Abstract
A wide variety of signaling transduction pathways contribute to tumorigenesis. Forkhead box Q1 (FOXQ1) is a member of the forkhead transcription factor family and its upregulation is closely correlated with tumor progression and prognosis of multiple cancer types, including colorectal cancer. However, the molecular mechanisms by which FOXQ1 promotes tumorigenesis, especially cancer cell invasion and metastasis in colorectal cancer, have not been fully elucidated. In the present study, we demonstrate that FOXQ1 is overexpressed in colorectal tumor tissues and its expression level is closely correlated with the stage and lymph node metastasis of colorectal cancer. In in vitro cultured SW480 colorectal cancer cells, knockdown of FOXQ1 expression by small interfering RNA greatly diminished the aggressive tumor behaviors of SW480 cells, including angiogenesis, invasion, epithelial-mesenchymal transition, and resistance to chemotherapy drug-induced apoptosis. Further mechanistic investigation showed that FOXQ1 silencing prevents the nuclear translocation of β-catenin, thus reducing the activity of Wnt signaling. Moreover, TGF-β1 induced the expression of FOXQ1 as well as the migration and invasion of SW480 cells, which was partially prevented following knockdown of FOXQ1. Our results demonstrate that FOXQ1 plays a critical role during the tumorigenesis of colorectal cancer and is a mediator of the crosstalk between Wnt and TGF-β signaling pathways. Our findings provide further insight into the cancer biology of colorectal cancer and suggest that FOXQ1 is a potential therapeutic target for the development of therapies for colorectal cancer.
Keywords: aggressive tumor behavior, colorectal cancer, epithelial-mesenchymal transition, FOXQ1, TGF-β1, Wnt signaling
Abbreviations
- μg
Microgramme
- μl
Microliter
- 5-FU
5-fluorouracil
- 7-AAD
7-aminoactinomycin D
- cDNA
Complementary DNA
- DAPI
4′,6-diamidino-2-phenylindole
- ddH2O
double distilled H2O
- DEPC
Diethy pyrocarbonate
- DMSO
Dimethyl sulfoxide
- EMT
Epithelial-Mesenchymal transition
- FOXQ1
Forkhead Box Q1
- L-OHP
Oxaliplatin
- MiRNA
MicroRNA
- MMP2
Matrix metalloproteinase-2
- NC-shRNA
Negative Control-shRNA
- PBS
Phosphate buffer solution
- PBS
phosphate buffered saline
- PKA
proteinkinase A
- PVDF
Polyvinylidene fluoride
- qRT-PCR
Quantitative real-time PCR
- RNAi
RNA interference
- SDS
Sodium dodecyl sulfonate
- shRNA
Short hairpin RNA
- TBS
Tris-buffered saline
- TEMED
Tetra methyl ethylene diamine
- TGF-β
Transformin growth β
- Tris
Trihydroxymethyl minomethane
- VEGF-A
Vascular endothelial growth factor-A
Introduction
Colorectal cancer is a malignant cancer frequently seen in the colon or rectum. Colorectal cancer has become the fourth leading cause of cancer-related death and the incidence of colorectal cancer is in a rising trend worldwide.1,2 Colorectal cancer originates from oncogenic transformation of intestinal epithelial cells, which acquire malignant characteristics that include loss control of cell proliferation, enhanced angiogenesis, sustained invasion, metastasis, and acquisition of drug resistance.3,4 A wide variety of signaling pathways and transcription factors have been found to be deregulated during the multistep processes of tumorigenesis of colorectal cancer, such as Wnt/β-catenin, Hedgehog, TGF-β/Smad, Snail, and Notch signaling pathways.5-7 It is, thus, widely accepted that colorectal cancer results from the accumulation of the alteration of multiple, not a single, signaling pathway. However, the coordination of these deregulated cancer signal pathways in the development, progression and maintenance of colorectal cancer is largely unknown.
Forkhead box Q1 (FOXQ1) is a new member of the forkhead (FOX) transcription factor family. Human FOXQ1 is an oncogene localized on chromosome 6p25.3.8 FOXQ1 protein has a conserved FOX DNA-binding domain and functions as a transcription factor. FOXQ1 has been shown to promote epithelial-mesenchymal transition (EMT) in breast cancer cells and non-small cell lung cancer cells by regulating the expression of E-cadherin, β-catenin and vimentin.9,10 FOXQ1 was reported to induce EMT and enhance the invasive capability of hepatocellular carcinoma cells by activating the expression of ZEB2 and Versican V1.11 In bladder cancer cells, small interfering RNA-mediated knockdown of FOXQ1 inhibited EMT and tumor invasion.12 Moreover, FOXQ1 is overexpressed and closely associated with EMT in cancer cells; accordingly, FOXQ1 has been proposed as an independent prognostic factor for non-small cell lung cancer.10 In colorectal cancer cells, FOXQ1 was found to be overexpressed and regulate p21CIP/WAF1 and TWIST1 to promote tumor growth and metastasis, respectively.13,14 These facts indicate that FOXQ1 plays an important role in the EMT, invasion and metastasis of many cancers, but the underlying molecular mechanisms remain to be determined.
Over-activation of Wnt/β-catenin signaling is one of the earliest events during the tumorigenesis of colorectal cancer15 and the majority of colorectal cancer patients demonstrate dysregulated Wnt signaling.16 Recent microarray studies using different colorectal cancer cell lines and clinical samples from colorectal cancer patients showed a strong positive correlation between FOXQ1 expression level and Wnt signaling activity.13,17 However, the molecular mechanism of the functional linkage between FOXQ1 and Wnt/β-catenin signaling is unknown. Moreover, TGF-β signaling has been shown to regulate the expression of FOXQ1 in epithelial cell differentiation and EMT.9,18 Very interestingly, interplay between Wnt and TGF-β signaling pathways in regulating colorectal cancer progression has been indicated by a recent study.19 These facts indicate that FOXQ1, Wnt and TGF-β signaling are functionally interconnected and suggest that FOXQ1 may function to mediate the crosstalk between Wnt and TGF-β signaling pathways during the tumorigenesis of colorectal cancer.
In the present study, FOXQ1 expression levels in tumor tissue, adjacent non-tumorous tissue and normal colorectal mucosa from 63 patients were assessed and the association of FOXQ1 level with tumor stage and metastasis of colorectal cancer was analyzed. Cell migration and invasion, and the activity of Wnt and TGF-β signaling in colorectal cancer SW480 cells following silencing of FOXQ1 by RNA interference (RNAi) was assayed.
Results
FOXQ1 is overexpressed and correlated with tumor stage and metastasis of colorectal cancer
To determine the expression status of FOXQ1 in colorectal cancers, we assessed the FOXQ1 protein levels in 62 groups of specimens with immunohistochemistry. Results showed that FOXQ1 was localized in the nucleus (Fig. 1A). Compared with that in adjacent non-tumorous tissues (Fig. 1B, 24.2%, 15/62) and distal normal colorectal mucosa samples (Fig. 1C, 6.5%, 4/62), FOXQ1 was overexpressed in colorectal cancer samples (Fig. 1A, 75.8%, 47/62) (P < 0.05) (Table 2). Further correlation analysis indicated that the expression of FOXQ1 was not correlated with the age or sex of the patients nor the size of the tumor (P > 0.05), but was closely associated with the stage of tumor and lymph node metastasis (P < 0.05) (Table 3). Thus, FOXQ1 is overexpressed in colorectal cancer and its level is associated with tumor stage and metastasis, suggesting that FOXQ1 plays an important in the late stage of the tumorigenesis of colorectal cancer.
Figure 1.
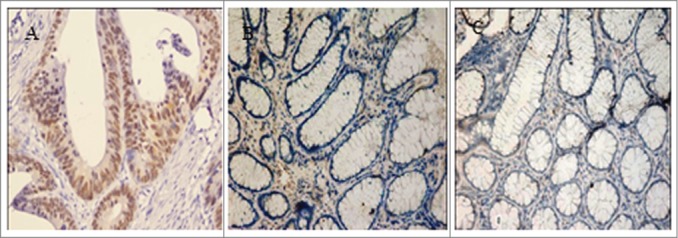
FOXQ1 was overexpressed in colorectal tumor cells. FOXQ1 was stained in colorectal cancer samples (A), adjacent nontumorous tissues (B), and distal normal colorectal mucosa samples (C). Images were acquired at 200X. Representative images are shown.
Table 2.
Comparison of FOXQ1 expression in tumor and normal tissues
| FOXQ1 |
|||
|---|---|---|---|
| Sample | Case | + | − |
| Colorectal cancer tissues | 62 | 47 | 15 |
| Adjacent nontumorous tissues | 62 | 16 | 46 |
| Normal colorectal mucosas | 62 | 4 | 58 |
| χ2a | 61.583 | ||
| χ2b | 31.008 | ||
| χ2c | 8.5846 | ||
Compared between normal and cancer tissues, P < 0.05.
Compared between cancer and adjacent tissues, P < 0.05.
Compared between normal and adjacent tissues, P < 0.05.
Table 3.
Correlation between FOXQ1 expression and clinical parameters of colorectal cancer patients
| FOXQ1 |
||||
|---|---|---|---|---|
| Index | Case | + | − | χ2 |
| Sex | 0.182 | |||
| Male | 36 | 28 | 8 | |
| Female | 26 | 19 | 7 | |
| Age (years) | 0.194 | |||
| ≥61 | 30 | 22 | 8 | |
| <61 | 32 | 25 | 7 | |
| Diameter of tumor (cm) | 0.721 | |||
| >5 | 20 | 17 | 3 | |
| ≤5 | 42 | 30 | 12 | |
| Tumor stage (Duke's) | 6.385* | |||
| A+B | 32 | 20 | 12 | |
| C+D | 30 | 27 | 3 | |
| Lymph node metastasis | 6.648* | |||
| Yes | 26 | 24 | 2 | |
| No | 36 | 23 | 13 | |
Compared between FOXQ1 positive and negative samples, P < 0.05.
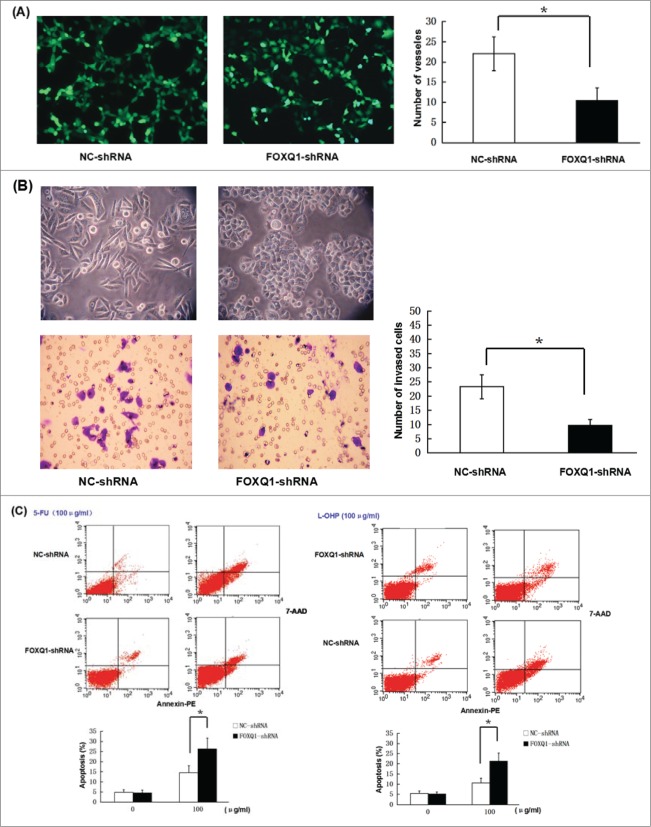
Knockdown of FOXQ1 expression inhibits the angiogenesis and invasion of SW480 cells while sensitizing SW480 cells to chemotherapeutic drugs
To study the function of FOXQ1 in the aggressive malignant behaviors of colorectal cancer, we knocked down FOXQ1 by transduction of specific shRNA in SW480 cells, a colorectal cancer cell line expressing a moderate level of FOXQ1.1 Three different shRNAs targeting different regions of FOXQ1 mRNA and a NC shRNA were used and the knockdown efficiency was measured. As shown in Fig. 2A, high efficiency of lentivirus-mediated transduction was obtained in SW480 cells. All three shRNAs decreased FOXQ1 mRNA (Fig. 2B, left panel) as well as protein levels (Fig. 2B, right panel). FOXQ1-sh1 showed the highest knockdown efficiency. After confirming the results with NC shRNA and non-transduced control cells (Fig. 2C), we selected FOXQ1-sh1 for further investigation, with the cells designated FOXQ1-shRNA cells compared with NC-shRNA cells.
Figure 2.
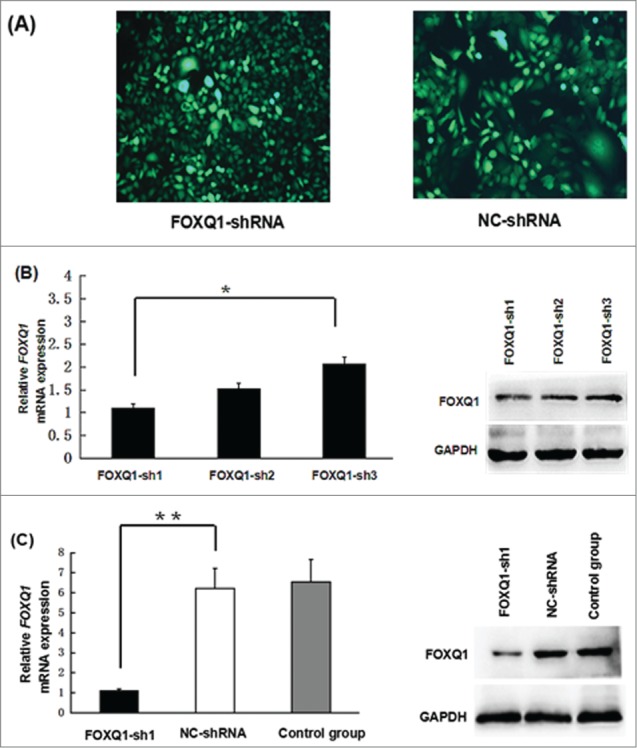
Knockdown of FOXQ1 expression by shRNA. (A) SW480 cells were transduced with lentivirus containing FOXQ1-sh1, -sh2, -sh3, or NC-shRNA for 72 hours. GFP signals were detected by fluorescence microscopy (100X) to indicate the transduction efficiency. (B) FOXQ1 mRNA (left panel) and protein (right panel) levels in SW480 cells transduced with FOXQ1-sh1, -sh2, -sh3, and NC-shRNA were measured by qRT-PCR and Western blot, respectively. (C) Knockdown efficiency of FOXQ1-sh1 was confirmed by qRT-PCR (left panel) and Western blot (right panel). The NC-shRNA transduced cells (NC-shRNA) and non-transduced cells (control group) were used as controls. The results are expressed as mean ± SD *P < 0.05, **P < 0.01.
The correlation between FOXQ1 expression and colorectal cancer stage/metastasis suggested that FOXQ1 might play a role in the process of EMT, cancer invasion and metastasis. To test this hypothesis, we performed an in vitro angiogenesis assay to measure the vessel formation of EA.hy926 cells treated with culture media from SW480 cells treated with FOXQ1-shRNA or NC-shRNA. In comparison to those in NC-shRNA culture media, the number of vessels was significantly decreased in EA.hy926 cells treated with FOXQ1-shRNA cell culture media, and most of the vessel formation was incomplete with reduced diameters. We next preformed Transwell assay to measure the invasive ability of SW480 cells following silencing of FOXQ1. The results showed that the number of cells migrating through the membrane was greatly reduced when FOXQ1 expression was knocked down, and the cells displayed epithelial-like morphology with tighter cell-cell junction adhesion compared with the NC-shRNA cells (Fig. 3B). Finally, we asked whether silencing of FOXQ1 sensitizes SW480 cells to chemotherapeutic drugs. We treated FOXQ1- and NC-shRNA cells with 100 μg/ml 5-FU or L-OHP for 24 hours and determined cell apoptosis by flow cytometry. Silencing of FOXQ1 did not induce apoptosis; 5-FU treatment led to apparent cell apoptosis. However, the apoptotic cells were significantly higher in FOXQ1-shRNA cells than that in NC-shRNA cells following 5-FU treatment (Fig. 3C, left panel). Similar results were obtained in FOXQ1- and NC-shRNA cells treated with L-OHP (Fig. 3C, right panel). These results showed that knockdown of FOXQ1 expression reduced the angiogenesis and invasion of SW480 cells, while decreased the resistance of SW480 cells to chemotherapy.
Figure 3.
Knockdown of FOXQ1 reduced the aggressive tumor behaviors of SW480 cells. (A) An in vitro angiogenesis assay measured the vessel formation of NC-shRNA-transduced EA.hy926 cells growing in FOXQ1- and NC-shRNA cell culture media after 48 hours. The number of vessels was counted by fluorescent microscopy (120X). (B) FOXQ1- and NC-shRNA cells seeded in the upper chamber of Transwell were examined by optical microscopy (120X) (upper panel). After 36 hours, the cells migrating through the insert membrane between the upper and lower chambers were fixed and stained by crystal violet. The number of migrated cells was counted by optical microscopy (120X) (lower panel). (C) FOXQ1- and NC-shRNA cells were treated with 5-FU (left panel) and L-OHP (right panel) at 100 μg/ml for 24 hours. Annexin and 7-AAD were used in flow cytometry to measure the percentage of apoptotic cells. The results are expressed as mean ± SD *P < 0.05, **P < 0.01.
Knockdown of FOXQ1 expression inhibits the nuclear translocation of β-catenin and activation of the Wnt signaling pathway
To elucidate the relationship between FOXQ1 and Wnt signaling pathway, the cellular localization of β-catenin in SW480 cells was examined following silencing of FOXQ1. As shown in Fig. 4A, β-catenin accumulated in the nucleus of NC-shRNA cells, indicating activation of the Wnt signaling pathway. In sharp contrast, the majority of β-catenin in FOXQ1-shRNA cells was retained in the cytoplasm, suggesting FOXQ1 promotes the activation of Wnt signaling. The protein level of β-catenin was not affected by FOXQ1 silencing, indicating that β-catenin is not under transcriptional control of FOXQ1 (Fig. 4B). To further assess the Wnt signaling activity, we examined the expression of Wnt signaling downstream target genes VEGF-A and MMP2, and EMT-associated markers E-cadherin, vimentin and N-cadherin. The mRNA levels of VEGF-A and MMP2, and mesenchymal markers vimentin and N-cadherin were significantly downregulated in FOXQ1-shRNA cells compared with those in NC-shRNA cells. However, the epithelial marker E-cadherin was dramatically upregulated in FOXQ1-shRNA cells compared with that in NC-shRNA cells (Fig. 4C, left panel). Similarly, Western blot analysis showed that the protein levels of VEGF-A, MMP2, vimentin and N-cadherin were decreased, while E-cadherin was increased following FOXQ1 knockdown by shRNA (Fig. 4C, right panel). These results imply that FOXQ1 might function through the Wnt signaling pathway to promote EMT, invasion and metastasis of colorectal cancer cells.
Figure 4.
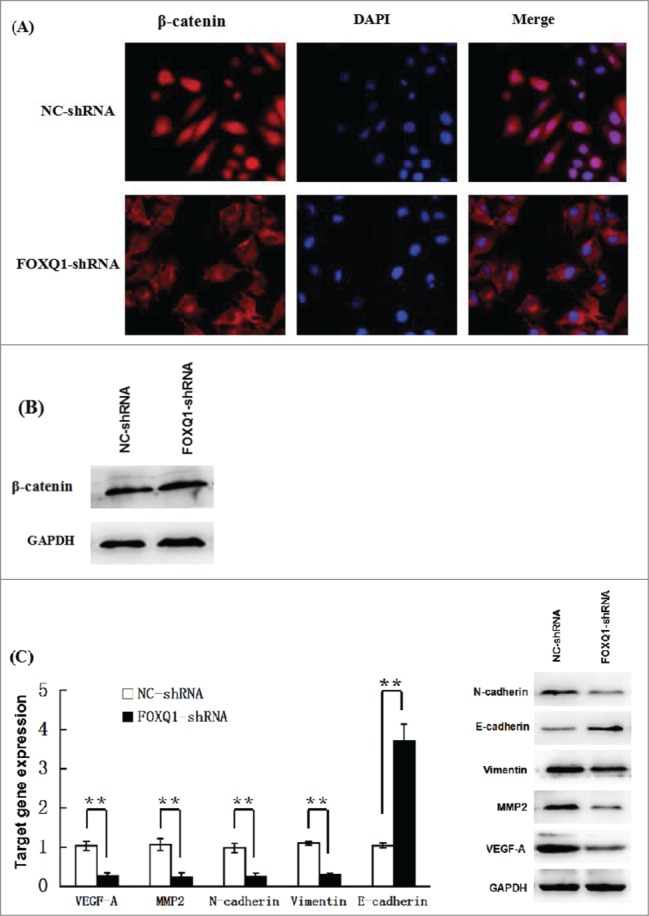
Knockdown of FOXQ1 expression suppressed Wnt signaling. (A) NC- and FOXQ1-shRNA cells were stained with fluorescent β-catenin antibody (red) and DAPI nuclear stain (blue). (B) Western blot analysis of the protein level of β-catenin in NC- and FOXQ1-shRNA cells. (C) Left panel, qRT-PCR analyses of the mRNA levels of Wnt downstream target genes and EMT associated markers in NC- and FOXQ1-shRNA cells; right panel, Western blot analyses of the protein levels of Wnt downstream target genes and EMT-associated markers in NC- and FOXQ1-shRNA cells. The results are expressed as mean ± SD **P < 0.01.
FOXQ1 mediates the crosstalk between Wnt and TGF-β signaling pathways
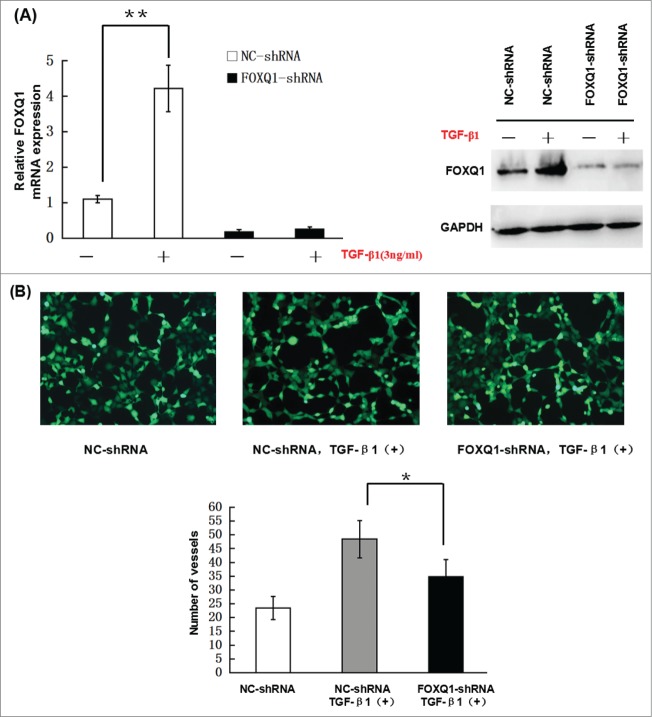
Previous studies showed that TGF-β induces FOXQ1 expression in epithelial cell differentiation and during EMT.20,21 To investigate whether TGF-β induces FOXQ1 expression in SW480 cells, we incubated NC- and FOXQ1-shRNA cells with 3 ng/ml TGF-β for 3 days and measured FOXQ1 mRNA and protein levels by qPCR and immunoblotting, respectively. In NC-shRNA cells, treatment with TGF-β resulted in an apparent increase in the mRNA (Fig. 5A, left panel) and protein (Fig. 5A, right panel) levels of FOXQ1, which were abolished by FOXQ1 shRNA in FOXQ1-shRNA cells, indicating that FOXQ1 is a downstream target of TGF-β signaling. To further investigate the role of FOXQ1 in TGF-β signaling in colorectal cancer cells, we examined the angiogenesis, invasion and EMT of NC-shRNA and FOXQ1-shRNA cells treated with TGF-β. In vitro angiogenesis, Transwell invasion and drug sensitivity assays were performed as described in Fig. 3 in NC- and FOXQ1-shRNA cells pre-treated with TGF-β1. The results showed that TGF-β1 stimulation of SW480 NC-shRNA cells drastically enhanced the vessel formation in EA.hy926 cells (Fig. 5B) and invasion of NC-shRNA cells in Matrigel (Fig. 5C), and reduced the sensitivity of these cells to 5-FU and L-OHP treatment (Fig. 5D). Further analyses demonstrated both the mRNA and protein levels of VEGF-A, MMP2, vimentin and N-cadherin were upregulated and E-cadherin was downregulated upon TGF-β1 stimulation (Fig. 5E). However, FOXQ1 shRNA significantly prevented the upregulation of VEGF-A, MMP2, vimentin and N-cadherin and downregulation of E-cadherin by TGF-β1 treatment. These results indicate that FOXQ1 mediates the crosstalk between the Wnt and TGF-β signaling pathways. However, in the absence of FOXQ1, TGF- β alone was still capable of inducing moderate aggressive behaviors in SW480 cells, indicating that additional signaling responsive to TGF-β stimulation contributes to the process as well. The identification and validation of these additional signaling pathways and their relationship with FOXQ1 await further investigation.
Figure 5.
(Continued).
Figure 5.
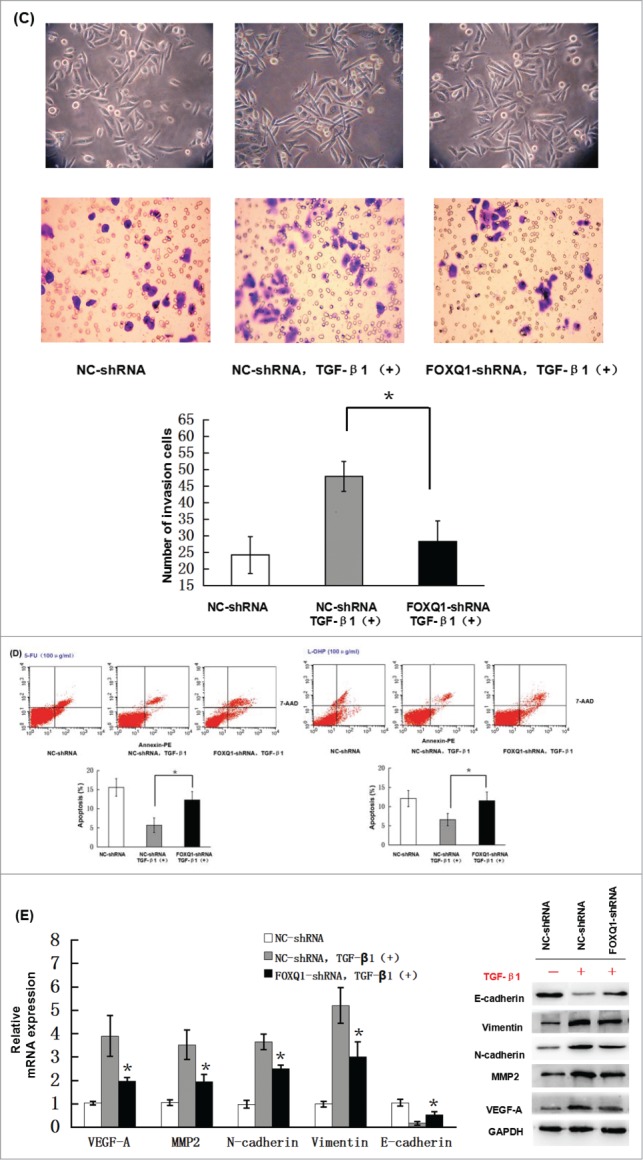
Knockdown of FOXQ1 partially inhibited TGF-β1-induced aggressive tumor behaviors in SW480 cells and the changes in expression of downstream genes. (A) NC- and FOXQ1-shRNA cells were incubated in growth media supplemented with TGF-β1 (3 ng/ml) for 3 days. FOXQ1 expression was measured by qRT-PCR and Western blot. (B) The culture supernatant from NC-shRNA cells cultured for 3 days or NC- and FOXQ1-shRNA cells cultured for 5 days containing TGF-β1 (3 ng/ml) was collected and used in an in vitro angiogenesis assay. The vessel formation was measured in NC-shRNA-transduced EA.hy926 cells. The number of vessels was counted by fluorescent microscopy (120X). (C) FOXQ1- and NC-shRNA cells were treated with TGF-β1 (3 ng/ml) for 5 days, and cell morphology was examined by optical microscopy (120X) (upper panel). For the invasion assay, FOXQ1- and NC-shRNA cells were pre-treated with TGF-β1 (3 ng/ml) for 3 days before being seeded in the upper chamber of Transwell. After 36 hours, the cells migrating through the insert membrane were fixed and stained by crystal violet. The number of migrated cells was counted by optical microscopy (120X) (lower panels). (D) For the drug sensitivity assay, FOXQ1- and NC-shRNA cells were incubated in TGF-β1-containing media (3 ng/ml) for 4 days, and 5-FU or L-OHP (100 μg/ml) was added to the cell culture for an additional 24 hours. Flow cytometry was performed to measure cell apoptosis. (E) Expression of Wnt downstream target genes and EMT-associated markers in TGF-β1 treated NC- and FOXQ1-shRNA cells were measured by qRT-PCR and Western blot. The results are expressed as mean ± SD *P < 0.05, **P < 0.01.
Discussion
In this study, we demonstrated that FOXQ1 is overexpressed and correlated with tumor stage and metastasis of colorectal cancer. Silencing of FOXQ1 by lentivirus-delivered shRNA inhibited the angiogenesis and invasion of SW480 colorectal cancer cells. Moreover, knockdown of FOXQ1 expression inhibited the nuclear translocation of β-catenin. Most importantly, it was discovered that FOXQ1 mediates the crosstalk between the Wnt and TGF-β signaling pathways. Our novel findings provide further insight into the cancer biology of colorectal cancer and will impact the development of small molecule anticancer drugs targeting the Wnt and TGF-β signaling pathways.
Colorectal cancer is a malignant cancer that affects millions of people worldwide each year. Metastasis, relapse and drug resistance are the current challenges for treatment of colorectal cancer.22 Radiation therapy is frequently used for local control of the tumor or reducing the tumor size for surgical resection. Chemotherapy agents, such as 5-FU and L-OHP, are usually offered to patients with advanced colorectal tumor, but the response rate is only approximately 40% and even lower for tumors of higher grade or cancer undergoing metastasis.23 In addition, the clinical efficacy of chemotherapy decreases with increasing patient age, and acute or chronic toxic effects are very common in patients undergoing chemotherapy.24 Molecular targeted therapy has become an alternative therapeutic approach or used as a combinational therapy with standard chemotherapy agents. For example, the recombinant monoclonal antibody against VEGF-A called bevacizumab, and chimeric monoclonal antibody against EGFR called cetuximab have been recently approved for clinical use in combination with 5-FU and irinotecan, respectively, to treat metastatic colorectal cancer.25 The development of additional targeted therapies may benefit a wider spectrum of patients and achieve better clinical outcomes.
It is now widely believed that FOXQ1 is an oncogene for a wide range of cancers. Dysregulated FOXQ1 leads to inactivation of E-cadherin and promotes EMT in various tumor types, including breast cancer, non-small cell lung cancer, and bladder cancer.9,10,26 Correlation between overexpression of FOXQ1 and colorectal cancer progression has been suggested by previous investigation in cancer cell lines.14 In the present study, 62 patients were recruited with different stages of colorectal cancer, and provided direct evidence demonstrating that the expression level of FOXQ1 is positively correlated with the stage of colorectal cancer and degree of lymph node metastasis. These results imply that FOXQ1 plays an important role in the EMT of colorectal cancer.
Very interestingly, our results indicate that FOXQ1 is a mediator between the Wnt and TGF-β signaling pathways. β-catenin is a critical player in Wnt signaling, which is involved in the pathogenesis of the majority of human malignancies, including colorectal cancer. β-catenin interacts with the transmembrane protein E-cadherin, which in turn stabilizes cell-cell adhesion and maintains epithelial barrier integrity.27 Activation of Wnt signaling induces the nuclear translocation of β-catenin, which not only initiates a cascade of downstream gene transcription, but also disrupts the β-catenin/E-cadherin complex and cell-cell adhesion to facilitate cell migration. In our study, diminished FOXQ1 expression by shRNA greatly inhibited the nuclear translocation of β-catenin; as a result, expression of Wnt target genes and EMT markers were downregulated and the aggressive tumor behaviors of SW480 cells were largely suppressed. Consistently, the expression of E-cadherin was significantly higher in SW480 FOXQ1-shRNA cells than that in the control cells. The excessive amount of E-cadherin could sequester β-catenin in the membrane-bound complex and prevent it from entering the nucleus. Formation of the β-catenin/E-cadherin complex might also restore cell-cell adhesion, as shown by the morphological changes observed in FOXQ1-shRNA cells. The mechanism by which FOXQ1 negatively regulates the expression of E-cadherin has been investigated in multiple studies. A transcriptional profiling study performed in a human noncancerous mammary epithelial cell line HMLER identified E-cadherin as a transcriptional target of FOXQ1, and direct interaction between FOXQ1 and the E-box in the E-cadherin promoter was demonstrated by luciferase reporter assays and chromatin immunoprecipitation.28,29 Whether FOXQ1 functions through a similar mechanism to modulate the E-cadherin transcription in colorectal cancer cells requires further investigation. Furthermore, a recent study suggested that FOXQ1 is as a direct transcriptional target of β-catenin and marker for Wnt signaling activity.17 This finding, along with ours, suggests that there might be a positive feedback loop in the regulation of Wnt signaling. Further investigation is required to dissect the signaling machinery underpinning the Wnt/FOXQ1-mediated EMT and tumor invasion.
TGF-β is a potent inducer of EMT and tumor invasion. Studies in many human cancers have suggested that FOXQ1 may function as a modulator of TGF-β signaling.9,18 Findings from our present study indicate that FOXQ1 functions downstream of TGF-β signaling and contributes to TGF-β on tumor progression. Our results showed that the TGF-β-induced aggressive tumor behaviors in SW480 cells could be partially rescued by loss of FOXQ1 expression. These results clearly indicate that FOXQ1 functions as a mediator in the crosstalk between the TGF-β and Wnt/β-catenin signaling pathways to promote progression of colorectal cancer. However, silencing FOXQ1 expression could not completely reverse the TGF-β-induced changes in SW480 cells, which suggests the contribution from additional molecules downstream of TGF-β signaling. Smad proteins are the downstream effectors of TGF-β and have been shown to be essential for the activation of Wnt signaling in TGF-β-induced mesenchymal stem cell proliferation.30 In addition, PI3K and PKA signaling pathways have been implicated in the process of mesenchymal stem cell differentiation that is induced by TGF-β.31 Involvement from these signaling pathways should be assessed and the potential interaction between FOXQ1 and these signaling pathways needs to be investigated.
Development of resistance to chemotherapy is a major mechanism for the low response rate and poor prognosis in patients with colorectal cancer. Our study indicates a critical role of FOXQ1 in regulating drug sensitivity of colorectal cancer cells to 5-FU and L-OHP. However, the signaling cascade that leads to changes in drug resistance remains unclear.
In conclusion, the present study demonstrates a novel function for FOXQ1, which acts as a mediator for TGF-β-induced aggressive tumor behaviors through modulation of Wnt/β-catenin signaling in colorectal cancer cells. This study provides additional evidence that FOXQ1 plays in important role in enhancing tumorigenicity and suggests that FOXQ1 is a potential therapeutic target for the development of therapies for colorectal cancer.
Materials and Methods
Patients
A total of 62 pairs of specimens were collected from colorectal cancer patients admitted to our hospital from March to September, 2012. Each pair of specimens included the tumor tissue, adjacent nontumorous tissue (2–5 cm from the tumor), and normal colorectal mucosa (≥8 cm from the tumor). There were 36 male and 26 female patients with an average age of 58.3 years (ranging from 31 to 83 years of age). According to the Dukes classification system, 34 patients were in stages A and B; 28 were in stages C and D. This study was performed in accordance with the Declaration of Helsinki (1964). Informed written consent was obtained from all patients.
shRNA and reagents
Lentiviruses carrying short hairpin RNA (shRNA) targeting FOXQ1 or non-targeting control (NC) were purchased from NeuronBiotech (Shanghai, China). The targeting sequences are: FOXQ1-sh1 (5′-CGCGGACUUUGCACUUUGA-3′), FOXQ1-sh2 (5′-CCAGCTCCTTCGCCATCGACA-3′), FOXQ1-sh3 (5′-GGCUGGCUUCAUCCACUGC −3′), and NC-shRNA (5′-TTCTCCGAACGTGTCACGT-3′). All reagents and primers used in quantitative real-time polymerase chain reaction (qRT-PCR) were purchased from Takara Bio (Dalian, China). FOXQ1 and β-catenin antibodies were purchased from Santa Cruz (Santa Cruz, CA, USA). E-cadherin, N-cadherin, vimentin, MMP2, c-Myc, VEGF-A and cyclin D1 antibodies were purchased from ProteinTech (Chicago, IL, USA). GAPDH antibody and horseradish peroxidase (HRP)-conjugated rabbit anti-mouse secondary antibody were purchased from Abgent (San Diego, CA, USA). Alexa Fluor 555-conjugated donkey anti-rabbit secondary antibody was obtained from Beyotime Biotechnology (Shanghai, China). Transwell plates and Matrigel were purchased from BD Biosciences (San Diego, CA, USA). The Streptavidin-Peroxidase (SP) staining kit and diaminobenzidine (DAB) substrate used in immunohistochemistry were purchased from CWBio (Beijing, China).
Immunohistochemistry
Formalin-fixed tissues were embedded in paraffin blocks and a series of sections (4 μm) were made. Rabbit anti-human FOXQ1 antibody was used in immunohistochemical staining (1:100 dilution) following the manufacturer's instruction. Stained tissue sections were examined under a Olympus light microscope (Olympus, USA) and scored based on the staining intensity: 0, no staining; 1, light yellow staining; 2, brownish yellow staining; and 3, brown staining. In addition, 5 fields were randomly selected from each section, and the percentage of positive area (area of positively stained cells) in the total area (area of total cells) was calculated. The sections were scored as follows: 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The scores of staining intensity and staining area were added up and the sections were classified as: negative (−), total score 0–1; weak positive (+), total score 2; positive (++), total score 3–5; and strong positive (+++), total score 6–7.
Immunofluorescent staining
Cells were seeded at 1 × 105 cells/ml on a glass slide. After treatment, cells were fixed with 4% cold paraformaldehyde for 20 min. Fixed cells were washed with phosphate buffered saline (PBS) 3 times, and permeabilized with 0.2% Triton for 10 min followed by PBS washing 3 additional times. Non-specific binding was blocked with 1% bovine serum albumin (BSA) for 30 min, and cells were incubated with rabbit anti-human β-catenin antibody (1:500 dilution) at 4°C overnight. After washing with PBS 3 times, cells were incubated with Alexa Fluor 555 donkey anti-rabbit secondary antibody (1:500 dilution) at room temperature for 2 hours followed by PBS washing 3 additional times. Next, DAPI staining was used to stain the nuclei and, after rinsing with ddH2O, the slide was mounted with glycerol and examined under a fluorescent microscope.
Lentiviral transduction of shRNA and selection of stable transfectants
SW480 cell line was obtained from the American Type Culture Collection (ATCC, Rockville, MD) and cultured in RPMI 1640 medium supplemented with 12% fetal bovine serum (Hyclone, Pittsburgh, PA, USA) at 37°C in a 5% CO2-humidified atmosphere. SW480 cells were seeded at 2 × 106 cells/ml in 6-well plates (1 ml/well). Culture media were changed after 14 hours, and lentiviruses carrying 3 different FOXQ1 targeting shRNA and NC shRNA were added to the cells at a multiplicity of infection of 30. Polybrene was added at a final concentration of 5 μg/ml in each well. Culture media were changed after 16 hours. Cells were examined under a fluorescent microscope after 72 hours to check the transduction efficiency. Transduced cells were transferred to flasks and puromycin was added to the cells at a final concentration of 2 μg/ml. After selecting for 2 weeks, puromycin was removed and total RNA and total proteins were isolated for further experiments.
Quantitative RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA using the Reverse Transcription Kit from Takara. After adjusting the cDNA concentration in all groups, quantitative RT-PCR (qRT-PCR) was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad) with SYBR Green. The PCR conditions were as follows: pre-denaturation at 95°C for 30 s; 35 cycles of denaturation (95°C for 5 s), annealing (55–60°C for 30 s) and extension (72°C for 1 min); final extension at 72°C for 10 min. The relative level of gene expression was calculated using the ΔΔCt method with normalization to GAPDH. All experiments were repeated 3 times. The primers used are listed in Table 1.
Table 1.
Primer sequences for qRT-PCR amplification of different genes
| Gene | Primer |
|---|---|
| GAPDH | Forward 5′-CTTTGGTATCGTGGAAGGACTC-3′ |
| Reverse 5′-GTAGAGGCAGGGATGATGTTCT-3′ | |
| FOXQ1 | Forward 5′-TGATTTCTTGCTATTGACCGATGC-3′ |
| Reverse 5′-GCCCAAGGAGACCACAGTTAGAG-3′ | |
| E-cadherin | Forward 5′-TGGCTTCCCTCTTTCATCTCC-3′ |
| Reverse 5′-TCATAGTTCCGCTCTGTCTTTGG-3′ | |
| N-cadherin | Forward 5′-CGTGAAGGTTTGCCAGTGTGA-3′ |
| Reverse 5′-CCTGGCGTTCTTTATCCCG-3′ | |
| Vimentin | Forward 5′-TCAATGTTAAGATGGCCCTTG-3′ |
| Reverse 5′-TGAGTGGGTATCAACCAGAGG-3′ | |
| MMP2 | Forward 5′-ACATCAAGGGCATTCAGGAGC-3′ |
| Reverse 5′-CACAGTCCGCCAAATGAACC-3′ | |
| VEGF-A | Forward 5′-TGCTGTGGACTTGAGTTGGGAG-3′ |
| Reverse 5′-CCTGGCCTTGCACATTCCTG-3′ |
Western blots
Total protein was extracted from cells or tissues using lysis buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM sodium chloride, 5 mM ethylenediaminetetraacetic acid, 1% Triton-X 100, 1% dithiothreitol, and 1% protease inhibitor cocktail (Roche). Equal amounts of protein extracts (50 μg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride membrane. Membranes were blocked with 5% w/v non-fat dry milk dissolved in Tris buffered saline plus Tween-20 (TBS-T; 0.1% Tween-20; pH 8.3) at room temperature for 2 hours. Different primary antibodies against GAPDH (1:2000), FOXQ1 (1:2000), E-cadherin (1:500), N-cadherin (1:500), vimentin (1:500), MMP2 (1:500), c-Myc (1:500), cyclin D1 (1:500) and VEGF-A (1:500) were added to the samples and incubated at 4°C overnight. β-actin was used as a loading control. After washing with TBS-T, membranes were incubated with HRP-labeled secondary antibodies for 2 hours at room temperature. Immunobands were visualized using an enhanced chemiluminescence kit (GE Healthcare, Waukesha, WI, USA) after exposure to X-ray films. The signal intensity of each band was measured by Fusion software (Vilber Lourmat, Marne-la-Vallée Cedex, France) to calculate protein levels. All experiments were repeated 3 times.
Transwell assay
Forty μl of 1:3 diluted Matrigel (50 mg/L) was added to the upper chamber of a Transwell plate (pore size of the membrane: 0.8 μm) and dried under UV light overnight. The upper chamber was rehydrated using serum free medium and 5 × 104 cells in 400 μl suspension were added. The lower chamber was filled with 600 μl complete medium. After incubation for 36 hours, cells and Matrigel were wiped from the upper chamber and the membrane was fixed with paraformaldehyde followed by crystal violet staining for 15 min. After air-drying, the membrane was mounted on a glass slide and examined under microscope.
In vitro angiogenesis assay
Culture supernatant from a 3-day culture of each group was mixed with an equal volume of fresh complete medium and added to 6-well plates seeded with EA.hy926 cells, which had been transduced with lentiviruses carrying NC shRNA at 2.5 × 105 cells/well. After incubation for 48 hours, angiogenesis was examined under a microscope.
Drug sensitivity test and flow cytometry
1 × 104 cells in 200 μl suspension from each group were seeded in 96-well plates. Five concentration gradients and duplicates for each were made by serial dilution of the initial cell suspension. Fluorouracil (5-FU) or oxaliplatin (L-OHP) was added at a final concentration of 100 μg/ml. After incubation for 24 hours, cells were harvested, counted, and analyzed by flow cytometry for apoptosis using 7-aminoactinomycin D (7-AAD) and Annexin V staining.
Statistical analysis
All data were analyzed using SPSS19.0 statistical software. Measurement data are expressed as mean ± standard deviation (SD). Comparison was made by the t test between 2 groups and by the one-way ANOVA among multiple groups. The results from immunohistochemistry were analyzed by the χ2 test. The results from qRT-PCR and Western blot were analyzed by the R test. A P value < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Professors Weixue Tang and Lixue Chen at the Experimental Research Center of the First Affiliated Hospital of Chongqing Medical University for providing the SW480 cells and the experimental platform.
References
- 1.Christensen J, Bentz S, Sengstag T, Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS One. 2013;8(3):e60051; PMID:23555880; http://dx.doi.org/ 10.1371/journal.pone.0060051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yee YK, Tan VP, Chan P, Hung IF, Pang R, Wong BC. Epidemiology of colorectal cancer in Asia. J Gastroenterol Hepatol. 2009; 24:1810-6; PMID:20002940; http://dx.doi.org/ 10.1111/j.1440-1746.2009.06138.x [DOI] [PubMed] [Google Scholar]
- 3.Ellis LM. Angiogenesis and its role in colorectal tumor and metastasis formation. Semin Oncol. 2004; 31:3-9; PMID:15696024; http://dx.doi.org/ 10.1053/j.seminoncol.2004.11.028 [DOI] [PubMed] [Google Scholar]
- 4.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009; 361:2449-60; PMID:20018966; http://dx.doi.org/ 10.1056/NEJMra0804588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand FE, Angus CW, Partis WJ, Sigounas G. Developmental pathways in colon cancer: crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle. 2012; 11:4344-51; PMID:23032367; http://dx.doi.org/ 10.4161/cc.22134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patai AV, Molnar B, Tulassay Z, Sipos F. Serrated pathway: alternative route to colorectal cancer. World J Gastroenterol. 2013; 19:607-15; PMID:23431044; http://dx.doi.org/ 10.3748/wjg.v19.i5.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheen YY, Kim MJ, Park SA, Park SY, Nam JS. Targeting the Transforming Growth Factor-beta Signaling in Cancer Therapy. Biomol Ther (Seoul). 2013; 21:323-31; PMID:24244818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieller A, Pasche B, Frank S, Glaser B, Kunz J, Witt K, Zoll B. Isolation and characterization of the human forkhead gene FOXQ1. DNA Cell Biol. 2001; 20:555-61; PMID:11747606; http://dx.doi.org/ 10.1089/104454901317094963 [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F, Ethier SP, Miller F, Wu G. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011; 71: 1292-301; PMID:21285253; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS One. 2012; 7:e39937; PMID:22761930; http://dx.doi.org/ 10.1371/journal.pone.0039937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia L, Huang W, Tian D, Zhang L, Qi X, Chen Z, Shang X, Nie Y, Wu K. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014; 59:958-73; PMID:24005989; http://dx.doi.org/ 10.1002/hep.26735 [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D, Wang H. Short hairpin RNA targeting FOXQ1 inhibits invasion and metastasis via the reversal of epithelial-mesenchymal transition in bladder cancer. Int J Oncol. 2013; 42:1271-8; PMID:23403865 [DOI] [PubMed] [Google Scholar]
- 13.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y, et al.. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010; 70:2053-63; PMID:20145154; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2161 [DOI] [PubMed] [Google Scholar]
- 14.Abba M, Patil N, Rasheed K, Nelson LD, Mudduluru G, Leupold JH, Allgayer H. Unraveling the role of FOXQ1 in colorectal cancer metastasis. Mol Cancer Res. 2013; 11:1017-28; PMID:23723077; http://dx.doi.org/ 10.1158/1541-7786.MCR-13-0024 [DOI] [PubMed] [Google Scholar]
- 15.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006; 25:7531-7; PMID:17143297; http://dx.doi.org/ 10.1038/sj.onc.1210059 [DOI] [PubMed] [Google Scholar]
- 16.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330-7; PMID:22810696; http://dx.doi.org/ 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen J, Bentz S, Sengstag T, Shastri VP, Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS One. 2013; 8:e60051; PMID:23555880; http://dx.doi.org/ 10.1371/journal.pone.0060051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feuerborn A, Srivastava PK, Kuffer S, Grandy WA, Sijmonsma TP, Gretz N, Brors B, Grone HJ. The Forkhead factor FoxQ1 influences epithelial differentiation. J Cell Physiol. 2011; 226:710-9; PMID:20717954; http://dx.doi.org/ 10.1002/jcp.22385 [DOI] [PubMed] [Google Scholar]
- 19.Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL, Eschrich SA, Yeatman TJ, Deane NG, Beauchamp RD. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology. 2012; 142:562-71.e2; PMID:22115830; http://dx.doi.org/ 10.1053/j.gastro.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuerborn A, Srivastava PK, Kuffer S, Grandy WA, Sijmonsma TP, Gretz N, Brors B, Gröne HJ. The Forkhead factor FoxQ1 influences epithelial differentiation; J. J Cell Physiol. 2011; 226(3):710-9; PMID:20717954; http://dx.doi.org/ 10.1002/jcp.22385 [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D, Wang H. Short hairpin RNA targeting FOXQ1 inhibits invasion and metastasis via the reversal of epithelial-mesenchymal transition in bladder cancer; J. Int J Oncol. 2013; 42(4):1271-78; PMID:23403865 [DOI] [PubMed] [Google Scholar]
- 22.Gill S, Blackstock AW, Goldberg RM. Colorectal cancer. Mayo Clin Proc. 2007; 82:114-29; PMID:17285793; http://dx.doi.org/ 10.1016/S0025-6196(11)60974-9 [DOI] [PubMed] [Google Scholar]
- 23.Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan P, Bridgewater J, et al.. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004; 350:2343-51; PMID:15175436; http://dx.doi.org/ 10.1056/NEJMoa032709 [DOI] [PubMed] [Google Scholar]
- 24.Sanoff HK, Carpenter WR, Sturmer T, Goldberg RM, Martin CF, Fine JP, McCleary NJ, Meyerhardt JA, Niland J, Kahn KL, et al.. Effect of adjuvant chemotherapy on survival of patients with stage III colon cancer diagnosed after age 75 years. J Clin Oncol. 2012; 30:2624-34; PMID:22665536; http://dx.doi.org/ 10.1200/JCO.2011.41.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng YD, Yang H, Chen GQ, Zhang ZC. Molecularly targeted drugs for metastatic colorectal cancer. Drug Des Devel Ther. 2013; 7:1315-22; PMID:24204124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011; 71:3076-86; PMID:21346143; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2787 [DOI] [PubMed] [Google Scholar]
- 27.Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR, Zhao Y, Harris DC, Zheng G. E-cadherin/beta-catenin complex and the epithelial barrier. J Biomed Biotechnol. 2011; 2011:567305; PMID:22007144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Meng F, Liu G, Zhang B. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011; 71(4):1292-301; PMID:21285253; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011; 71(8):3076-86; PMID:21346143; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2787 [DOI] [PubMed] [Google Scholar]
- 30.Jian H, Shen X, Liu I, Semenov M, He X, Wang XF. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006; 20:666-74; PMID:16543220; http://dx.doi.org/ 10.1101/gad.1388806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S. TGF-beta regulates beta-catenin signaling and osteoblast differentiation in human mesenchymal stem cells. J Cell Biochem. 2011; 112:1651-60; PMID:21344492; http://dx.doi.org/ 10.1002/jcb.23079 [DOI] [PMC free article] [PubMed] [Google Scholar]