms associated with cell cycle regulation and cell death (Table 3). Functional annotation analyses identified 227 (of 1229) genes hyperacetylated by SAHA and 38 (of 1162) deacetylated genes as linked to cell cycle regulation terms, and 99 hyperacetylated and 36 deacetylated genes as associated with cell death terms (Table 3 and Supplementary Table 3). Thus, in contrast to the histone acetylomic analysis comparing early regenerating to sham-operated liver, more pro-apoptotic genes were hyperacetylated than deacetylated by SAHA in early regenerating liver.
Table 3.
Summary of genes differentially acetylated 12 h after PH by SAHA vs. Vehicle (FDR<0.1)
| Total (unique) | Cell proliferation- and cell death-associated GO terms enriched for 2 | Number of genes linked to3: | ||
|---|---|---|---|---|
| Effect of SAHA | genes1 | differentially acetylated genes (FDR<0.1) | Cell proliferation4 | Cell Death4 |
| Decreased Acetylation | 1198 (1162) | none | 38 | 36 |
| Increased Acetylation | 1257 (1229) | pathways in cancer, programmed cell death, regulation of apoptosis, apoptosis, regulation of programmed cell death, regulation of cell death, cell cycle control, colorectal cancer, cell death, non-small cell lung cancer, oncogenesis, death, positive regulation of apoptosis, apoptosis, positive regulation of programmed cell death, positive regulation of cell death, cell cycle, cell proliferation and differentiation | 227 | 99 |
1In SAHA- vs. vehicle-treated replicates.
2Identified using DAVID bioinformatics database (see references 11–12) from GOTERM_BP_ALL, PANTHER_BP_ALL, KEGG_PATHWAY, PANTHER_PATHWAY on significantly de- or hyper-acetylated genes, respectively.
3Based on examination of all terms into which DAVID sorted differentially acetylated genes.
4See Supplementary Table 3 for specific genes.
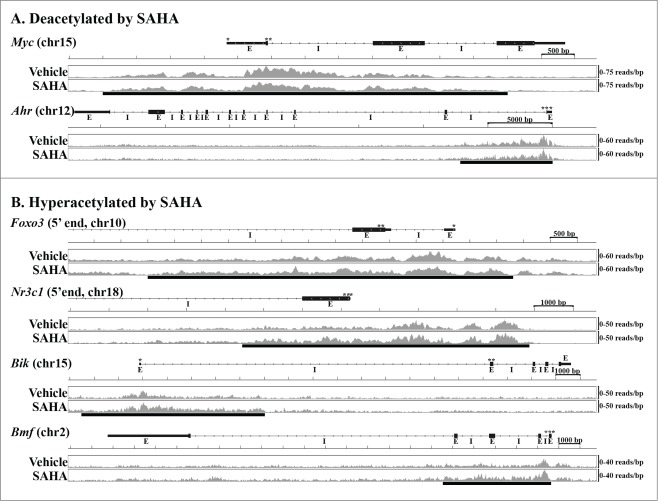
Next, gene-specific differences in liver histone acetylation during early regeneration in SAHA- and vehicle-treated mice (Supplementary Table 2) were compared to those observed between regenerating and sham-operated liver (Supplementary Table 1). Using q < 0.1 to identify differentially acetylated genes in each experiment, this analysis showed that SAHA reversed PH-induced deacetylation of 57 genes and prevented regenerative hyperacetylation of 80 genes (Supplementary Table 4). To further evaluate the impact of Zn-HDAC inhibition on early regenerative changes in liver histone acetylation, a similar evaluation was conducted using a threshold of q < 0.2 for identification of differentially acetylated genes in each experiment. In this case, the results showed SAHA-dependent reversal of regenerative deacetylation of 157 genes and disruption of hyperacetylation of 116 genes (Supplementary Table 4). Further examination revealed that among the cell proliferation- and cell death-associated genes differentially acetylated during normal early regeneration (Table 2), SAHA reversed PH-induced changes in liver histone acetylation of 17 that were normally deacetylated and 12 that were hyperacetylated (Table 4). Similarly, evaluation of the impact of SAHA on regenerative histone hyper- and de-acetylation of the over-connected genes identified by the interactome analyses (Figs. 2A, B) showed that Zn-HDAC inhibition disrupts PH-induced effects on acetylation of many of the most over-connected hyperacetylated (e.g., Ahr, Egfr, Myc, Pik3r1, and Tcf7l2) and deacetylated (e.g., ChREBP/Mxlpil, Dbp, Foxo1, Foxo3, Nr3c1, Srebf1)) genes (Supplementary Table 4). Taken together, these data show that SAHA-mediated Zn-HDAC inhibition reverses regenerative changes in both liver histone de- and hyperacetylation, and they identify several specific targets of such regulation. Examples of gene-specific, SAHA-dependent disruption of PH-induced changes in liver histone acetylation are illustrated in Figure 3.
TABLE 4.
Cell-Proliferation & Cell-Death Associated Genes1 with PH-Induced Changes in Histone Acetylation that are Reversed by SAHA
| Hyper-acetylated by PH and Deacetylated by SAHA2 |
Deacetylated by PH and Hyper-acetylated by SAHA2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene3 | FDR4 PH/Sham | FDR5 SAHA/Veh | Effect of PH on Expression6 | Effect of SAHA on Expression6 | Gene3 | FDR4 PH/Sham | FDR5 SAHA/Veh | Effect of PH on Expression6 | Effect of SAHA on Expression6 |
| Adipor2 | 0.04 | 0.2 | no effect | no effect | Bcar1 | 0.01 | <0.001 | no effect | increased |
| Dap | 0.04 | <0.001 | decreased | no effect | Bik | 0.04 | <0.001 | decreased | increased |
| Egfr | 0.04 | 0.2 | no effect | no effect | Bmf | <0.001 | <0.001 | decreased | increased |
| Gadd45g | <0.001 | <0.001 | increased | no effect | Csk | 0.05 | <0.001 | no effect | no effect |
| Igf1 | 0.05 | 0.009 | no effect | no effect | Ddah1 | 0.09 | <0.001 | no effect | no effect |
| Lcn2 | <0.001 | 0.003 | increased | no effect | Dpm1 | 0.09 | 0.02 | no effect | no effect |
| Myc | 0.03 | 0.02 | increased | decreased | Ern1 | 0.08 | 0.1 | no effect | increased |
| Nlrp12 | <0.001 | 0.002 | increased | no effect | Foxo1 | 0.03 | 0.2 | no effect | no effect |
| Pik3r1 | 0.03 | 0.2 | increased | no effect | Foxo3* | 0.06 | 0.04 | decreased | increased |
| Prox1 | <0.001 | <0.001 | no effect | no effect | Gclc | 0.08 | <0.001 | no effect | no effect |
| Sik1 | <0.001 | <0.001 | increased | no effect | H2-K1 | 0.03 | <0.001 | increased | no effect |
| Tcf7l2 | 0.09 | 0.06 | no effect | no effect | Jmy | 0.05 | 0.1 | no effect | increased |
| Ndfip1 | <0.001 | 0.03 | decreased | no effect | |||||
| Nr3c1 | 0.006 | <0.001 | decreased | no effect* | |||||
| Sesn2 | 0.002 | 0.003 | no effect | no effect | |||||
| Tmbim6 | 0.02 | 0.006 | decreased | no effect | |||||
| Traf4 | 0.09 | 0.009 | no effect | increased | |||||
1Identified using DAVID bioinformatics database 2as described in text; 3Listed by “Official gene symbol” (genes in bold are over-connected in the interactome analysis in Figures 2A-B). See 4Supplementary Table 1 and 5Supplementary Table 2. 6Using p < 0.05; see Figure 4A and Supplementary Figure 3.
Figure 3.
The Influence of SAHA on Histone Acetylation in Early Regenerating Liver. Examples of gene-specific patterns of histone acetylation in regenerating liver from animals treated with SAHA or vehicle control are shown for specific genes identified as (A) deacetylated (Myc, Ahr) or (B) hyperacetylated (Foxo3, Nr3c1, Bik, Bmf) 12 hours after PH. UCSC gene maps (transcription (*) and translation (**) start sites, exons (E), and introns (I) as designated) are aligned with abundance of immunoprecipitated sequence (sequence reads per bp with scale indicated to the right) integrated from livers of 4 replicates each treated with SAHA or vehicle. Sequence abundance images were generated as in Figure 1, and bars below these data indicate specific sequence(s) identified as differentially acetylated.
Examination of the concordance between histone acetylation and mRNA expression in SAHA- vs. vehicle-treated regenerating liver
Finally, the correlation between (a) SAHA-mediated reversal of regenerative changes in liver histone acetylation and (b) corresponding changes in mRNA expression of the cell cycle and cell death-associated genes listed in Table 4 was assessed. This analysis defined the subset of these genes that demonstrate concordant changes in histone acetylation (i.e., hyper- or de-acetylation) and mRNA expression (i.e., increased or decreased expression, respectively). For example, the anti-proliferative/pro-apoptotic genes Foxo3, Bik, and Bmf, whose liver histone acetylation (Fig. 1) and corresponding mRNA expression (Fig. 4A and 13) are normally suppressed during early regeneration, were each hyperacetylated and induced by SAHA 12 hours after PH (Figs. 3, 4, Table 4). Hepatic expression of several of the other cell cycle- and cell death-associated genes deacetylated during normal regeneration, including Bcar1, Ern1, Jmy, and Traf4, were also induced but that of others, including Csk, Ddah1, Dpm1, Foxo1, Gclc, Ndfip1, Nr3c1, Sesn2, and Tmbim6, were not significantly affected by SAHA at this time point (Table 4 and Supplementary Fig. 3). Conversely, Myc, which is normally hyperacetylated (Fig. 1) and induced (Fig. 4A and 17) by PH, was deacetylated and suppressed by SAHA (Figs. 3, 4, Table 4), but expression of other cell cycle-associated genes normally hyperacetylated in regenerating liver, including Adipor2, Dap, Egfr, Gadd45g, Igf1, Lcn2, Nlrp12, Pik3r1, Prox1, Sik1, and Tcf7l2, were not significantly altered by SAHA (Table 4 and Supplementary Fig. 3). Thus, SAHA causes coincidental, concordant effects on the expression of some but not all genes whose PH-induced regulation of histone acetylation is disrupted by this anti-regenerative Zn-HDAC inhibitor. These effects include disruption of both the hyperacetylation and induction of mRNA expression of Myc and the deacetylation and suppression of expression of Foxo3, Bik, and Bmf in early regenerating liver (Fig. 4A).
Figure 4.
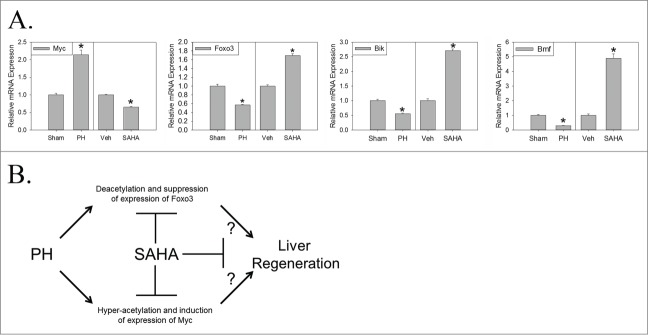
Hepatic mRNA Expression of Differentially Acetylated Genes during and Model of Epigenetic Regulation of Liver Regeneration. (A) Relative mRNA expression (± standard error) 12 hours after PH or sham surgery (indexed to sham; *P < 0 .05 for PH vs. sham; n = 6 replicates per experimental group) or 12 hours after PH in vehicle- (Veh) or SAHA-treated mice (indexed to Veh; *P < 0 .05 for SAHA vs. vehicle; n = 6 replicates per experimental group). (B) Proposed model of epigenetically regulated Myc/Foxo3 switch in regenerating liver (known positive (→) and negative (—|) regulation and hypothesized (?) regulation as indicated; see text for additional discussion).
Discussion
We recently described the regulation and functional importance of Zn-HDACs during experimental liver regeneration.2 Based on those findings, we undertook the analyses here to characterize regenerative patterns of change in liver histone acetylation during early regeneration and determine the effect of Zn-HDAC inhibition on such patterns. To our knowledge, this is the first report to ever describe the results of acetyL-histone ChIP-Seq analyses of experimental liver regeneration. Those results define distinct, gene-specific patterns of pro-regenerative hyper- and de-acetylation in liver after PH. Several of the hyperacetylated genes are known to be induced or activated during or to promote liver regeneration (Table 2), with others reported to promote cell proliferation in other settings but not yet investigated in regenerating liver. Of note, Cdkn1a, which is hyperacetylated 12 hours after PH (Fig. 1), encodes a CDKI whose expression is also known to be up regulated during early liver regeneration.7 Similarly, many of the genes identified as deacetylated are known to be downregulated during liver regeneration and to suppress cell proliferation during regeneration or in other models (Table 2). Based on these findings, studies to evaluate the time course of patterns of change in liver histone acetylation throughout experimental regeneration should now be conducted both to define the dynamic nature of such epigenetic regulation and as a prelude to investigating whether disruption of such regulation mediates impaired regeneration in SAHA-treated and other experimental models. Ultimately, those efforts could inform consideration of the importance of epigenetic regulation of liver regeneration in human liver diseases.
The data reported here show that an anti-regenerative treatment regimen of SAHA reverses PH-induced changes in both the deacetylation of anti-proliferative and pro-apoptotic genes and the hyperacetylation of pro-regenerative genes (Table 4). While the former result (SAHA-dependent gene-specific hyperacetylation) could directly result from the Zn-HDAC inhibitory activity of SAHA, the latter must occur by other indirect mechanisms. One possibility is that SAHA might directly promote hyperacetylation and thereby prevent transcriptional downregulation of specific Zn-HDAC isoforms that are normally suppressed during early regeneration (and relatively resistant to SAHA-mediated inhibition). With this in mind, it is intriguing that HDAC11 expression is downregulated during normal liver regeneration 2 and that the studies here show that SAHA induces HDAC11 hyperacetylation in regenerating liver (Supplementary Tables 2, 3). Zn-HDAC inhibition might also promote hyperacetylation and induce the expression of other SAHA-resistant HDACs, e.g., NAD-dependent sirtuins. Interestingly, a recent report demonstrated the anti-regenerative activity of the sirtuin Sirt1,18 and the data reported here show Sirt1 is also hyperacetylated by SAHA during early regeneration (Supplementary Tables 2, 3). Based on these considerations, future analyses should examine whether induction of HDAC11 or Sirt1 contribute to SAHA's anti-regenerative activity, and, if so, attempt to define the specific mechanisms that mediate this effect.
To begin to investigate the functional consequences of SAHA-dependent inhibition of PH-induced changes in liver histone acetylation, we compared gene-specific patterns of such change to corresponding effects on hepatic mRNA expression. The results showed concordant effects of PH vs. sham surgery and SAHA vs. vehicle administration on alterations in histone acetylation and mRNA expression of Myc (whose PH-induced hyperacetylation and induction is suppressed by SAHA; Figs. 1, 3, 4) and Foxo3 (whose deacetylation and suppression of expression is reversed by SAHA; Figs. 1, 3, 4). SAHA also prevented the PH-induced deacetylation and suppression of Bik and Bmf (Figs. 3, 4), which, like Foxo3, promote apoptosis. Interestingly, Myc has been reported to antagonize the anti-proliferative/pro-apoptotic activity of Foxo319,20 and, similarly, Foxo3 to disrupt Myc function 21 in various models. A recent study reported that Myc might be dispensable for recovery of liver mass after PH; however, that report also showed a reduction in PH-induced hepatocellular proliferation in liver-specific Myc null mice.22 Nevertheless, the data reported here suggest that a Myc/Foxo3 switch is epigenetically regulated during normal liver regeneration, and that disruption of such regulation might contribute to impaired regeneration in SAHA-treated animals (Fig. 4B).
Finally, the findings here have provocative implications with respect to our own and other previous studies identifying alterations in metabolism that occur in response to hepatic insufficiency as the source of essential signals that promote liver regeneration.23 Although the specific molecular mechanisms that couple metabolism to liver regeneration require further elucidation, several indirect observations suggest epigenetic regulation of histone acetylation as an attractive candidate. For example, supplemental glucose affects patterns of histone acetylation in cell culture,24-26 PH-induced hypoglycemia occurs during experimental liver regeneration, and glucose supplementation inhibits regeneration.7,23,27 In addition, HDAC5 undergoes PH-induced nuclear localization in regenerating liver,2 and this Zn-HDAC also exhibits hypoglycemia-induced nuclear localization and regulates FOXO target gene expression in other models.28 Finally, recent studies suggest that specific metabolites modulate isoform-specific HDAC activity in vivo.29 Together, these considerations suggest that investigating patterns of liver histone acetylation in experimental models in which PH-induced alterations in metabolism are disrupted and regeneration is impaired could further elucidate epigenetic mechanisms linking metabolism and established pro-regenerative signaling pathways to liver regeneration. The long-term goal of such effort should be translation of the findings into clinical trials investigating metabolic strategies with which to promote hepatic regeneration and thereby improve patient outcomes in acute and chronic human liver diseases.
Materials and Methods
Animal husbandry and surgery
PH or sham surgery was performed on mice as described in Supplementary Material and previously.5,7,30-32 Some mice were treated with SAHA (or vehicle control) as previously described.2 All experiments were approved by the Washington University Animal Studies Committee and conducted in accordance with institutional guidelines and the criteria outlined in the “Guide for Care and Use of Laboratory Animals” (NIH publication 86-23 revised 1985).
Ac-H3K9 ChIP-Seq
Ac-H3K9 ChIP of livers from n = 3-4 replicates in each experimental group was performed as described in Supplementary Material and previously.2 DNA recovered from the acetyL-histone enriched chromatin and the corresponding input samples were submitted to the Washington University Genome Technology Access Center for blunt ending, adaptor ligation, size selection, and amplification according to established protocols. These libraries were sequenced using the Illumina HiSeq-2500 as single 50 bp reads. Raw data were de-multiplexed and aligned to the most recent mouse reference genome assembly using Novoalign (Novocraft; Selangor, Malaysia). Sequence peaks were identified using MACS software.33 Determination of significant differences in the abundance of peak sequences between experimental groups was performed with DiffBind, an open source Bioconductor package that utilizes edgeR software for statistical analysis of replicated sequence count data.34,35 Genes identified as differentially acetylated were analyzed by gene ontology (GO) and other classification schema using the DAVID bioinformatics database (http://david.abcc.ncifcrf.gov/.36,37). Patterns of interaction between differentially acetylated genes within experimental groups (i.e., interactome analyses) were examined using MetaCoreTM from GeneGo (Thomson Reuters).
With respect to the analyses described above, Benjamini and Hochberg false discovery rate (FDR) thresholds (q values) were employed to either (a) identify DNA sequences (and corresponding genes) as significantly differentially-acetylated between replicates of regenerating vs. sham-operated- or vehicle- vs. SAHA-treated regenerating-liver (using the data analysis pipeline described above) or (b) identify gene term categories, using the DAVID bioinformatics database and the ontology classification schema enumerated in the Tables, in which genes identified as differentially acetylated in (a) were significantly enriched. Thus, Tables 1 and 3 list the number of genes (in column 2) identified as significantly hyper- or de-acetylated in regenerating vs. sham-operated (or vehicle- vs. SAHA-treated) liver using an FDR threshold of q<0.1, and the ontology terms (column 3) identified by DAVID as significantly enriched for those genes, also using q<0.1. Supplementary Tables 1 and 2 list the specific FDR q values calculated for each sequence (and associated gene) on which the differential acetylation analysis was performed. The FDR q values in Table 4 (columns 2 and 3) are also listed in Supplementary Tables 1, 2, respectively. In Supplementary Tables 3, 4, the FDR thresholds refer to the comparison described above in (a) for identification of sequences (and corresponding genes) as significantly hyper- or de-acetylated between replicates of experimental groups (i.e., regenerating vs. sham-operated- and/or vehicle- vs. SAHA-treated) as described.
qRT-PCR: Hepatic mRNA expression in livers from n = 6 animals in each experimental group was determined using semi-quantitative real-time reverse-transcription polymerase-chain-reaction (qRT-PCR) as described in Supplementary Material and previously.5,7,31,38,39 Gene-specific oligonucleotide primers are listed in Supplementary Material.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
These studies were supported by funding from the Children's Discovery Institute of Washington University and St. Louis Children's Hospital (DAR), the Washington University Medical School Department of Pediatrics (DAR), the Washington University Medical School Digestive Disease Research Core Center (DDRCC, supported by NIH-NIDDK P30-DK52574), and an unrestricted donation from Karsyn's Kause Foundation (DAR), and by core facilities of the Washington University Medical School DDRCC.
References
- 1. Rudnick DA. Liver regeneration: the developmental biologists approach. In: Orlando G, Lerut JP, Soker S, Stratta RJ, eds. Regenerative Medicine Applications in Organ Transplantation. 1st ed Waltham, MA: ElsevierAcademic Press, 2014, 353-74. [Google Scholar]
- 2. Huang J, Barr E, Rudnick DA. Characterization of the regulation and function of zinc-dependent histone deacetylases during rodent liver regeneration. Hepatology 2013 May; 57(5):1742-51; http://dx.doi.org/ 10.1002/hep.26206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 1995; 57:289-300 [Google Scholar]
- 4. Liao Y, Shikapwashya ON, Shteyer E, Dieckgraefe BK, Hruz PW, Rudnick DA. Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice. J Biol Chem 2004 Oct 8; 279(41):43107-16; PMID:15265859; http://dx.doi.org/ 10.1074/jbc.M407969200 [DOI] [PubMed] [Google Scholar]
- 5. Shteyer E, Liao Y, Muglia LJ, Hruz PW, Rudnick DA. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology 2004 Dec; 40(6):1322-32; PMID:15565660; http://dx.doi.org/ 10.1002/hep.20462 [DOI] [PubMed] [Google Scholar]
- 6. Kienzl-Wagner K, Moschen AR, Geiger S, Bichler A, Aigner F, Brandacher G, Pratschke J, Tilg H. The role of lipocalin-2 in liver regeneration. Liver Int 2014 Jul 10; PMID:25040147; http://dx.doi.org/ 10.1111/liv.12634 [DOI] [PubMed] [Google Scholar]
- 7. Weymann A, Hartman E, Gazit V, Wang C, Glauber M, Turmelle Y, Rudnick DA. p21 is required for dextrose-mediated inhibition of mouse liver regeneration. Hepatology 2009 Mar 19; 50:207-15; PMID:19441104; http://dx.doi.org/ 10.1002/hep.22979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelsen CJ, Rickheim DG, Tucker MM, Hansen LK, Albrecht JH. Evidence that cyclin D1 mediates both growth and proliferation downstream of TOR in hepatocytes. J Biol Chem 2003 Feb 7; 278(6):3656-63; PMID:12446670; http://dx.doi.org/ 10.1074/jbc.M209374200 [DOI] [PubMed] [Google Scholar]
- 9. Moreno-Carranza B, Goya-Arce M, Vega C, Adan N, Triebel J, Lopez-Barrera F, Quintanar-Stéphano A, Binart N, Martínez de la Escalera G, Clapp C. Prolactin promotes normal liver growth, survival, and regeneration in rodents: effects on hepatic IL-6, suppressor of cytokine signaling-3, and angiogenesis. Am J Physiol Regul Integr Comp Physiol 2013 Oct 1; 305(7):R720-6; PMID:23948778; http://dx.doi.org/ 10.1152/ajpregu.00282.2013 [DOI] [PubMed] [Google Scholar]
- 10. Paranjpe S, Bowen WC, Tseng GC, Luo JH, Orr A, Michalopoulos GK. RNA interference against hepatic epidermal growth factor receptor has suppressive effects on liver regeneration in rats. Am J Pathol 2010 Jun; 176(6):2669-81; PMID:20395437; http://dx.doi.org/ 10.2353/ajpath.2010.090605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci U S A 2007 Oct 23; 104(43):17081-6; PMID:17940036; http://dx.doi.org/ 10.1073/pnas.0704126104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monga SP, Pediaditakis P, Mule K, Stolz DB, Michalopoulos GK. Changes in WNTbeta-catenin pathway during regulated growth in rat liver regeneration. Hepatology 2001 May; 33(5):1098-109; PMID:11343237; http://dx.doi.org/ 10.1053/jhep.2001.23786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurinna S, Stratton SA, Tsai WW, Akdemir KC, Gu W, Singh P, Goode T, Darlington GJ, Barton MC. Direct activation of forkhead box O3 by tumor suppressors p53 and p73 is disrupted during liver regeneration in mice. Hepatology 2010 Sep; 52(3):1023-32; PMID:20564353; http://dx.doi.org/ 10.1002/hep.23746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW, Barton MC. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology 2013 May; 57(5):2004-13; PMID:23300120; http://dx.doi.org/ 10.1002/hep.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sacilotto N, Espert A, Castillo J, Franco L, Lopez-Rodas G. Epigenetic transcriptional regulation of the growth arrest-specific gene 1 (Gas1) in hepatic cell proliferation at mononucleosomal resolution. PLoS One 2011; 6(8):e23318; PMID:21858068; http://dx.doi.org/ 10.1371/journal.pone.0023318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin J, Hong IH, Lewis K, Iakova P, Breaux M, Jiang Y, Sullivan E, Jawanmardi N, Timchenko L, Timchenko NA. Cooperation of CEBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology 2014 Jul 9; PMID:25043739; http://dx.doi.org/ 10.1002/hep.27295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fausto N, Webber EM. Control of liver growth. Crit Rev Eukaryot Gene Expr 1993; 3(2):117-35; PMID:8324292 [PubMed] [Google Scholar]
- 18. Garcia-Rodriguez JL, Barbier-Torres L, Fernandez-Alvarez S, Juan VG, Monte MJ, Halilbasic E, et al. . SIRT1 controls liver regeneration by regulating BA metabolism through FXR and mTOR signaling. Hepatology 2013 Dec 12; 59:1972-83; PMID:24338587; http://dx.doi.org/ 10.1002/hep.26971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chandramohan V, Mineva ND, Burke B, Jeay S, Wu M, Shen J, Yang W, Hann SR, Sonenshein GE. c-Myc represses FOXO3a-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor. J Cell Biochem 2008 Aug 15; 104(6):2091-106; PMID:18393360; http://dx.doi.org/ 10.1002/jcb.21765 [DOI] [PubMed] [Google Scholar]
- 20. Chandramohan V, Jeay S, Pianetti S, Sonenshein GE. Reciprocal control of Forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27Kip1 levels. J Immunol 2004 May 1; 172(9):5522-7; PMID:15100294; http://dx.doi.org/ 10.4049/jimmunol.172.9.5522 [DOI] [PubMed] [Google Scholar]
- 21. Peck B, Ferber EC, Schulze A. Antagonism between FOXO and MYC regulates cellular powerhouse. Front Oncol 2013; 3:96; PMID:23630664 ; http://dx.doi.org/ 10.3389/fonc.2013.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanders JA, Schorl C, Patel A, Sedivy JM, Gruppuso PA. Postnatal liver growth and regeneration are independent of c-myc in a mouse model of conditional hepatic c-myc deletion. BMC Physiol 2012; 12:1; PMID:22397685; http://dx.doi.org/ 10.1186/1472-6793-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang J, Rudnick DA. Elucidating the metabolic regulation of liver regeneration. Am J Pathol 2014 Feb; 184(2):309-21; PMID:24139945; http://dx.doi.org/ 10.1016/j.ajpath.2013.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. . Regulation of cellular metabolism by protein lysine acetylation. Science 2010 Feb 19; 327(5968):1000-4; PMID:20167786; http://dx.doi.org/ 10.1126/science.1179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009 May 22; 324(5930):1076-80; PMID:19461003; http://dx.doi.org/ 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rathmell JC, Newgard CB. Biochemistry. A glucose-to-gene link. Science 2009 May 22; 324(5930):1021-2; PMID:19460991; http://dx.doi.org/ 10.1126/science.1174665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rudnick DA, Davidson NO. Functional relationships between lipid metabolism and liver regeneration. Int J Hepatol 2012 Jan 26; 2012:549241; PMID:22319652; http://dx.doi.org/ 10.1155/2012/549241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 2011 May 13; 145(4):607-21; PMID:21565617; http://dx.doi.org/ 10.1016/j.cell.2011.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. . Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013 Jan 11; 339(6116):211-4; PMID:23223453; http://dx.doi.org/ 10.1126/science.1227166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang J, Glauber M, Qiu Z, Gazit V, Dietzen DJ, Rudnick DA. The influence of skeletal muscle on the regulation of liver:body mass and liver regeneration. Am J Pathol 2012 Feb; 180(2):575-82; PMID:22155110; http://dx.doi.org/ 10.1016/j.ajpath.2011.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gazit V, Weymann A, Hartman E, Finck BN, Hruz PW, Tzekov A, Rudnick DA. Liver regeneration is impaired in lipodystrophic fatty liver dystrophy mice. Hepatology 2010 Dec; 52(6):2109-17; PMID:20967828; http://dx.doi.org/ 10.1002/hep.23920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gazit V, Huang J, Weymann A, Rudnick DA. Analysis of the role of hepatic PPARgamma expression during mouse liver regeneration. Hepatology 2012 Jun 18; 56:1489-98; PMID:22707117; http://dx.doi.org/ 10.1002/hep.25880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9(9):R137; PMID:18798982 ; http://dx.doi.org/ 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010 Jan 1; 26(1):139-40; PMID:19910308; http://dx.doi.org/ 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. . Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012 Jan 19; 481(7381):389-93; PMID:22217937; http://dx.doi.org/ 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4(1):44-57; PMID:19131956 [DOI] [PubMed] [Google Scholar]
- 37. Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009 Jan; 37(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turmelle YP, Shikapwashya O, Tu S, Hruz PW, Yan Q, Rudnick DA. Rosiglitazone inhibits mouse liver regeneration. FASEB J 2006; 20:2609-11; PMID:17077279; http://dx.doi.org/ 10.1096/fj.06-6511fje [DOI] [PubMed] [Google Scholar]
- 39. Clark A, Weymann A, Hartman E, Turmelle Y, Carroll M, Thurman JM, Holers VM, Hourcade DE, Rudnick DA. Evidence for non-traditional activation of complement factor C3 during murine liver regeneration. Mol Immunol 2008 Jun; 45(11):3125-32; PMID:18452991; http://dx.doi.org/ 10.1016/j.molimm.2008.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2011 Jan; 29(1):24-6; PMID:21221095; http://dx.doi.org/ 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013 Mar; 14(2):178-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.