Abstract
The Arabidopsis root epidermal cells decide their fates (root-hair cell and non-hair cell) according to their position. SCRAMBLED (SCM), an atypical leucine-rich repeat receptor-like kinase (LRR RLK) mediates the positional information to the epidermal cells enabling them to adopt the proper fate. Via feedback regulation, the SCM protein accumulates preferentially in cells adopting the root-hair cell fate. In this study, we determine that TRY, but not the related factor CPC, is responsible for this preferential SCM accumulation. We observed severe reduction of SCM::GUS expression in the try-82 mutant root, but not in the cpc-1 mutant. Furthermore, the overexpression of TRY by CaMV35S promoter caused an increase in the expression of SCM::GUS in the root epidermis. Intriguingly, the overexpression of CPC by CaMV35S promoter repressed the expression of SCM::GUS. Together, these results suggest that TRY plays a unique role in generating the appropriate spatial expression of SCM.
Keywords: Arabidopsis, CAPRICE, epidermal patterning, LRR RLK, root hair, SCRAMBLED, TRITYCHON
Lateral inhibition is used to specify 2 cell fates in the Arabidopsis root epidermis: root-hair cells and non-hair cells.1 The non-hair-cell fate is promoted by an activator complex that includes WEREWOLF (WER), a R2R3 MYB transcription factor, GLABRA3 (GL3), a bHLH MYC transcription factor, and TRANSPARENT TESTA GLABRA1 (TTG1), a WD40-repeat protein, which together stimulates the transcription of the GLABRA2 (GL2) gene.2,3 GL2 represses the expression of ROOT HAIR DEFECTIVE6 (RHD6) that is required for the transcription of downstream root-hair genes, and thereby directs cells to differentiate into non-hair cells.4 The non-hair-cell fate activator complex also induces the transcription of the root-hair-cell fate activators, CAPRICE (CPC) and TRIPTYCHON (TRY). CPC and TRY are single R3 MYB transcription factors, and they are small enough to diffuse freely into neighboring cells via plasmodesmata.5-8 Like WER, CPC and TRY are able to bind to GL3 which disrupts the WER/GL3/TTG1 non-hair-cell fate activator complex.9,10 It has been experimentally shown that CPC and WER compete for GL3 binding quantitatively.3 Therefore, an epidermal cell with a higher WER/CPC ratio acquires functional activator complex, whereas an epidermal cell with a lower WER/CPC ratio fails to form a functional activator complex, and adopts the root-hair-cell fate. Without the guidance of a positional cue, a similar lateral inhibition mechanism appears to occur stochastically during trichome patterning in the epidermis of Arabidopsis leaves.1 In the root epidermis, the WER/CPC ratio is determined in a position-dependent manner. A root epidermal cell adjacent to 2 underlying cortical cells (H cell) has a lower WER/CPC ratio and differentiates into root-hair cells, on the other hand, the WER/CPC ratio is higher in a root epidermal cell in contact with one cortical cell (N cell) and, so adopts the non-hair-cell fate.11-14 Accordingly, the CPC and WER proteins were found predominantly in the nuclei of the H cells and N cells, respectively.8,15,16 SCRAMBLED (SCM), an atypical leucine-rich repeat receptor-like kinase (LRR RLK), mediates the positional information by reducing the expression of WER in the H cells.17,18 Moreover, the SCM proteins exhibit preferential accumulation in the H cells than the N cells, due to a feedback regulation.19
In our previous study, we found that SCM expression in the root epidermis is negatively regulated by WER and GL3, and positively regulated by TRY.19 However, we were unable to define the possible effect of CPC in this study, due to the use of the unusual cpc-3 mutant allele, which does not affect root epidermal patterning. The cpc-3 allele does bear a missense mutation (Glu26Lys) in the CPC gene, but it alters root hair formation only in the cpc-3 try-82 double mutant.19 Therefore, to accurately assess the possible role of CPC in SCM feedback regulation, we sought to examine the effect of cpc-1, the strong allele.5
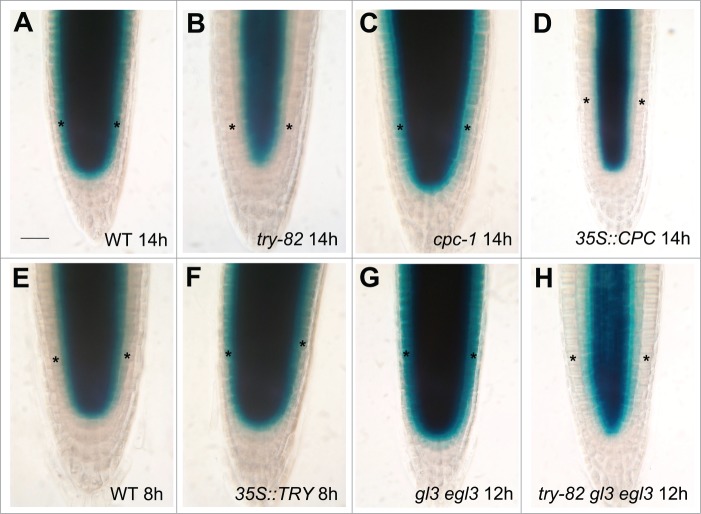
In the root epidermis of try-82 mutant plants, SCM::GUS expression was reduced severely, relative to the wild-type (Fig. 1A and B). However, SCM::GUS expression was not detectably altered in the cpc-1 mutants (Fig. 1C). To confirm that TRY is involved in increasing SCM expression, we examined the effect of overexpressing TRY on SCM expression. Following the introduction of the SCM::GUS reporter transgene into the 35S::TRY line by crossing, we observed that overexpression of TRY (using CaMV35S promoter) increased SCM::GUS expression in the root epidermis (Fig. 1E and F). Thus, we conclude that TRY, but not CPC, acts as a positive regulator of SCM expression in the epidermis of Arabidopsis roots.
Figure 1.
Expression of SCM::GUS reporter gene in the root of wild-type and various epidermal cell fate mutants. Arabidopsis seeds were sterilized and germinated.21 Four-day-old roots harboring SCM::GUS reporter gene were stained with GUS staining solution at 37°C for indicated time as described in previous work.19 (A) The wild-type root stained for GUS for 14 hours, (B) the try-82 root stained for GUS for 14 hours, (C) the cpc-1 root stained for GUS for 14 hours, (D) the 35S::CPC root stained for GUS for 14 hours, (E) the wild-type root stained for GUS for 8 hours, (F) the 35S::TRY root stained for GUS for 8 hours, (G) the gl3 egl3 root stained for GUS for 12 hours and (H) the try-82 gl3 egl3 root stained for GUS for 12 hours. Asterisks indicate the epidermis of the roots. The scale bar represents 50 μm.
CPC and TRY both repress GL2 expression by inhibiting the activity/formation of the WER/GL3/TTG1 complex, consistent with the observation that the wer mutant is epistatic to the cpc mutant with respect to GL2 expression.20 However, we discovered that, with respect to SCM expression, the try-82 mutant is epistatic to the gl3 egl3 mutant (Fig. 1H) because the try-82 gl3 egl3 triple mutant exhibited severely reduced SCM::GUS expression in the root epidermis, similar to the try-82 single mutant (Fig. 1H), whereas SCM::GUS expression in the epidermis of the gl3 egl mutant was up-regulated as described in previous study (Fig. 1G).19 Because the effect of try-82 was epistatic to gl3 egl3 rather than additive, it may be that the WER/GL3/TTG1 negatively regulates SCM expression by inhibiting TRY function. We further analyzed the effect of overexpression of CPC by the CaMV35S promoter and we found that it repressed SCM::GUS expression in the epidermis (Fig. 1D). This confirms that CPC is not the positive regulator of SCM expression. The ability of overexpressed CPC proteins to alter the SCM::GUS expression may be due to inhibition of TRY as competitive analogs of TRY.
It is notable that our results uncover a distinction in the action of TRY and the WER/GL3/TTG1 complex on different part of this gene regulatory network. Specifically, it appears that WER/GL3/TTG1 complex and TRY regulate SCM expression in a different way from GL2 expression. Considering that TRY is presumed to be a transcriptional repressor, a novel SCM expression inhibitor may exist in the epidermis whose expression is suppressed by TRY and induced by the WER/GL3/TTG1 complex.
In this study, we describe a TRY-dependent, but CPC-independent feedback regulation of SCM expression in the Arabidopsis root epidermis. Although CPC and TRY are closely related proteins, some differences in their functions have been previously identified. For example, the leaves of the cpc-1 mutant develop more trichomes over the entire epidermal surface; whereas, the try-82 mutant leaves produce increased trichomes in clusters.6 EGL3, a redundant homolog of GL3, also has different characteristics than its closely related GL3. The GL3 proteins move from the H cells to neighboring N cells via plasmodesmata, but the EGL3 proteins do not move between the root epidermal cells, and they appear to trap the CPC proteins in the nuclei of the H cells.8,22 Thus, the TRY-specific SCM feedback regulation reported here can be considered as an example of a gene duplication leading to the acquisition of novel functions during plant evolution.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the US. National Science Foundation (grant no. IOS-1121602 to J.S. and S.H.K).
References
- 1. Ryu KH, Zheng X, Huang L, Schiefelbein J. Computational modeling of epidermal cell fate determination systems. Curr Opin Plant Biol 2013; 16:5-10; PMID:23287386; http://dx.doi.org/ 10.1016/j.pbi.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 2. Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003; 130:6431-9; PMID:14627722; http://dx.doi.org/ 10.1242/dev.00880 [DOI] [PubMed] [Google Scholar]
- 3. Song SK, Ryu KH, Kang YH, Song JH, Cho YH, Yoo SD, Schiefelbein J, Lee MM. Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol 2011; 157:1196-208; PMID:21914815; http://dx.doi.org/ 10.1104/pp.111.185785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLOS Genet 2012; 8:e1002446; PMID:22253603; http://dx.doi.org/ 10.1371/journal.pgen.1002446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 1997; 277:1113-6; PMID:9262483; http://dx.doi.org/ 10.1126/science.277.5329.1113 [DOI] [PubMed] [Google Scholar]
- 6. Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 2002; 21:5036-46; PMID:12356720; http://dx.doi.org/ 10.1093/emboj/cdf524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu S, Koizumi K, Macrae-Crerar A, Gallagher KL. Assessing the utility of photoswitchable fluorescent proteins for tracking intercellular protein movement in the Arabidopsis root. PLOS One 2011; 6:e27536; PMID:22132108; http://dx.doi.org/ 10.1371/journal.pone.0027536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang YH, Song SK, Schiefelbein J, Lee MM. Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol 2013; 163:193-204; PMID:23832626; http://dx.doi.org/ 10.1104/pp.113.221028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 2002; 129:5409-19; PMID:12403712; http://dx.doi.org/ 10.1242/dev.00111 [DOI] [PubMed] [Google Scholar]
- 10. Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 2004; 55:389-98; PMID:15604688; http://dx.doi.org/ 10.1007/s11103-004-0893-8 [DOI] [PubMed] [Google Scholar]
- 11. Cormack RGH. Investigations on the development of root hairs. New Phytol 1935; 34:30-54; http://dx.doi.org/ 10.1111/j.1469-8137.1935.tb06826.x [DOI] [Google Scholar]
- 12. Bunning E. Uber die Differenzierungsvorgange in der Cruciferenwurzel. Planta 1951; 39:126-53; http://dx.doi.org/ 10.1007/BF01910114 [DOI] [Google Scholar]
- 13. Dolan L, Duckett C, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig RS, Roberts K. Clonal relations and patterning in the root epidermis of Arabidopsis. Development 1994; 120:2465-74. [Google Scholar]
- 14. Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 1994; 166:740-54; PMID:7813791; http://dx.doi.org/ 10.1006/dbio.1994.1352 [DOI] [PubMed] [Google Scholar]
- 15. Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 2005; 132:5387-98; PMID:16291794; http://dx.doi.org/ 10.1242/dev.02139 [DOI] [PubMed] [Google Scholar]
- 16. Ryu KH, Kang YH, Park YH, Hwang I, Schiefelbein J, Lee MM. The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 2005; 132:4765-75; PMID:16207757; http://dx.doi.org/ 10.1242/dev.02055 [DOI] [PubMed] [Google Scholar]
- 17. Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 2005; 307:1111-13; PMID:15618487; http://dx.doi.org/ 10.1126/science.1105373 [DOI] [PubMed] [Google Scholar]
- 18. Kwak SH, Schiefelbein J. The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev Biol 2007; 302:118-31; PMID:17027738; http://dx.doi.org/ 10.1016/j.ydbio.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 19. Kwak SH, Schiefelbein J. A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr Biol 2008; 18:1949-54; PMID:19097902; http://dx.doi.org/ 10.1016/j.cub.2008.10.064 [DOI] [PubMed] [Google Scholar]
- 20. Lee MM, Schiefelbein J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 2002; 14:611-8; PMID:11910008; http://dx.doi.org/ 10.1105/tpc.010434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schiefelbein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 1990; 2:235-43; PMID:12354956; http://dx.doi.org/ 10.1105/tpc.2.3.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 2005; 132:291-8; PMID:15590742; http://dx.doi.org/ 10.1242/dev.01565 [DOI] [PubMed] [Google Scholar]