Abstract
Bacteriophage λ has four adjacent genes - S, R, Rz and Rz1 - dedicated to host cell lysis. While S, encoding the holin and antiholin, and R, encoding the endolysin, have been intensively studied, the products of Rz and Rz1 have not been characterized at either the structural or functional levels. Rz1 is an outer membrane lipoprotein and our results indicate that Rz is a type II signal anchor protein. Here we present evidence that an Rz-Rz1 complex that spans the periplasm carries out the final step in the process of host lysis. These results are discussed in terms of a model where endolysin-mediated degradation of the cell wall is a prerequisite for conformational changes in the Rz-Rz1 complex leading to the juxtaposition and fusion of the IM and OM. Fusion of the two membranes removes the last physical barrier to efficient release of progeny virions.
Introduction
Rz and Rz1, and their equivalents, are unique in biology. They are the only genes that share the same DNA in different reading frames (Fig. 1A) and are associated with the same phenotype. Moreover, nonsense mutations in either Rz or Rz1 abolish lysis and plaque-formation if moderate concentrations of divalent cations are present, making them the only lambda genes with lethal or conditionally-lethal phenotypes on wt E. coli whose function remains undefined (Zhang and Young, 1999). Recently, a comprehensive bioinformatic search found Rz/Rz1 equivalents in nearly all phages of Gram-negative hosts (Summer et al., 2007). The diversity of Rz-Rz1 equivalents was striking: 37 unrelated sequence families, including 8 families with the embedded structure found in lambda (8 families), but also families in which Rz1 extends beyond Rz (overlapped structure; 23 families) and others where the two genes are completely separated (6 families).
Figure 1. The Rz/Rz1 genes.
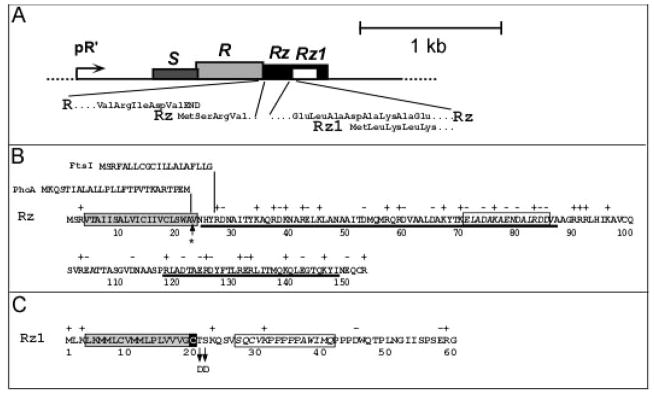
A. The lambda lysis cassette. The four lysis genes are shown drawn to scale downstream of the lambda late promoter, pR′. The insets show the initial codons of Rz, beginning at the end of the R endolysin gene, and Rz1, which is embedded entirely within Rz in the +1 reading frame. Modified with permission from Summer et al. (2007).
B. Rz amino acid sequence. The TMD is boxed in gray and predicted alpha-helical regions are underlined. Above the Rz sequence, the sequences of the PhoA secretory signal and the FtsI N-terminal TMD are shown above the junctions where they are substituted in the PhoA-Rz and FtsI-Rz hybrid constructs. The asterisk indicates the position of a predicted signal peptidase I cleavage site (Bendtsen et al., 2004). The boxed italic text here and in panel C indicates the polypeptide sequence used for raising antibodies.
C. Rz1 amino acid sequence. The lipoprotein (signal peptidase II) signal sequence predicted by LipoP (Juncker et al., 2003) is boxed in gray, with the processed Cys residue highlighted in black. The positions of the two Asp substitutions that redirect Rz1 to the cytoplasmic membrane are indicated by arrows.
The phenotype associated with Rz (or Rz1) mutations has been known for nearly three decades, but the molecular role of these two proteins in host lysis has not been elucidated. Although it was suggested that Rz might encode a murein-specific endopeptidase detected in lambda lysates (Taylor, 1971; Young et al., 1979; Bienkowska-Szewczyk and Taylor, 1980), neither protein has been associated with an enzymatic activity. Rz encodes a 153 aa polypeptide with a hydrophobic N-terminus (Fig. 1B) that is predicted to be either a secretory signal or N-terminal transmembrane domain (TMD) by sequence analysis algorithms. No studies describing its sub-cellular localization under normal levels of expression have been reported. Rz1 spans only 60 codons, with a predicted signal peptidase II cleavage site at Cys20 (Fig. 1C) (Hanych et al., 1993b; Zhang and Young, 1999); its product was identified as a 6 kDa outer membrane lipoprotein by palmitate-labeling in vivo (Kedzierska et al., 1996).
There is some genetic evidence that the Rz and Rz1 proteins interact. First, the Rz/Rz1 equivalents from phage P2, lysB/lysC, complement defects in the lambda genes, but only as a cognate pair(Markov et al., 2004). In addition, yeast two-hybrid analysis of a library of phage T7 genes found multiple positives between clones with the last 10 codons of 18.7, the T7 Rz1 equivalent, and the last 50 codons of 18.5, the Rz equivalent (Bartel et al., 1996). These data suggest that Rz and Rz1 interact in a C-terminus to C-terminus fashion, which may account for the architectures of the embedded and overlapped Rz/Rz1 genes, in that these unusual arrangements minimize the likelihood of recombinational separation of the interacting domains (Summer et al., 2007).
The discovery of a new class of proteins, the spanins, which are functionally equivalent to Rz/Rz1 pairs, provides new insight into Rz/Rz1 function (Summer et al., 2007). Spanins have a lipoprotein signal peptide as well as a C-terminal TMD and, thus, should provide a physical connection between the inner and outer bacterial membranes. The ability of the T1 spanin gene to complement the Rz-Rz1 lysis phenotype provides further support to the notion that Rz and Rz1 interact and, in fact, indicates that Rz-Rz1 complexes span the periplasm and connect the inner (IM) and outer (OM) membranes.
Recently, Rz-Rz1 equivalents were identified in the tectivirus PRD1, and in the same study, a role for the Rz-Rz1 proteins in lysis was proposed (Krupovic et al., 2008). In this scheme, it was suggested that Rz-Rz1 interact not only with each other but also, as a complex, with the holin, to transmit the mechanical stress of the holin-mediated lesion in the cytoplasmic membrane to the outer membrane, resulting in its disruption.
Here we describe experiments that unambiguously resolve the sub-cellular localization of the λ Rz and Rz1 proteins when their genes are expressed from the λ late promoter, assess the ability of Rz-Rz1 to form complexes, and examine the requirement of holins for Rz/Rz1 function. The results are discussed in terms of a model for the participation of these proteins in a hitherto unrecognized final stage of the bacteriophage infection cycle.
Results
Analysis of membranes in lambda inductions
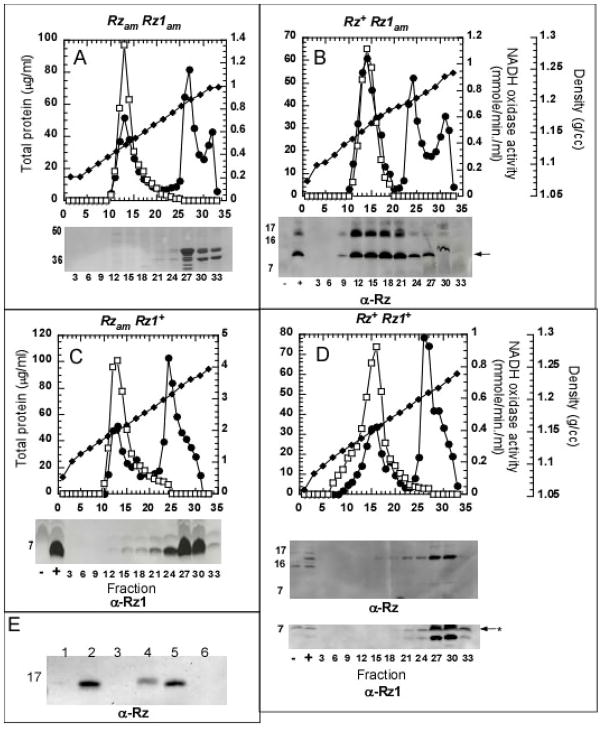
Accepted methods for separating the IM and OM by isopycnic gradient centrifugation involve first converting the cells to spheroplasts and then disrupting the cells by sonication (Osborn and Munson, 1974). Failure to convert cells to spheroplasts or having the spheroplasts undergo lysis before mechanical disruption leads to poor separation of the IM and OM fractions. In our hands (and as reported previously (Reader and Siminovitch, 1971)), stable spheroplasts could not be formed from cells expressing the λ holin gene. To avoid this problem, we examined the sub-cellular distribution of Rz and Rz1 expressed at their normal, physiological levels using a set of isogenic prophages carrying a null mutation in the S holin gene. As a control, isopycnic sucrose gradient analysis was performed for an induced lysogen with null mutations in S, Rz and Rz1 (λ901RzamRz1am) (Fig. 2A). Total protein, NADH oxidase (an IM-marker), and major OM proteins (OMPs) were normally distributed in two major peaks at the densities expected of IM and OM fractions. When isogenic Rz+ lysogens were induced, Rz appeared as a 17 kDa membrane-associated protein (Fig. 2E). This species could be extracted with detergent but not by sodium carbonate, as expected for proteins integrated into the cytoplasmic membrane. When membranes from these induced lysogens were fractionated by isopycnic sucrose gradient centrifugation, Rz was found in the IM fraction, distributed identically to NADH oxidase activity (Fig. 2B). When parallel analyses were performed on membranes from an induced λ901RzamRz1+ lysogen, Rz1, the lipoprotein, was found to be located in the OM fractions (Fig. 2C). However, in the induction of the isogenic Rz+Rz1+ lysogen, the distribution of the Rz protein in particular changed dramatically, co-fractionating with Rz1 (Fig. 2D).
Figure 2. The subcellular localization of Rz and Rz1 at physiological levels of expression.
Panels A – E: Membranes from an induced culture of RY17299 carrying isogenic λ901 prophages with the indicated RzRz1 alleles were analyzed by (Panels A – D) sucrose density gradient or by carbonate extraction (Panel E), as described in Materials and Methods. Symbols: total protein, filled circles; NADH oxidase activity, open boxes; density, filled diamonds. In all cases, the abscissa of the gradient distributions and gel or immunoblot insets is sample number, from the top of the gradient. Panel A: Coomassie-blue stained SDS gel showing major OMPs distributed in fractions 27–33. Panels B and D: representative fractions immunoblotted with anti-Rz antibody, with selected MW standards indicated to the left. +, −: total membrane samples from induced lysogens with wt (+) or null (−) Rz or Rz1 genes, as indicated. Arrow in panel B indicates the position of putative Rz degradation product (see text). Panel D also has an immunoblot using anti-Rz1 with the postion of a reproducible background band indicated by an asterisk. Lanes in Panel E: 1, soluble fraction from French-press disrupted cells; 2, total membrane fraction from disrupted cells; 3, soluble fraction from carbonate extraction; 4, insoluble fraction from carbonate extraction; 5, soluble fraction from 1% EBB extraction; 6, fraction insoluble in 1% EBB.
The Rz and Rz1 proteins form a complex
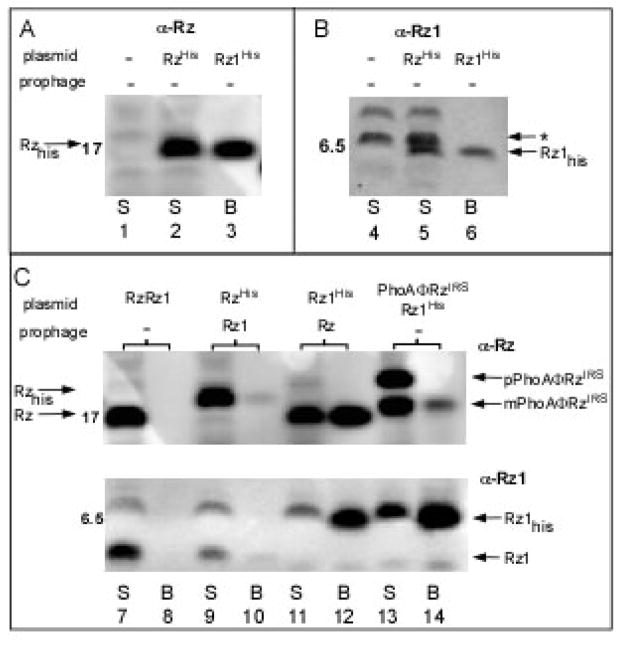
While there is genetic and phylogenetic evidence to suggest that Rz and Rz1 interact to form a complex, the physical association of these proteins has never been demonstrated. To show that Rz and Rz1 co-purify, we constructed alleles of Rz and Rz1 encoding C-terminal hexahistidine tagged products, confirmed that each was functional by complementation (data not shown), and prepared detergent-solubilized extracts from cells expressing the gene pairs Rzhis/Rz1 or Rz/Rz1his. Complexes were isolated by immobilized metal affinity chromatography (IMAC) and then analyzed by SDS-PAGE and Western blotting. As can be seen in Fig. 3C (lanes 11 and 12), the majority of Rz is bound to the affinity resin only when the Rz gene is co-expressed with Rz1his. Surprisingly, the converse is not true; when Rz1 is co-expressed with Rzhis, no Rz1 is retained by the affinity resin, Fig. 3C (lanes 9 and 10). However, in this experiment, only a small fraction of the Rzhis protein present in the extracts binds to the resin. Since Rzhis is quantitatively bound to the resin in the absence of Rz1, Fig. 3A (lanes 2 and 3), we interpret this to mean that, in the Rzhis/Rz1 complex, the oligohistidine tag of Rzhis is masked by Rz1 and is not free to interact with the IMAC resin. Further evidence that Rz and Rz1 interact derives from the observation that when the Rz gene is expressed in the absence of Rz1, the Rz protein is subject to a specific proteolytic event resulting in a ~12kDa product recognized by antisera raised against a C-terminal epitope (Fig 2B, arrow). This cleavage product is not detected in cells expressing the Rz/Rz1 gene pair, suggesting that the protease-sensitive site in Rz is not exposed in Rz/Rz1 complexes (Fig. 2D).
Figure 3. Rz and Rz1 form a complex.
Induced RY17299 lysogens carrying single isogenic λ901 prophages and plasmids carrying various Rz and Rz1 alleles were converted to spheroplasts, lysed, and subjected to batch affinity fractionation with magnetic Talon beads. The Rz or Rz1 proteins present in each case are shown above each lane. Total spheroplasts (S) and proteins bound to the beads (B) were analyzed by SDS-PAGE and Western blotting with the antibodies indicated above (panels A,B) or to the side (panel C) of each panel. Panels A and B: Lanes 1 and 4, RzamRz1am lysogen with pRE; Lanes 2 and 3, RzamRz1am lysogen with pRE-RzHIS; Lanes 5 and 6, RzamRz1am lysogen with pRE-Rz1HIS; Panel C; Lanes 7 and 8, RzamRz1am lysogen with pRE-RzRz1; Lanes 9 and 10, RzamRz1 lysogen with pRE-RzHIS; Lanes 11 and 12, RzRz1am lysogen with pRE-Rz1HIS; Lanes 13 and 14, RzamRz1am lysogen with pRE-PhoAΦRzIRSRz1HIS. The position of relevant MW standards is indicated on the left of each panel. An asterisk indicates the position of a reproducible background band. The bands representing Rz, RzHIS, Rz1, Rz1HIS as well as mature (mPhoAΦRzIRS) and precursor (pPhoAΦRzIRS) forms of the PhoA-Rz chimera are indicated with arrows.
Rz/Rz1 complexes must connect the IM and OM to be functional
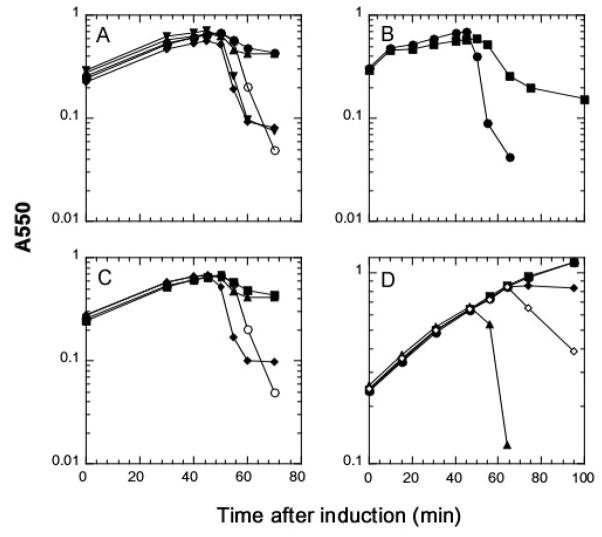
To provide evidence that the proposed connection between the IM and OM is required for the function of the Rz/Rz1 complex, we tested the ability of Rz and Rz1 derivatives with altered topological determinants to replace the wild type proteins. First, the N-terminal TMD of Rz was replaced with the canonical signal sequence of the soluble periplasmic protein PhoA. Roughly half of the PhoAss-Rz chimera accumulated as a processed species that formed a complex with Rz1 (Fig. 3, lanes 13–14), but could not functionally replace Rz (Fig. 4A). In fact, the phoAss-Rz gene exhibited dominant-negative character, in that its expression in an induced Rz+Rz1+ lysogen partially blocked lysis in the presence of 10mM MgCl2 (Fig. 4B). In contrast, replacement of the TMD of Rz with that of FtsI, a type II membrane protein involved in cell division (Guzman et al., 1997), produced a chimera that functioned similarly to Rz in cell lysis (Fig. 4A). Next, we altered the Rz1 sequence at the two codons immediately distal to the Cys20 residue at which the lipoprotein precursor cleavage and processing occurs, replacing Thr21Ser22 with Asp-Asp. As predicted from the specificity of the LOL system (Terada et al., 2001), this alteration redirected Rz1 to the IM (Fig. 5). Importantly, this Rz1 derivative was no longer functional (Fig. 4C).
Figure 4. Rz1 function requires its OM localization signal.
Growth of the indicated cultures after lysogenic (Panels A, B, C) and/or IPTG (Panels B, D) induction was monitored as described in Materials and Methods. In all panels, the open symbols indicate standard LB media while the filled symbols indicate LB supplemented with 10 mM MgCl2. Panel A: MC4100(λ900RzamRz1+) carrying pRE(○, ●); pRE-RzIRS (▼); pRE-PhoAΦRzIRS (▲); pRE-FtsIΦRzIRS (◆). Panel B: MC4100(λ900) carrying pJF118EH (●), pJF-PhoAΦRzIRS (■). Panel C: MC4100(λ900Rz+Rz1am) carrying pRE (○,■); pRE-Rz1AU1 (◆); pRE-Rz1AU1T21D,S22D (▲). Panel D: RY17299lacIQ carrying pRW (●); pRE-RzRz1 (■); pRW-LyzC13S,C44S (◇, ◆); pRW-RzRz1-LyzC13S,C44S (▲).
Figure 5. The non-functional Rz1Au1T21DS22D mutant localizes to the inner membrane.
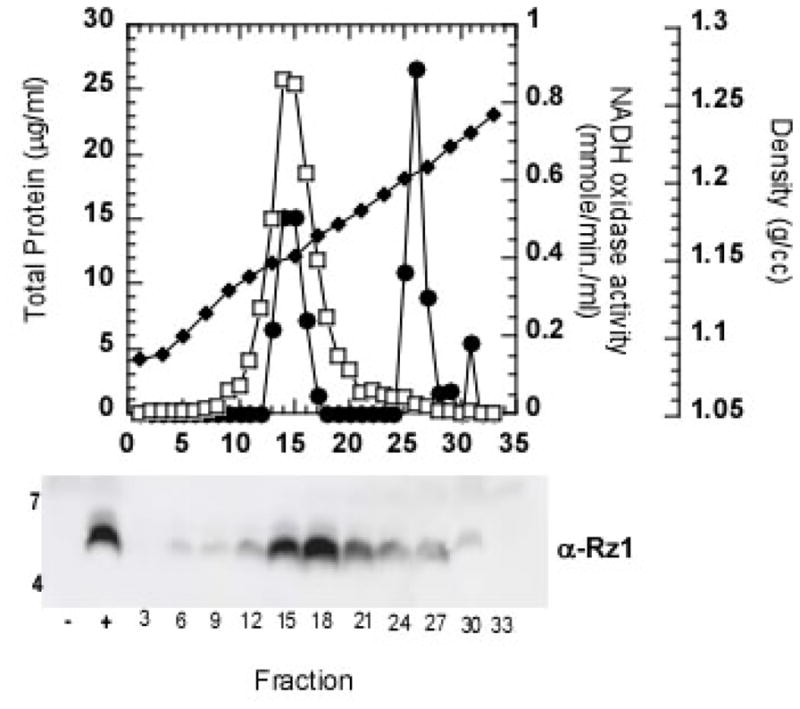
Membranes from induced RY17299(λ901RzamRz1am) cells carrying the plasmid pRE-Rz1AU1T21D,S22D were analyzed by sucrose gradient and immunoblot, as described for Figure 1.
The function of the Rz-R1 complex is independent of the holin protein
Many dsDNA phage encode SAR-endolysins which are capable of eliciting host lysis in the absence of a holin (Xu et al., 2004). In our studies of Lyz, the SAR-endolysin of bacteriophage P1, we observed that when E. coli expressed the lyz gene in media containing 10mM MgCl2, they were converted to spherical forms rather than undergoing overt lysis (Fig. 4D). This sensitivity to Mg+2 clearly mirrored the phenotype of Rz− or Rz1− mutants of bacteriophage λ. Strikingly, cells expressing the Rz/Rz1 gene pair in addition to lyz no longer showed a Mg+2-dependent lysis defect (Fig. 4D). In fact, cultures expressing the three genes showed an earlier and sharper lysis than those expressing lyz alone. These results demonstrate that Rz-Rz1 function, and by extension, also the function of spanins, is independent of holin function.
Discussion
Rz and Rz1 localization
Here, we demonstrate that, at physiological levels of expression, Rz and Rz1 are localized to the IM and OM, respectively, with their C-terminal domains predicted to lie in the periplasm. Importantly, when Rz and Rz1 are co-expressed, a large fraction of Rz redistributes to the OM fractions in isopycnic sucrose gradients, likely due to an association between the Rz and Rz1 proteins. This result is at variance with several other reports showing that Rz and Rz1 accumulate in fractions of density intermediate between the IM and OM (Hanych et al., 1993a; Kedzierska et al., 1996; Taylor et al., 1996). However, in all but one case, these experiments were done using either induced S+ lysogens or over-expression of the Rz or Rz1 genes from T7 promoters. Both approaches are problematical. As noted above, separation of the membranes by isopycnic gradient centrifugation depends on spheroplast formation, and stable spheroplasts cannot be obtained if the lambda holin is expressed. In the one experiment using a thermally induced λcI857Sam7 lysogen, Rz1 was found to be restricted to the OM fraction (Taylor et al., 1996). Significantly, in the same report, Rz1 was found in all cellular fractions if it was over-produced from a hyper-expression T7 plasmid vector.
It is also worth noting that a previous study claimed that the IM and OM completely lose separability in induced lambda lysogens, with essentially all the membranes converted to an intermediate density fraction (Kucharczyk et al., 1991). The change was shown to occur progressively during the infection cycle and to be dependent on late gene expression. However, recent work has shown that the S holin is triggered to hole formation by small decreases in membrane potential (Gründling et al., 2001), which will occur regardless of which method is used to harvest cells prior to spheroplast formation. Thus, these findings are also likely to be artifacts of unsuccessful spheroplasting, especially in view of our results showing that the membranes retain distinct densities in inductions of Sam7 lysogens (Fig. 2).
Structure and topological requirements of Rz-Rz1 complexes
We have shown that Rz-Rz1his complexes can be isolated from detergent-solubilized spheroplasts (Fig. 3) and that these complexes appear to mask a proteolysis-sensitive site in the C-terminal third of the Rz periplasmic domain (Fig. 2B). Based on the 12 kDa size of the proteolysis fragment, the protease-sensitive site (ca. residues 90 – 115) is, according to secondary structure analysis, in an unstructured region separating two α-helices that occupy most of the Rz periplasmic domain (Fig. 1B). Since protection of Rz from proteolysis did not require the co-expression of a functional holin/endolysin gene pair, the interaction between Rz and Rz1 can occur in vivo in the presence of the normal complement of peptidoglycan. These observations are also consistent with results from yeast two-hybrid assays that indicated the C-termini of the Rz and Rz1 equivalents of bacteriophage T7 interact (Bartel et al., 1996).
Significantly, the function of this complex is lost if either protein is improperly localized (Fig. 4). Redirecting Rz1 to the IM by changing the lipoprotein sorting signal eliminated function (Fig. 4C). Moreover, when the C-terminal domain of Rz is exported to the periplasm using a cleavable signal sequence instead of its normal type II signal anchor domain, it also loses function (Fig. 4A), and, in this case, actually antagonizes the function of the wild type protein (Fig. 4B). In contrast, Rz function was maintained when its N-terminus was replaced with the type II signal anchor of FtsI (Fig. 4A), indicating that being physically tethered to the IM is necessary and sufficient for Rz to carry out its normal function.
A model for the role of Rz-Rz1 in lysis
Our findings are inconsistent with a recent model for the function of Rz/Rz1-like proteins stemming from the study of bacteriophage PRD1 (Krupovic et al., 2008). In this scheme, the Rz/Rz1 complex transmits mechanical stress from holin-mediated lesion formation in the IM to the OM, resulting in its disruption. Here, we show that holin-independent cell lysis brought about by SAR endolysins is blocked by millimolar concentrations of divalent cations and that this blockage is relieved by co-expression of a non-cognate Rz/Rz1 lysis gene pair. This would appear to rule out any direct role for holin proteins in the function of the Rz/Rz1 complex. Note that this does not rule out an effect of the converse: i.e., effects of Rz/Rz1 on holin function. Indeed missense changes in lysC, the Rz1 gene of phage P2, significantly delay the onset of lysis (Markov et al., 2004). However, without information about the expression of the nearby P2 holin gene under these conditions, these results cannot yet be interpreted in terms of effects of a mutant Rz-Rz1 complex on the IM, and thus potentially, on holin function.
The simplest interpretation of the results reported here might be that the Rz-Rz1 complex improves the efficiency of endolysin degradation of the murein layer. Since the OM is covalently attached to the cell wall by >105 oligopeptide linkages between Lpp and the murein, a complex that spans the periplasm could conceivably push the IM away from the murein layer. This might make endolysin-mediated murein degradation more efficient, similarly to the requirement for sucrose-mediated plasmolysis in spheroplast formation (Birdsell and Cota-Robles, 1967). We think this is unlikely to be the basis of Rz-Rz1 function. With phage lambda (R. Young, unpublished; also, see Fig. 3 of Gründling et al. (2001)) and with cells undergoing lysis via a SAR endolysin (M. Xu, unpublished observations), cells pass through a brief spherical stage just before lysis. In the absence of Rz-Rz1 function and the presence of moderate levels of divalent cations, they progress no further. It is difficult to conceive of murein structures uniformly distributed in the spherical cell periplasm that are somehow resistant to the endolysin, especially considering the diversity of enzymatic activities that are manifested among phage endolysins.
An alternative model, which we favor, is that the Rz-Rz1 complex mediates a third and final step in host lysis. In this view, the first step is the temporally-programmed permeabilization of the cytoplasmic membrane by the holin, resulting in the release of a cytoplasmic endolysin or the activation of a SAR endolysin. The second stage is the endolysin-dependent degradation of the murein layer. We propose that this is followed by a third stage involving the fusion of the IM and OM mediated by the Rz-Rz1 complexes. This would cause the cytoplasm and the environment to become topologically equivalent and, thus, all conceivable barriers to the release of the progeny virions would be eliminated. We suggest that the initial Rz-Rz1 complexes are metastable but are competent to undergo conformational changes that bring the two membranes in contact and facilitate their fusion, analogous to the function of SNARE complexes in eucaryotic systems (Jahn and Scheller, 2006). However, premature membrane fusion is prevented since the peptidoglycan meshwork would interfere with the proposed conformational changes and, obviously, prevent close apposition of the IM and OM. Although this final step is not required in typical laboratory medium lacking sufficient divalent cations to stabilize the OM, the lack of Rz or Rz1 function is noticeable as a diminution in the sharpness of lysis profiles, not only in lysis mediated by SAR endolysins (Fig. 4D) but also with the canonical lambda SR holin-endolysin lysis (e.g., see Fig. 4 of Summer et al. (Summer et al., 2007)), The fact that genes for Rz-Rz1 equivalents are found in nearly all phages of Gram-negative hosts (Summer et al., 2007), indicates that in the osmotic and ionic conditions found in nature, the OM is a significant barrier even after holin-endolysin mediated destruction of the cell wall.
The model predicts that the Rz-Rz1 complex must exist in at least two conformations, with the final form more conformationally stable but inhibited from formation as long as the murein layer is present. How the peptidoglycan barrier interferes with the conformational change is unclear, but the simplest notion would be that the final complexes are too large to be formed as long as the meshwork of the murein is still present. It should be noted that membrane-fusion actitivity for Rz1 has been suggested before, based on studies with purified, SDS-solubilized Rz1 protein and artificial liposomes (Bryl et al., 2000). However, it is unclear whether any small, proline-rich, N-terminally lipidated polypeptide might not show this effect, since no controls with mutant Rz1 proteins lacking the putative activity were undertaken. Indeed, our results indicate that Rz1 acting alone has no biological activity. Our model can be tested in a number of ways, including, most directly, by high-resolution cryo-electron microscopy, which may allow visualization of the Rz-Rz1-dependent membrane fusion event.
Finally, we note that while the three steps of phage lysis mediated by holins, endolysins and Rz-Rz1 complexes form a sequential pathway, in which holin function is required for endolysin function, which is in turn required for Rz-Rz1 function, they are mechanistically independent (i.e., do not require heterotypic interactions with each other). This likely accounts for the remarkable diversity and mosaicism found in phage lysis cassettes, which are composed of many unrelated families of holins and Rz-Rz1 proteins, and at least three types of endolysins (Wang et al., 2000; Young, 2002; Young and Wang, 2006; Summer et al., 2007). If there is no direct interaction between the three components of the pathway that assault the three elements of the cell envelope, there would be no restriction on recombinational assortment of the three functions, other than the recently discovered requirement that pinholins must serve SAR endolysins(Park et al., 2007).
Materials and Methods
Bacterial growth and induction
The bacterial strains, bacteriophage, and plasmids used in this study are listed in Table 1. Bacterial cultures were grown in standard LB media supplemented with MgCl2 (10 mM), ampicillin (100 μg/ml), kanamycin (40 μg/ml), and chloramphenicol (10 μg/ml) when appropriate. Culture growth and lysis profiles were monitored as previously described (Smith et al., 1998). Briefly, overnight cultures were diluted 300:1 and grown with aeration at 30°C for lysogenic cultures and 37°C for non-lysogens. Lysogens were thermally induced at A550 ~0.3 by aeration at 42°C for 20 min, followed by continued growth at 37°C. When indicated, isopropyl β-D-thiogalactosidase (IPTG) was added to a final concentration of 1mM.
TABLE 1.
Phages, strains, lysogens and plasmids
| Genotypes and relevant features | Sources, references1 | |
|---|---|---|
| Phage designation | ||
| λ143 | λimm434 cI Ram5 | Laboratory stock |
| λ800Δ (SR) | λΔstf tfa)::cat cI857 Δ(SR); forms lysis-defective, CamR prophage | (Smith et al., 1998) |
| λ900 | λΔ(stf tfa)::cat cI857bor::kan; carries CamR and KanR; wt RzRz1 | (Zhang and Young, 1999) |
| λ900RzQ100am Rz1+ | (Zhang and Young, 1999) | |
| λ900Rz+ Rz1W38am | (Zhang and Young, 1999) | |
| λ901 | Sam7 recombinant, isogenic to λ900 | |
| λ901RzQ100amRz1+ | ||
| λ901Rz+Rz1W38am | ||
| λ901RzQ100am Rz1W38am | ||
| E. coli K-12 Strains | ||
| XL-1 Blue | recA endA1 gyrA96 thi1 hsdR17 supE44 relA1 lac [F′ proAB lacZΔM15::Tn10] | Stratagene |
| MC4100 | F− araD139 Δ[argF-lac]U169 rpsL150 relA1 flb5301 deoC1 ptsF25 rbsR | (Silhavy et al., 1984) |
| MG1655 | F− ilvG rph1 rfb50 | Laboratory stock |
| RY17299 | MG1655 ΔtonA | Laboratory stock |
| RY17299 lacIq1 | (Park et al., 2006) | |
| RY17299(λ901 Rz+Rz1+) | Lysogen carrying λ900Sam7Rz+Rz1+ prophage | |
| RY17299(λ901 RzamRz1+) | ||
| RY17299(λ901Rz+Rz1am) | ||
| RY17299(λ901RzamRz1am) | ||
| MC4100(λ900) | (Zhang and Young, 1999) | |
| MC4100(λ900RzamRz1+) | (Zhang and Young, 1999) | |
| MC4100(λ900Rz+Rz1am) | (Zhang and Young, 1999) | |
| Plasmids | ||
| pQ | pSC101 origin with low-copy mutation; Q cloned under Plac/ara-1 promoter | (Gründling et al., 2001) |
| pJF118EH | ColE1 origin, Ptac, lacIq and ampR; medium copy Ptac expression vector | (Fürste et al., 1986) |
| pJFLyz | pJF118EH carrying P1 lyz, nucleotides 20225–21079 of the P1 genome | (Xu et al., 2005) |
| pJF-PhoAΦRzIRS | phoAΦRz from pRE-PhoAΦRz cloned into pJF118EH | |
| pJF-PhoA | pJF118EH carrying phoA | (Xu et al., 2004) |
| pJF-FtsIcmyc | pJF118EH carrying ftsIcmyc | (Xu et al., 2004) |
| pRE | Derivative of pJF118EH with lacIQ and Ptac promoter replaced with pR′ promoter region from λ; transcriptionally activated by lambda Q | (Park et al., 2006) |
| pRE-RzRz1 | Rz+Rz1+ cloned in pRE | |
| pRE-Rz | Rz+ cloned in pRE with Rz1W38am null mutation | |
| pRE-Rz1 | Rz1+ cloned in pRE | |
| pRE-RzHIS | pRE-Rz with GGHHHHHH inserted after codon 153 of Rz | |
| pRE-RzIRS | RzIRS cloned in pRE | |
| pRE-FtsIΦRzIRS | Codons 24 to 40 of ftsI replacing codons 4–26 of Rz | |
| pRE-Rz1AU1 | Rz1AU1 cloned in pRE | |
| pRE-PhoAΦRzIRS | Codons 1–22 of Rz replaced by codons 1–26 of phoA, encoding the signal sequence | |
| pRE-Rz1AU1T21D,S22D | pRE-Rz1 carrying the T21D and S22D mutations | |
| pRE-PhoAΦRzIRSRz1HIS | Rz1HIS inserted downstream of PhoAΦRz | |
| pRE-Rz1HIS | pRE-Rz1 with GGHHHHHH inserted after codon 60 of Rz1 | |
| pRW | Derivative of pRE with bor gene flanking multiple cloning site, to provide downstream homology for recombination | (Zheng et al., 2008) |
| pRW-LyzC13S,C44S | P1 lyzC13S, C44S cloned into pRW | G. Kuty, unpublished |
| pRW-RzRz1-LyzC13S,C44S | RzRz1 inserted downstream of lyzC13S, C44S |
Unless otherwise specified, constructs were made for this study.
The RzRz1 genes are either expressed in their native context in the λ genome from prophage induction or from derivatives of the medium-copy plasmid pRE carrying the λ late promoter, pR′. In all cases, the λ late gene activator Q is either supplied in trans from an induced prophage or from pQ, a very low copy vector carrying Q cloned under the control of the Plac/ara-1 promoter (Lutz and Bujard, 1997; Gründling et al., 2001).
DNA techniques
Isolation of plasmid DNA, DNA amplification by PCR, DNA transformation, site-directed mutagenesis and DNA sequencing were performed as described previously (Tran et al., 2005). Sequences of primers used in cloning and sequencing are available upon request.
Phage manipulation
The phage λSam7 and its isogenic Rz and Rz1 variants listed in Table 1 were obtained by crossing the Rz and/or Rz1 mutant alleles onto λ, as previously described (Zhang and Young, 1999; Gründling et al., 2000b). Lysogens were selected, their allele and single copy status confirmed by sequencing and PCR, respectively, as previously described (Tran et al., 2005; Zheng et al., 2008).
Plasmid construction
All DNA manipulations were performed according to standard techniques described previously (Tran et al., 2005). The plasmids pRE-RzRz1 and pRE-Rz have nucleotides 45950 – 46427 of λ, carrying the Rz ribosome binding site (RBS) through the end of Rz1, inserted between the KpnI and BamHI sites of pRE (Table 1). In addition, pRE-Rz contains a silent mutation in the Rz reading frame, which converts codon 38 of the Rz1 reading frame to an amber codon (Zhang and Young, 1999). The plasmid pRE-Rz1 has nt 46173 – nt 46368 of λ inserted between the BamHI and HindIII sites of pRE. Derivatives of these plasmids carrying alleles modified with sequences encoding IRS, AU1 and HIS tags, as well as additional derivatives with hybrid constructs, are described in Table 1.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were performed as described previously (Gründling et al., 2000a; Bernhardt et al., 2002). SeeBlue Plus2 (Invitrogen) prestained standard was included as a molecular mass standard. Briefly, proteins were transferred to PVDF membrane (Pall Life Sciences) using a Hoefer TE unit at 0.1 mA overnight at 4° C. Antibodies (Sigma Genosys) were generated in rabbits against the synthetic peptides CELADAKAENDALRDD, corresponding to the Rz residues 71–85 (modified with an N-terminal Cys for conjugation) and SQCVKPPPPPAWIMQ, corresponding to Rz1 residues 27–41 (Fig. 1BC), and used at a dilution of 1:1000. The secondary antibody, goat-anti-rabbit-HRP (Thermo Scientific), was used at a dilution of 1:5000. Chemiluminescence was detected using a Bio-Rad XR Gel Doc system.
Sodium carbonate extraction
A 200 ml culture of RY17299(λ901Rz+Rz1am) was induced, aerated 1 h, and harvested by centrifugation in a JA-10 rotor at 8,000 × g for 10 min. The cells were resuspended in 1 ml of French press buffer (100mM Na2HPO4, 100mM KCl, 5 mM EDTA, pH 8.0) supplemented with 5 μl Protease Inhibitor Cocktail (Sigma-Aldrich P8465). The cells were disrupted by passage through a French pressure cell (Spectronic Instruments, Rochester, N.Y.) at 16,000 lb/in2, and cleared of unbroken cells by centrifugation in a Damon-IEC Spinette clinical centrifuge at 1,000xg for 10 min. Membranes from three equal portions were pelleted in a TLA 100.3 rotor at 150,000 × g for 1 h at 4 °C. One pellet was resuspended in 1 ml of 1M Na2 CO3, pH 11, incubated on ice for 30 min, and fractionated into peripheral (supernatant) and integral (pellet) protein fractions by centrifugation in a TLA 100.3 rotor at 150,000 × g for 1 h. A second pellet was resuspended in 1 mL of 1% Empigen BB, 100 mM NaCl, 50 mM Tris-Cl, pH 8.0, incubated at room temperature with gentle agitation for 1 h, and centrifuged in a TLA 100.3 rotor at 150,000 × g for 1 h. As a control, the final membrane pellet was resuspended in 1 ml of French press buffer and then centrifuged at 150,000 × g for 1 h as before. In each case, the insoluble material was resuspended in 1X SDS-PAGE buffer and the soluble material was mixed with an equal volume of 2X SDS-PAGE buffer. All samples were heated to 100°C for 5 min before analysis by SDS-PAGE and Western blotting.
Subcellular localization of Rz and Rz1
Fifty min after thermal induction, 250 ml cultures of the indicated strains were rapidly cooled to 4° C by swirling the growth flasks in an ice-H2O bath. All subsequent steps carried out at 4° C. Cells were harvested by centrifugation at 8,000 × g for 15 min in a Beckman JA-10 rotor. The supernatant was completely removed and the cells converted to spheroplasts as described in Osborn and Munson (1974). The spheroplasts were transferred to a Beckman Ultra-Clear SW28 centrifuge tube and ruptured by sonication using a Sonicator Cell Disruptor W-375 (60% output, 3 × 15 sec bursts separated by 1 minute intervals to prevent heating). For harvesting of membranes and fractionation by density gradient, all sucrose solutions were prepared w/w and contained 5 mM EDTA, pH7.5. The lysate was layered on a two-step gradient consisting of 12 ml of 18% and 3 ml of 55% sucrose and centrifuged in a Beckman SW28 rotor at 100,000 × g for 2 h. The total membrane fraction was layered on a 5-step sucrose gradient and centrifuged as described (Osborn and Munson, 1974). Gradients were collected in 1 ml fractions from the top using a Model 640 ISCO Density Gradient Fractionator. The protein concentration of individual fractions was determined with the Bio-Rad protein assay (Bio-Rad) using BSA as the standard according to the manufacturer’s instructions. NADH oxidase activity assays were performed as described previously (Owen et al., 1982). The refractive index of alternate fractions was determined with a refractometer (Spectronic Instruments, 334610). Approximately 800 μl of every third gradient fraction was diluted to a final sucrose concentration of less than 10% with 1 mM EDTA, pH7.5 and centrifuged in a Beckman 70.1Ti rotor at 100,000 × g for 2 h at 4°C. Membrane pellets were resuspended in 60 μl of 1X SDS-PAGE buffer and boiled for 5 min. The distribution of the major OMPs was determined by SDS-PAGE and Coomassie blue staining.
Isolation of Rz-Rz1 complexes
Twenty-five ml cultures of lysogenic cells harboring the indicated plasmids were harvested at 50 min after induction by centrifugation in an Eppendorf 5702 centrifuge at 2,800 × g for 15 min. The cells were converted to spheroplasts and lysed as described (Broome-Smith and Spratt, 1986; Tran et al., 2005), except that a 50 μl aliquot of spheroplasts was reserved for SDS-PAGE analysis and the Protease Inhibitor Cocktail used (Sigma-Aldrich, P8849) did not contain EDTA. A 500 μl volume of lysate was incubated with 50 μl of Dynabeads TALON (Invitrogen) at 4°C for 2 h with gentle agitation. The beads were then collected magnetically and washed three times with lysis buffer containing 1 mg/ml BSA (MP Biomedicals). The beads were resuspended in 50 μl of 1X SDS-PAGE buffer and boiled for 5 min. All samples were cleared of beads and debris by brief centifugation in a tabletop microcentrifuge and analyzed by SDS-PAGE and Western blotting.
Acknowledgments
The clerical assistance of Daisy Wilbert is gratefully acknowledged. Support for this work came from PHS grant GM27099, from the Welch Foundation, and, for the efforts of ES, NSF grant EF0523951.
References
- Bartel PL, Roecklein JA, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. The Escherichia coli FKBP-type PPIase SlyD is required for the stabilization of the E lysis protein of bacteriophage ϕX174. Mol Microbiol. 2002;45:99–108. doi: 10.1046/j.1365-2958.2002.02984.x. [DOI] [PubMed] [Google Scholar]
- Bienkowska-Szewczyk K, Taylor A. Murein transglycosylase from phage λ lysate: purification and properties. Biochim Biophys Acta. 1980;615:489–496. doi: 10.1016/0005-2744(80)90515-x. [DOI] [PubMed] [Google Scholar]
- Birdsell DC, Cota-Robles EH. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J Bacteriol. 1967;93:427–437. doi: 10.1128/jb.93.1.427-437.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome-Smith JK, Spratt BG. A vector for the construction of translational fusions to TEM beta-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- Bryl K, Kedzierska S, Laskowska M, Taylor A. Membrane fusion by proline-rich Rz1 lipoprotein, the bacteriophage lambda Rz1 gene product. Eur J Biochem. 2000;267:794–799. doi: 10.1046/j.1432-1327.2000.01051.x. [DOI] [PubMed] [Google Scholar]
- Fürste JP, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Gründling A, Bläsi U, Young R. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J Biol Chem. 2000a;275:769–776. doi: 10.1074/jbc.275.2.769. [DOI] [PubMed] [Google Scholar]
- Gründling A, Bläsi U, Young R. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J Bacteriol. 2000b;182:6082–6090. doi: 10.1128/jb.182.21.6082-6090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, Manson MD, Young R. Holins kill without warning. Proc Natl Acad Sci U S A. 2001;98:9348–9352. doi: 10.1073/pnas.151247598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Weiss DS, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanych B, Kedzierska S, Walderich B, Taylor A. Molecular cloning, overexpression of Rz lysis gene of phage λ and subcellular localization of its protein product. In: de Pedro MA, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York and London: Plenum Press; 1993a. pp. 269–276. [Google Scholar]
- Hanych B, Kedzierska S, Walderich B, Uznanski B, Taylor A. Expression of the Rz gene and the overlapping Rz1 reading frame present at the right end of the bacteriophage lambda genome. Gene. 1993b;129:1–8. doi: 10.1016/0378-1119(93)90689-z. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska S, Wawrzynow A, Taylor A. The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene. 1996;168:1–8. doi: 10.1016/0378-1119(95)00712-1. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Cvirkaite-Krupovic V, Bamford DH. Identification and functional analysis of the Rz/Rz1-like accessory lysis genes in the membrane-containing bacteriophage PRD1. Mol Microbiol. 2008;68:492–503. doi: 10.1111/j.1365-2958.2008.06165.x. [DOI] [PubMed] [Google Scholar]
- Kucharczyk K, Laskowska E, Taylor A. Response of Escherichia coli cell membranes to induction of lambda cl857 prophage by heat shock. Mol Microbiol. 1991;5:2935–2945. doi: 10.1111/j.1365-2958.1991.tb01853.x. [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucl Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov D, Christie GE, Sauer B, Calendar R, Park T, Young R, Severinov K. P2 growth restriction on an rpoC mutant is suppressed by alleles of the Rz1 homolog lysC. J Bacteriol. 2004;186:4628–4637. doi: 10.1128/JB.186.14.4628-4637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Munson R. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 1974;31:642–653. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- Owen P, Graeme-Cook KA, Crowe BA, Condon C. Bacterial membranes: preparative techniques and criteria of purity. In: Hesketh TR, Kornberg HL, Metcalfe JC, Northcote D, Pogson CI, Tipton KF, editors. Techniques in Lipid and Membrane Chemistry B4/I. Amsterdam: Elsevier/North Holland Scientific Publishers Ltd; 1982. pp. 1–69. [Google Scholar]
- Park T, Struck DK, Dankenbring CA, Young R. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J Bacteriol. 2007;189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Struck DK, Deaton JF, Young R. Topological dynamics of holins in programmed bacterial lysis. Proc Natl Acad Sci U S A. 2006;103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader RW, Siminovitch L. Lysis defective mutants of bacteriophage lambda: On the role of the S function in lysis. Virology. 1971;43:623–637. doi: 10.1016/0042-6822(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Bacterial strains. In: Silhavy TJ, Berman ML, Enquist LW, editors. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. pp. xi–xiii. [Google Scholar]
- Smith DL, Chang CY, Young R. The λ holin accumulates beyond the lethal triggering concentration under hyper-expression conditions. Gene Expr. 1998;7:39–52. [PMC free article] [PubMed] [Google Scholar]
- Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J Mol Biol. 2007;373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- Taylor A. Endopeptidase activity of phage λ-endolysin. Nature New Biol. 1971;234:144–145. doi: 10.1038/newbio234144a0. [DOI] [PubMed] [Google Scholar]
- Taylor A, Kedzierska S, Wawrzynów A. Bacteriophage λ lysis gene product modified and inserted into Escherichia coli outer membrane: Rz1 lipoprotein. Microbial Drug Resistance. 1996;2:147–153. doi: 10.1089/mdr.1996.2.147. [DOI] [PubMed] [Google Scholar]
- Terada M, Kuroda T, Matsuyama SI, Tokuda H. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J Biol Chem. 2001;276:47690–47694. doi: 10.1074/jbc.M109307200. [DOI] [PubMed] [Google Scholar]
- Tran TAT, Struck DK, Young R. Periplasmic domains define holin-antiholin interactions in T4 lysis inhibition. J Bacteriol. 2005;187:6631–6640. doi: 10.1128/JB.187.19.6631-6640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- Xu M, Struck DK, Deaton J, Wang IN, Young R. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc Nat Acad Sci, U S A. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. [PubMed] [Google Scholar]
- Young R, Wang IN. Phage lysis. In: Calendar R, editor. The Bacteriophages. Oxford: Oxford University Press; 2006. pp. 104–126. [Google Scholar]
- Young R, Way S, Yin J, Syvanen M. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J Mol Biol. 1979;132:307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- Zhang N, Young R. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol Gen Genet. 1999;262:659–667. doi: 10.1007/s004380051128. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Dankenbring CA, Young R. Evolutionary dominance of holin lysis systems derives from superior genetic malleability. Microbiology. 2008 doi: 10.1099/mic.0.2008/016956-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]