Abstract
Mucins are important components that exert a variety of functions in cell-cell interaction, epidermal growth factor receptor signaling, and airways protection. In the conducting airways of the lungs, mucins are the major contributor to the viscoelastic property of mucous secretion, which is the major barrier to trapping inhaled microbial organism, particulates, and oxidative pollutants. The homeostasis of mucin production is an important feature in conducting airways for the maintenance of mucociliary function. Aberrant mucin secretion and accumulation in airway lumen are clinical hallmarks associated with various lung diseases, such as asthma, chronic obstructive pulmonary disease, cystic fibrosis, emphysema, and lung cancer. Among 20 known mucin genes identified, 11 of them have been verified at either the mRNA and/or protein level in airways. The regulation of mucin genes is complicated, as are the mediators and signaling pathways. This review summarizes the current view on the mediators, the signaling pathways, and the transcriptional units that are involved in the regulation of airway mucin gene expression. In addition, we also point out essential features of epigenetic mechanisms for the regulation of these genes.
Keywords: mucus, transcription, lung, epigenetics, differentiation
INTRODUCTION
The conducting airways of the lung are not just passive conduits for the passage of air between the lungs and environment. Cells lining the airway epithelium help to humidify the air as well as to trap environmental pollutants, particles, and pathogens to prevent their entry into the sterile areas of the lower airspace (1, pp. 443–538). To accomplish this function, secretory cells in the airways secrete a viscous mucus layer that helps to trap incoming particles, whereas coordinated beating of cilia from ciliated cells moves them cephalad, where the particles are eventually either expectorated or swallowed into the gastrointestinal (GI) tract (2).
The mechanism of airway mucus production and secretion has gained greater attention in recent years because of its function in mechanical host defense and its pathological ability to plug the airways and impair gas exchange. Asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis (CF) are three of the more prevalent and well-known lung diseases associated with mucus hypersecretion and accumulation in airways. Previously, research in airway obstruction focused on bronchoconstriction and dynamic airway collapse as the primary causes of airway narrowing. More recently, postmortem studies and biopsies of airways in patients with more severe forms of asthma and COPD have revealed that mucus plugging may be a more significant cause of airway narrowing than previously recognized (2, 3, 3a). Some investigators have questioned whether mucus hypersecretion is of clinical significance in diseases such as COPD when other variables such as age, smoking, and FEV1 (as a rough measurement of lung function) are controlled for (suggesting that excess secretions of mucus are merely a marker for a more severe underlying disease rather than a true cause of physiological impediment) (4). Other studies show that mucus secretion is predictive of mortality, but more so in patients with lower baseline lung function (5). This would suggest that mucus secretions have more physiological effects on gas exchange when it occurs on airways already narrowed from other causes such as bronchoconstriction, dynamic airway collapse, mucosal edema, and subepithelial fibrosis.
Although mucus is composed of various components, including water, glucose, various ionic solutes, and small antimicrobial protein peptides, it is the large, heavily glycosylated proteins called mucins that are primarily responsible for giving mucus their viscoelastic properties. Twenty mucin genes have been identified. Among these, 11 are expressed, either at the message and/or the protein level, in the lungs (6). The rest are found secreted in other body fluids such as saliva, GI secretions, and uterine secretions. To understand the cellular mechanism of mucus secretion, one can study the various steps of their synthesis: transcription of the mucin genes (MUC) into mRNAs, translation into polypeptides and intracellular packaging, glycosylation of the polypeptide backbones in the organelles, and, finally, secretion into the airways. Because of their large size and their complex carbohydrate backbones, mucins are difficult to purify. The protein backbones of various mucins (termed apomucins) have been used to generate antibodies, which have been used successfully in some studies, particularly those involving immunostaining of goblet cells (7). However, more detailed protein studies of mucins are somewhat limited by the facts that mucins exhibit poor migration in various gel matrices and that antibodies sometimes cannot bind to the native protein when their epitopes are hidden by the complex glycosylated backbones. Thus, relatively more data have accumulated on the regulation of these genes at the mRNA level in terms of both their transcriptional and posttranscriptional mechanisms.
This review focuses on the more common mucin genes found in the lungs, particularly those of the secreted gel-forming mucins. We focus on the intracellular signal transduction and transcriptional and posttranscriptional mechanisms that govern their expression. We touch briefly on what upstream signals from cytokines and/or ligands affect these mucins as relating to the intracellular mechanisms, but an in-depth examination of the multitude of mediators reported that can induce or repress mucins is beyond the scope of this review. Readers are referred to another excellent review on these upstream events (6).
MUCINS EXPRESSED IN THE LUNGS
Mucin proteins are synthesized by two types of secretory cells in the lungs: submucosal glandular cells and the surface mucous/goblet cells that line the epithelium. These cells can usually be seen histologically by Alcian blue–periodic acid–Schiff (PAS) staining because of the storage of these heavily glycosylated proteins in cytoplasmic secretory granules (8). Normal airway epithelium contains a paucity of surface goblet cells and a moderate number of submucosal glands. But in inflammatory airway diseases such as asthma and COPD, the surface goblet cells expand considerably (mucous cell hyperplasia/metaplasia), and submucosal gland frequency and size increase (9). The increase in surface goblet cells and sub-mucosal glands in these diseases may be what causes the excessive mucus secretion seen in these disorders (2, 3, 5).
The 11 mucins that have been described to express in the lungs are MUC1, -2, -3, -4, -5AC, -5B, -6, -7, -8, -13, and -19 (6). Among these, MUC5AC and MUC5B are felt to be the predominant mucins in airway mucus because they are the only ones consistently found at the protein level in mucus and in sputum (10–12). Thus, most of the data on mucins in the respiratory tracts have focused on these two mucins. However, the other mucins are expressed at the mRNA level, both in vitro and in vivo, on airway epithelial cells (13, 14). The significance of these other mucins in the contribution to airway mucus rheology and goblet/mucous cell metaplasia is not as clear as that of these two gel-forming mucins.
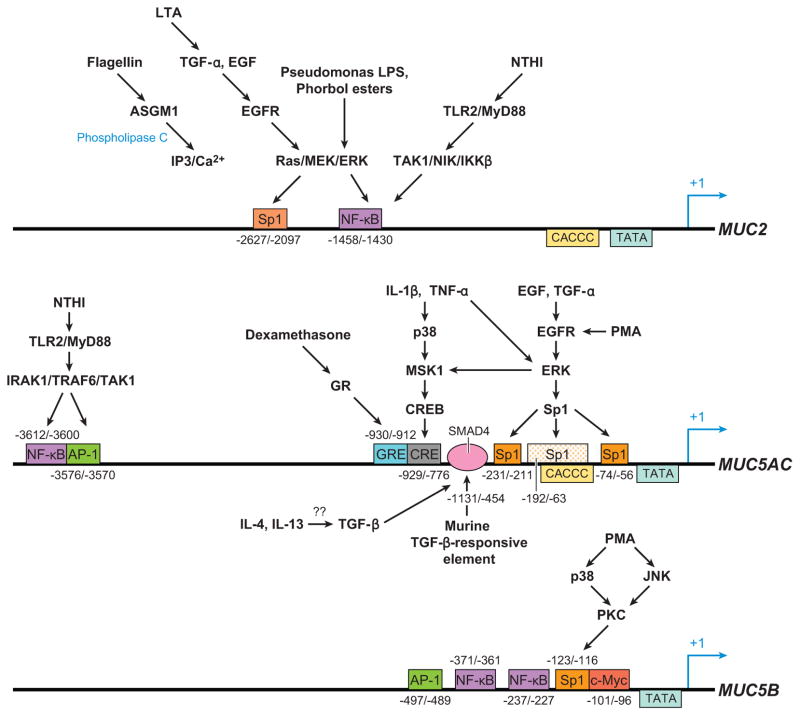
The stimuli that upregulate mucin gene expression can be broadly categorized into (a) inflammatory cytokines, (b) bacterial products, (c) growth factors, (d) environmental chemicals or pollutants, and (e) miscellaneous chemical agents. Among these, inflammatory cytokines can be subdivided further into proinflammatory [tumor necrosis factor-α (TNF-α), interleukin (IL)-1β], TH1 [interferon-γ (IFN-γ)], TH17 (IL-17A), and TH2 (IL-4, IL-9, IL-13) cytokines. Cytokines and bacterial products have been the most extensively studied because of their secretion into airways affected by lung diseases such as asthma, COPD (inflammatory cytokines), and CF (bacterial products) (6). Another focus of research has been growth factors, especially retinoic acids (RA) and ligands that activate epidermal growth factor receptor (EGFR) and cell proliferation, repair, and differentiation (15, 16). Likewise, environmental toxic agents such as cigarette smoke and ozone are important pathophysiological causes of diseases of COPD and emphysema, so their effects on airway epithelial cells and mucins have been explored. And finally, certain chemical agents such as phorbol esters that activate specific intracellular signaling cascades have been used to delineate specific signal transduction mechanisms that affect mucin gene expression (17–19). Despite the large number of heterogeneous mediators involved, patterns have emerged owing to the identification of a few specific signal transduction pathways and transcription mechanisms that are critical to mucin gene expression (Figure 1).
Figure 1.
A rough outline of 5′ promoter regulatory regions for MUC2, MUC5AC, and MUC5B. Although this picture is not comprehensive, some specific pathways are provided to illustrate the stimulus and signal transduction pathways that activate certain transcription factors to their binding sites on the mucin promoters. The mapping of specific regulatory regions for MUC5B to their stimulus has not been elucidated in detail, but some putative sites from promoter sequence analysis are provided.
MUC2 EXPRESSION AND REGULATION
MUC2 was one of the first gel-forming mucins to be characterized in detail. It is located next to the MUC5AC gene on chromosome 11p15.5. MUC2 is approximately 17 kb in size, with 48 exons, and the protein product is more than 5100 amino acids long (20). Its promoter region was the first of the mucins to be elucidated (21). MUC2 is expressed primarily in the small intestine and colon of the GI tract (22), but its mRNA has also been detected in the airways of the lungs (23). However, there have been questions raised about how much MUC2 contributes to airway mucus secretion in normal and hypersecretory states. In the few biochemical studies of expectorated mucus done to date, the amount of MUC2 present is very small compared with MUC5AC and MUC5B (24–26). Also, although MUC2 expression is increased in the airways of patients with asthma, the localization of MUC2 to surface goblet cells stained by Alcian blue–PAS staining is not as clear cut as the localization of MUC5AC in the goblet cells of the lung (27–29). Some investigators have postulated that MUC2 is not detected in expectorated mucus because its high insolubility makes it harder to assay (30). Others have also noted that MUC2 expression occurs early during goblet cell hyperplasia/metaplasia and precedes that of MUC5AC expression, suggesting MUC2 to be an early marker for mucous cell differentiation (13). Numerous inflammatory mediators can stimulate MUC2 expression in airway epithelial cells in culture. Many of these same mediators, interestingly, can also upregulate MUC5AC (31). So far, two main classes of mediators that upregulate MUC2—bacterial products and cytokines—have been described in detail. Bacterial gene products include microbial membrane components [lipopolysaccharide (LPS), lipoteichoic acid (LTA), and flagellin]. Cytokines include mainly the proinflammatory cytokines IL-1β and TNF-α.
MUC2 Expression Regulated by Mitogen-Activated Protein Kinase/Nuclear Factor-κB Pathways
The lungs of CF patients are chronically colonized and infected with bacteria, particularly gram-negative bacteria such as Pseudomonas aeruginosa and gram-positive bacteria such as Staphylococcus aureus. Because goblet cell hyperplasia is abundant in CF, investigators hypothesized that the bacterial products from the chronic infection could induce epithelial cells to express mucin genes and differentiate into goblet cells. Studies to test this hypothesis first started with MUC2 (Figure 1). Examination of the promoter region of MUC2 revealed a putative nuclear factor-κB (NF-κB) binding region at −1452/−1441 from the transcriptional start site (32). NF-κB activation in airway epithelial cells is a common event during inflammatory conditions. Utilizing bacterial extracts from P. aeruginosa and Staphylococcus aureus, investigators found that that LPS and LTA can induce MUC2 expression in the colon epithelial cell line HM3 (32, 33). Further studies showed that both signaled through the activation of Ras-MEK1/2-ERK1/2 pathways. In the case of LPS, ERK phosphorylation leads to the activation of the 90-kDa ribosomal S6 kinase (pp90sk), which subsequently activates NF-κB and promotes its translocation into the nucleus. There, NF-κB binds to a cis element at −1458/−1430 of the MUC2 gene to promote transcription (32). LTA does not act directly on Ras but instead first activates ADAM10, a matrix metalloprotease (MMP), to cleave membrane-bound pro-EGF (epidermal growth factor) to release free EGF ligands. These EGF ligands then bind to EGFR, which then activates RAS-MEK1/2-ERK1/2, as does LPS, to cause NF-κB activation of MUC2 transcription (33). P. aeruginosa also has a protein called flagellin that can activate airway epithelial cells and MUC2 expression (34). Although flagellin normally binds to TLR5 and activates NF-κB in airway cells (35), the induction of MUC2 appears to work through the binding of flagella by ASGM1, a membrane ganglioside. After binding ASGM1, ATP is released and binds to an extracellular nucleotide receptor that signals through phospholipase C activation and the generation of inositol triphosphate and Ca2+. As do LPS and LTA, the signal then converges on the mitogen-activated protein kinase (MAP kinase) ERK and leads to its activation and subsequent downstream transcriptional activation of MUC2 (34) (Figure 1).
MUC2 Expression Regulated by the MyD88/Nuclear Factor-κB Pathway
Bacterial products usually act on airway epithelial cells by binding to Toll-like receptors (TLRs) on their surface membranes. There are twelve known TLRs that recognize different components of bacteria, fungi, and/or viruses. Signal transduction events after the binding of microbial products to these receptors frequently converge downstream on the activation of NF-κB (36). However, TLR signaling can activate NF-κB through two parallel routes: the MyD88-dependent and -independent pathways (36). In the above studies (32–34), MUC2 activation by LPS, LTA, and flagellin was suspected not to involve the TLRs at all. LPS, for example, is usually bound to a serum protein called LPS binding protein (LBP) that is then brought to the TLR4 by either a membrane-bound or a soluble receptor called CD14 (37). In the studies above (32–34), however, neutralizing anti-CD14 antibodies could not inhibit Pseudomonas’s LPS stimulation of MUC2, suggesting that another receptor besides TLR4 is used (32). Likewise, LTA was believed to bind to surface platelet-activating factor receptors (PAFRs), and the flagellin was thought to involve the ASGM1 instead of TLRs (33, 34). Nevertheless, the involvement of TLRs through the MyD88-dependent pathway has been reported for MUC2 activation. Lysates from nontypeable hemophilus influenza (NTHI) (a pathogen common in COPD patients) were able to upregulate MUC2 expression through NF-κB activation. Its mechanism involves binding to TLR2, which leads to the activation of the MyD88-TAK-NIK pathway and subsequent IKK phosphorylation (38). Interestingly, this same study also showed that part of the upregulation of MUC2 depended on a non-TLR/MyD88 pathway that involved transforming growth factor (TGF)-β/Smad signaling.
MUC2 Expression Regulated by Inflammatory Cytokines
Like bacterial products, inflammatory cytokines induce MUC2 gene expression in airway epithelial cells. IL-1β, TNF-α, and IL-4 thus far have been identified as having this activity. IL-1β and TNF-α are common proinflammatory cytokines that are elevated in a variety of both airway- and non-airway-related lung disorders and do not necessarily have any specific association with mucous hypersecretory diseases. Nevertheless, airway diseases with mucous cell hyperplasia, particularly COPD, have elevated levels of these cytokines in the blood as well as in the lungs (39, 40). In addition, both IL-1β and TNF-α induce mucous cell hyperplasia in vivo in mouse models (41, 42). Both cytokines can also activate NF-κB signaling in airway epithelial cells (43, 44), which, as shown above, can lead to the activation of MUC2 expression. Thus, these cytokines may play some role in affecting the expression of mucin genes and mucous cell differentiation.
TNF-α induces the expression of MUC2 in NCI-H292 cells (31, 45). The detailed mechanism on how this induction occurs is not clear, although TNF-α induction did not require new protein synthesis (45), and it did not increase the activity of a 2.8-kb promoter fragment of MUC2 (31). This suggests that TNF-α induces MUC2 by a transcriptional mechanism but that the TNF-responsive element may be beyond the 2.8 kb of the transcriptional start site. IL-1β also stimulates MUC2 transcription in NCI-H292 cells (47). In one report, a retinoic acid receptor (RAR)-α antagonist could inhibit the induction of MUC2 by IL-1β and TNF-α, suggesting that IL-1β and TNF-α act partly through RAR pathways (48). Another report revealed that IL-1β’s stimulation of MUC2 occurs through the protein kinase C (PKC) and ERK pathways (49). In addition, this report also revealed that phosphoinositol 3-kinase (PI3K) inhibitors could partly block the induction of MUC2 without affecting ERK phosphorylation (49). This finding suggests that IL-1β’s effect occurs through a parallel PI3K/AKT pathway (49). IL-4 can induce MUC2 expression in NCI-H292 cells as well as in colon cell lines (50, 51). IL-4 can induce goblet cell hyperplasia in mouse in vivo. However, this in vivo effect of IL-4 showed that only Muc5ac and not Muc2 was induced (50, 52).
MUC2 Expression Regulated by Epidermal Growth Factor Ligands and Retinoic Acid
Growth factors are important for stimulating the growth and differentiation of airway epithelial cells. Among them, EGF family ligands and vitamin A (all-trans RA) are of primary importance in stimulating mucociliary differentiation of airway epithelial cells in culture (53, 54). Not surprisingly, these ligands regulate mucin gene expression as well. EGF family ligands include EGF, TGF-α, amphiregulin- and heparin-bound EGF, betacellulin, epiregulin, epigen, and the neuregulins. They bind to a class of receptors called ErbB receptors, ErbB1–4; the ErbB1 receptor is also known by its more common name, EGFR (55). EGFR signaling regulates MUC2 expression. Treatment of NCI-H292 cells with either EGF or TGF-α can stimulate MUC2 transcription. This transcriptional up-regulation was mediated through the activation of RAS/RAF/ERK pathways. Promoter analysis revealed that the EGFR-responsive element was localized to −2627/−2097 of the MUC2 promoter and that the putative transcription factor responsible for activation was Sp1 (31) (Figure 1). As discussed below, this EGFR/RAS/RAF/ERK/Sp1 pathway is also important for MUC5AC regulation.
All-trans RA (ATRA or RA) is another growth factor that is important in regulating mucin gene expression. There are reports of MUC2 induction by RA in human cultured airway epithelial cells, and RAR-α is apparently the primary mediator of this effect (13, 56). Interestingly, RA treatment of primate airway epithelial cells appears to inhibit MUC2 expression, suggesting that some species differences may exist as to RA’s effect on mucin (56, 57).
MUC5AC EXPRESSION AND REGULATION
MUC5AC is located on chromosome 11p15.5, in between MUC2 and MUC5B. Its cDNA is approximately 17.5 kb in length, with a central large exon, similar to MUC5B, that is approximately 10.5 kb long. Motifs flanking this central exon are cysteine-rich, von Willabrand–like domains (58–60). MUC5AC is expressed primarily on surface mucous cells in the stomach, lungs, endocervix, and conjuntiva (61). In the lungs, goblet cell hyperplasia/metaplasia of surface epithelial cells in inflammatory diseases correlate closely with increases in the expression of MUC5AC messages (62, 63). Although surface goblet cells express other mucin genes (14, 64–66), MUC5AC is probably this cell type’s most specific marker. Numerous reports have shown that inflammatory cytokines (e.g., TNF-α, IL-1β, IL-4, IL-6, IL-9, IL-13, IL-17), bacterial products (e.g., LPS, LTA, peptidoglycans, flagellin), growth factors (e.g., EGF, TGF-α, RA, thyroid hormones), proteases [e.g., neutrophil elastase (NE)], environmental pollutants (e.g., cigarette smoke extracts, acrolein, ozone), and viral mediators (e.g., respiratory syncytial virus, rhinovirus) can prominently stimulate MUC5AC expression either in vitro or in vivo on airway epithelial cells (Figure 1, middle).
MUC5AC Expression Regulated by MAP Kinases
Like MUC2, MUC5AC is upregulated by many cytokine mediators and bacterial products. TNF-α and IL-1β induce MUC5AC in various airway epithelial cell lines (31, 48, 67–69). One mechanism for TNF-α’s induction of MUC5AC involves signaling through MAP kinases ERK and p38 downstream to produce cyclic AMP–responsive element binding protein (CREB) activation. A CREB-responsive cis element on the MUC5AC promoter is present and is localized to the −776 to −929 region (68). Phorbol esters such as phorbol 12-myristate 13 acetate (PMA) can also activate MUC5AC expression through ERK pathways, but the downstream effects of PMA involve EGFR and Sp1 rather than CREB (18). A bacterial cytoplasmic protein from NTHI called P6 can activate MUC5AC expression. This mechanism appears to involve the binding of P6 to TLR2, which subsequently passes on the signal through the MyD88/TRAF6 pathway to p38 and NF-κB activation (70).
Regulation of MUC5AC Expression by Nuclear Factor-κB
Examination of the promoter region of MUC5AC reveals several putative NF-κB binding sites (58). Studies have confirmed that NF-κB is a major transcription factor that regulates MUC5AC gene expression. Some of the signal transduction events from MAP kinases lead downstream to NF-κB activation. The MyD88 pathway induced by NTHI leads to NF-κB-mediated transcription of MUC5AC (48). Pseudomonas extracts can induce MUC2 and MUC5AC, likely by NF-κB mechanisms (58). Although the investigators (58) did not definitively link the pseudomonas induction of MUC5AC to NF-κB, the presence of NF-κB sites on MUC5AC promoter strongly suggests that they are the putative pseudomonas-responsive elements. The TNF-α induction of MUC5AC noted above leads to CREB-mediated transcriptional activation of MUC5AC (68). However, another group has shown that TNF-α can also induce MUC5AC expression in NCI-H292 cells through NF-κB. Adenoviral delivery of an IKKB mutant could block MUC5AC’s stimulation by TNF-α, both in vitro in human cells as well as in vivo in mice (41).
Several groups have reported that NE can induce MUC5AC in airway epithelial cells (71–73). One group found that the mechanism involved activation of the ERK and NF-κB (71), although other groups reported different mechanisms such as mRNA stability and EGFR activation (72, 73). The differences in signaling and mechanistic steps are difficult to sort out because different cell systems and treatments were used. However, there are compelling data that reactive oxygen species (ROS) formation is required upstream for NE-mediated MUC5AC expression (74–76). The generation of ROS by NE appears to be mediated by different types of NADPH oxidases (77, 78) and may also require PKC activation (74). This may mean that ROS act upstream of NF-κB in MUC5AC expression because ROS activation of NF-κB in airway epithelial and smooth muscle cells has been observed (79–81).
Transcriptional Activation of MUC5AC Through Epidermal Growth Factor Receptor Signaling Pathways
As noted above, the ErbB family receptors play important roles in growth, repair, and tumorigenesis in epithelial cells. Signaling through EGFR (the ErbB1 receptor) has long been known to play an important role in modulating the expression of mucin genes (82, 83). Many stimuli, including cigarette smoke, NE, phorbol esters, eosinophil products, bacterial products, and human airway trypsin-like protease, can upregulate MUC5AC through EGFR (18, 73, 74, 84–86). EGFR is expressed on airway epithelial cells, and in many cases its ligands, such as TGF-α and amphiregulin, are also present on the membrane in their inactive forms (55). Stimuli work by cleaving these ligands to their active forms, which then act in an autocrine or a paracrine manner to upregulate MUC5AC (18, 73, 74). As noted for NE, ROS are an important mechanism for stimulating MUC5AC. Cigarette smoke is also an important environmental stimulus for generating ROS (87). One group has shown that ROS generation from NE and cigarette smoke leads to EGFR activation, through the activation of TNF-α-converting enzyme (TACE), to cleave pro-TGF-α from the membrane into the active soluble form of TGF-α (74, 88). Because dual oxidases are expressed in airway epithelial cell surface, dual oxidase-1 (Duox-1) may be involved in TACE activation (78). Other enzymes besides TACE that can also cleave pro-EGFR ligands to activate MUC5AC include MMPs, dis-integrin and metalloproteinase domain proteins (ADAMs), and tissue kallikrein (TK) (89, 90). Downstream of EGFR, the mechanism that leads to MUC5AC expression can involve ERK activation with subsequent binding of transcription factors Sp1 (18) and Fra-2 to the MUC5AC promoter (91). However, ROS generated by cigarette smoke can also induce MUC5AC by a non-EGFR, non-ERK pathway that involves JNK activation and subsequent binding of the JunD transcription factor to the promoter (91).
TH2 Cytokines’ Effects on MUC5AC: The Importance of Injury and Repair Mechanisms
Among the inflammatory cytokines that have been associated with mucous cell hyperplasia, the TH2 cytokines, such as IL-4, IL-9, and IL-13, have been most frequently noted to induce MUC5AC gene expression both in vitro and in vivo (6). This observation is consistent with the close association of mucous cell hyperplasia/metaplasia seen in allergic airway diseases such as asthma. IL-9 induces MUC5AC/Muc5ac both in vitro and in vivo (92–96), although few data exist as to the intracellular mechanisms of its action. IL-9’s effect on MUC5AC may be mediated through IL-13 (95, 96), although other studies dispute the role of IL-13 (92, 94). IL-9’s ability to directly induce MUC5AC gene expression on airway epithelial cells is controversial (92, 97). Under one scenario, IL-9’s ability to directly induce mucous cells in culture may require a coexisting degree of injury and repair (98). IL-9 may have some role in inducing MUC5AC expression, although its requirement in mucous cell differentiation in the mouse is not absolute (99).
IL-4 and IL-13 are the TH2 cytokines most frequently associated with MUC5AC/Muc5ac induction. The IL-4 and IL-13 receptors are heterodimers and share a common alpha receptor (IL-4Rα), which may explain why much of their cellular effector functions are similar. After receptor binding, these cytokines’ signals are transduced through the Janus kinase (JAK) pathways that ultimately lead to STAT6 nuclear translocation and the transcriptional regulation of various genes by STAT6. IL-4 and IL-13 stimulate MUC5AC expression both in vitro and in vivo (50, 52, 100). Although the predominant signaling for both these two cytokines may be through IL-4Rα, the association between IL-4’s MUC5AC induction is less clear than for IL-13. IL-4’s effect on mucous cell metaplasia/hyperplasia and MUC5AC/Muc5ac has been predominantly observed in vivo (50, 52), whereas in vitro experiments with defined IL-4 administration to airway epithelial cells have produced conflicting results (97, 100–103). Hence, IL-4’s role in inducing mucous cell differentiation may be due to indirect mechanisms from the inflammatory cells and mediators it recruits to the lung rather than a direct effect on airway epithelial cells.
IL-13 has a greater association with mucous cell differentiation in asthma. Although there are also conflicting data, there is more abundant and stronger evidence for IL-13’s role in directly inducing airway epithelial cells to upregulate MUC5AC/Muc5ac and differentiate into mucous cells (97, 103, 104). However, IL-13’s ability to induce mucous cell formation and MUC5AC in culture appears to require a much more prolonged exposure (4 days to 2 weeks) than is typical for most cytokine responses (100, 101). Because IL-13’s ability to induce STAT6 phosphorylation occurs very rapidly, with nuclear translocation seen within 30 min, direct transcriptional activation of MUC5AC by STAT6 seems unlikely (105). Examination of the promoter region of MUC5AC in mouse and humans also does not reveal clear-cut putative STAT6 binding sites, and other putative transcription factor binding sites such as SMAD4 and HIF-1α are more likely the direct inducers of Muc5ac transcription (106). Despite its importance in inducing MUC5AC expression, the EGFR pathway does not appear to be involved in IL-13’s effects (107). STAT6 likely is an upstream signal for IL-13 that induces other transcriptional signals and/or the release of other soluble factors that then act downstream on MUC5AC expression and mucous cell differentiation.
One reason why IL-13 requires a prolonged exposure time to affect MUC5AC expression is because of coexisting injury and repair of the airway. Indeed, epithelial denudation and sloughing have long been observed in the respiratory tracts of asthmatic patients (108). IL-13 induces proliferative response in various airway epithelial-type cells, although it does not directly damage epithelial cells, and it is not clear whether this proliferation is related to mucous cell formation (103, 104). However, recent evidence suggests that other airway epithelial cells can transdifferentiate (i.e., exhibit metaplasia) into mucous cells (109, 110). Allergen-mediated inflammation appears to recruit Clara cells to differentiate into mucous cells, although it is not clear whether IL-13 is the agent involved (109). It is also unclear if such a finding in mouse airways can be translated to humans.
Ciliated cells, long thought to be terminally differentiated, can also transdifferentiate to repopulate the airway epithelium during injury (111). One recent report, using a Sendai virus infection model in mouse, suggested how IL-13, EGFR, and injury and repair could work in concert. The Sendai virus in this model causes significant injury to the airways of mice so that a repair response is triggered. In the C57B6 strain of mice, this injury leads to persistent goblet cell metaplasia/hyperplasia along with IL-13 upregulation. The investigators noted that this injury leads to hyperplasia of the ciliated cells that was dependent on EGFR and PI3K signaling. However, this ciliated hyperplasia could not lead to goblet cells directly; rather, IL-13 was needed to induce these ciliated cells to trans-differentiate into goblet cells (110).
Negative Regulators of MUC5AC Expression and of Mucous Cell Differentiation
Fewer data have accumulated on the mechanisms whereby MUC5AC and mucous cell differentiation can be downregulated. Some anti-inflammatory pharmaceutical agents have been shown to inhibit MUC5AC expression. Macrolide antibiotics, felt to have some intrinsic anti-inflammatory effects independent of its bactericidal actions, can inhibit MUC5AC expression (112). Glucocorticoids, a mainstay pharmacological agent to treat asthma and COPD, can inhibit MUC5AC expression by transcriptional repression (113). A transcription factor named Foxa2 (HNF-3β) appears to repress the formation of mucous cells and MUC5AC expression. Conditional knockout of the Foxa2 gene in mice leads to impaired alveolarization and goblet cell hyperplasia. Mucous cell differentiation and upregulation of Muc5ac lead to the downregulation of Foxa2 (114). IL-13 and EGFR signaling can also downregulate Foxa2. Some evidence indicates that Foxa2 can promote Clara cell formation (115). These data are consistent with the above data regarding the ability of Clara cells to transdifferentiate into mucous cells, and Foxa2 may be the key transcription factor that modulates this process.
MUC5B EXPRESSION AND REGULATION
MUC5B is located next to MUC5AC on the chromosome 11. The gene structure reveals that MUC5B is approximately 39 kb long, with 49 exons, and produces a polypeptide estimated to be approximately 5701 amino acids. The central large exon region is 10,713 bp long and includes the entire tandem repeat region. In intron 36, a variable number of tandem repeats (VNTR) polymorphism is noted among different individuals, but so far no linkage to specific genetic disorders has been identified (116). In the 5′ region upstream of the transcriptional start site, putative binding sites for AP-1, Sp1, NF-κB, and other transcription factors exist (116). Less is known about the mechanisms of MUC5B regulation because fewer studies of MUC5B have been performed than for MUC5AC.
In adult tissues, MUC5B is localized almost exclusively to submucosal glands in the airways. In fetal development, however, MUC5B is coexpressed with MUC5AC on surface epithelial cells from week 13 onward. At approximately week 23, its expression on the surface is reduced, and its expression in the glands is more predominant (14). When primary airway epithelial cells are plated in cultures and mucociliary differentiation is initiated in air-liquid interface and RA treatment, upregulation and coexpression of both MUC5AC and MUC5B occur, suggesting that cultured cells show some similarity to surface airway cells during fetal development (13). In addition, pathological examination of certain human lung diseases such as emphysema and usual interstitial pneumonia has shown that MUC5B messages can be upregulated in surface airway epithelial cells in association with mucous cell hyperplasia/metaplasia (116). In mouse models of asthma, airway epithelial cells have also shown upregulation of Muc5b in association with mucous cell hyperplasia (64). Cumulatively, these data suggest that, although MUC5AC/Muc5ac is the predominant marker for surface goblet cells, MUC5B/Muc5b may also be upregulated and be a disease marker in certain circumstances. This coexpression of MUC5B and MUC5AC in surface goblet cells is similar to that seen in the fetal lung (14). This suggests that injury may induce some type of progenitor cell proliferation or transdifferention of differentiated cells that mimics the behavior of airway epithelial cells in fetal lung development.
Upregulation of MUC5B Expression in Airway Epithelial Cells
Because MUC5B is upregulated in airway epithelial cells in inflammatory lung diseases, it is natural to suspect that inflammatory cytokines stimulate MUC5B expression as they do for MUC5AC. In a study on primary human airway epithelial cultures, a panel of 20 cytokines (IL-1α, IL-1β, IL-2 to -18, and TNF-α) revealed that IL-6 and IL-17 were the only cytokines able to upregulate MUC5B and MUC5AC (97). Other agents that can upregulate MUC5B expression in airway epithelial cultures include RA, rhinovirus 14 infection, alpha defensins (human neutrophil peptides 1–3), and PMA (19, 118, 119). One report showed that IL-13 could upregulate MUC5B, although other studies have shown no effect (97, 120).
In vivo, upregulation of MUC5B/Muc5b could be seen on surface airway goblet cells in a mouse model of asthma as well as in human airway sections from patients with inflammatory lung disease (64, 116). MUC5B is a major component of secreted mucus and, in some reports, was found in concentrations approximately ten times that of MUC5AC (11, 12). In one report, patients with a rare pulmonary disorder known as diffuse panbronchiolitis—a progressive airway disorder found mostly in Asian countries (particularly Japan) and characterized by airway narrowing, mucous hyperplasia, and distal airspace enlargement—showed high levels of MUC5B expression on the surface goblet cells (121). Examination of the genomic DNA of patients with this disease revealed a high correlation between specific polymorphisms in the promoter region of MUC5B and MUC5B expression and severity of the disease (121). The etiology of this disease is not clear, but associations with specific bacterias such as P. aeruginosa have been noted clinically, and the disease is amenable when treated with macrolide antibiotics such as erythromycin. CF is also associated with significant goblet cell hyperplasia, and P. aeruginosa chronically colonizes the airways and causes clinical exacerbations. However, the relationship between Pseudomonas and the mucous cell hyperplasia/metaplasia between these very different pulmonary disorders is not clear.
Signal Transduction Pathways in the Regulation of MUC5B Expression
The upregulation of MUC5B by IL-17A on primary airway epithelial cultures appeared to be partly indirect and due to the release of IL-6 and autocrine stimulation. IL-6’s upregulation of MUC5B appears to depend on the ERK MAP kinase pathway because the inhibitor U0126 could partly block its stimulation (97). PMA is capable of inducing MUC5B expression. This chemical ligand is an activator of PKC. A study showed that activation of MUC5B by PMA required the p38 and JNK pathways, which led to transcriptional activation of the MUC5B promoter by Sp1 (19) (Figure 1, bottom). This mechanism is somewhat different from the PMA induction of MUC5AC, which involved the EGFR-mediated pathway, which also led to Sp1 activation of gene transcription (18). The nucleotide UTP can induce both MUC5B secretion as well as MUC5B expression. UTP stimulation of MUC5B expression depends on MAP kinase pathways because inhibitors to ERK could block UTP-dependent induction. Transduction of signals from the surface P2Y receptors through a G protein–coupled intermediate receptor through ERK pathways is thought to lead to MUC5B upregulation by UTP (123).
REGULATION OF OTHER MUCIN GENES IN THE LUNGS
MUC1, a membrane-bound mucin with a cytoplasmic tail, was the first mucin molecule to be cloned. It is expressed in secretory epithelial cells of the airways, GI tract, and female reproductive tract and is associated with goblet cells (124). Its function is not well defined yet, but it appears to play a role in antiadhesion of tumor cells or bacteria (124, 125). It can mediate signal transduction responses through Ras, β-catenin, and p53. Signaling through these pathways may affect the apoptotic state of the cell, which may explain the greater expression of MUC1 in neoplastic cells (126). MUC1’s expression in airway epithelial cells can be induced by NE and CpG DNA from bacteria (127, 128). Otherwise, it is regulated by inflammatory mediators in noncancerous airway cells.
MUC3 is primarily an intestinal mucin, and it is not expressed in the fetal or normal adult lung (28, 29). However, some studies have shown that MUC3 is expressed at detectable levels in lung adenocarcinomas and bronchoalveolar carcinomas (129). Airway cultures of primary nasal and airway epithelial cells appear to upregulate MUC3 during differentiation (130).
MUC4 is another membrane-bound mucin. Its expression in airway epithelial cells appears to be nonspecifically localized to basal, ciliated, and goblet cells (29). Mucociliary differentiation of cultured primary airway epithelial cells increases MUC4 expression (130). IL-4, IL-9, and NE have also been reported to increase expression of MUC4 in airway epithelial cells. In the case of the IL-4 and IL-9, transcriptional activation was believed to be the mechanism (131, 132), whereas for NE increase in mRNA stability occurred (133).
MUC6, a secreted mucin, is similar to MUC3 in that it is expressed primarily in intestinal cells, except in some cases in which it is upregulated in lung carcinoma cells (129). MUC7 is expressed in the lungs primarily in the serous cells of the submucosal glands (134). Its expression increases in response to RA in culture, and it can be upregulated by inflammatory cytokines (IL-1β, TNF-α, IL-4, and IL-13), EGF, and bacterial products such as LPS (135). Analysis of the promoter region of MUC7 revealed that a region −138/+30 containing AP-1 and NF-κB sites is critical to basal expression as well as the responsiveness of MUC7 to TNF-α. Mutagenesis experiments further elucidated that the AP-1 sites are important for MUC7’s basal expression whereas NF-κB is important for the responsiveness of MUC7 to TNF-α (136). MUC8 is a secreted mucin that is also primarily localized to the submucosal glands of the trachea (137). Its expression, however, does not appear to be related to differentiation (130). Although MUC11 and MUC13 have been localized to the lung, few data exist regarding their regulation. MUC19 is a recently identified secreted gel-forming mucin. Its expression is localized mainly to the mucous cells of the salivary glands and submucosal glands of the trachea (138). The extent to which MUC19 secretion is significant to airway mucus rheology remains to be defined.
EPIGENETIC MECHANISMS: A RELATIVELY UNEXPLORED AREA OF GENE REGULATION IN MUCIN BIOLOGY
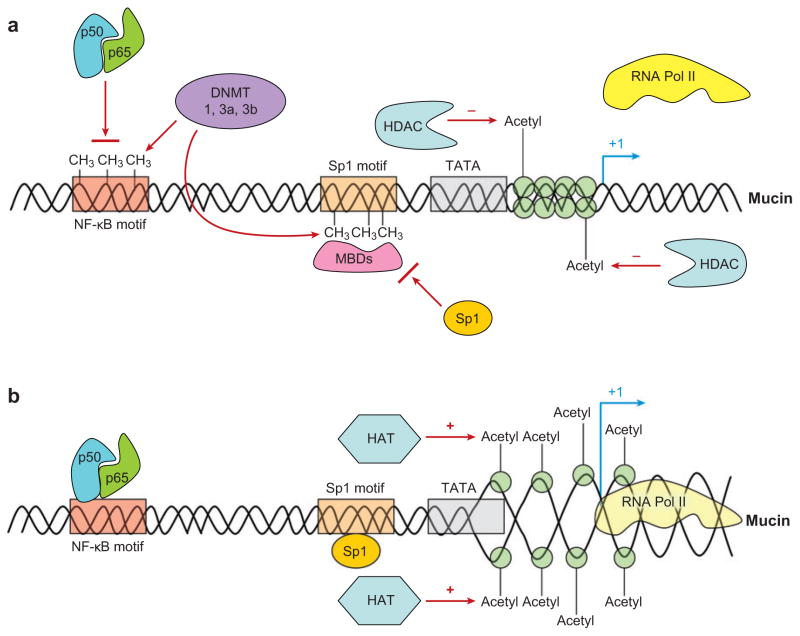
Beyond the typical transcriptional and post-transcriptional processes reviewed here, another mode whereby mucin genes may be regulated is through epigenetic mechanisms (139). Epigenetics is defined as heritable changes in gene function that are not associated with actual changes in genomic DNA sequences. In practical terms, it is usually used to describe the turning on and off of genes due to DNA methylation, histone modifications, and microRNA silencing. DNA methylation involves the addition of a methyl group to the cytosine nucleotide of cytosine-guanine (CG) dinucleotide sequences in the genomic DNA (Figure 2). This methylation tends to inhibit the binding of transcription factors and to turn off gene expression. In addition, methylation can also repress transcription indirectly by promoting the binding of a family of proteins called methyl binding proteins (MBDs), which act as repressors of transcription (140). A clustering of CG dinucleotide sequences in a region of DNA is referred to as a CpG island. These CpG islands tend to play important regulatory roles when they occur in the promoter regions of genes. The chemical addition of a methyl group to the DNA backbone is usually catalyzed by enzymes called DNA methyltransferases or methylases. One example of this type of methylase is the enzyme called maintenance methylase enzyme or DNA methyltransferase 1 (DNMT1). This enzyme acts during DNA replication in cell division to reproduce the methylation imprinting of the old DNA strand on the newly synthesized DNA strand. Thus, this methylase only replicates the existing methylation and does not add de novo methylation to previously unmethylated DNA elements.
Figure 2.
Theoretical mechanism of how epigenetic mechanisms can regulate mucin gene transcription. (a) Mucin gene expression is low. DNA methylation decreases the binding of transcription factors either directly or through methyl binding proteins (MBDs). DNA methyltransferases (DNMT) catalyze the transfer of methyl groups de novo to CpG sites. Nucleosome structure is relatively closed by histones deacetylated by histone deacetylases (HDAC). (b) Mucin gene expression is high. Loss of DNA methylation allows transcription factors to bind to their putative binding sites. Acetylation of histones by histone acetylases (HAT) leads to the opening of nucleosomes and increased transcription. NF-κB, nuclear factor-κB; RNA Pol II, RNA polymerase II.
Other DNA methylases include the DNMT2, -3a, and -3b. These DNA methylases, unlike maintenance methylase, can de novo methylate previously unmethylated DNA regions. How these methyltransferases bind with specificity to methylate some regions of DNA and not others is not entirely clear. Also, although methylation and demethylation of genes do occur, specific DNA demethylating enzymes have not been conclusively identified. Biochemically, there is evidence that DNA demethylating enzymes exist in nuclear extracts. Some reports suggest that the methyl binding protein MBD2 has demethylating enzymatic activities, but other studies have disputed this contention (140a, 140b). In either case, methylation and demethylation in DNA promoter regions around specific genes can turn on and off gene expression, especially affecting the overall level of expression. Changes in the total levels of DNA methyltransferases are believed to alter the methylation levels of specific genes (Figure 2). For example, knockdown experiments of DNA methyltransferases with siRNA and inhibition with the inhibitor 5′-azacytidine can decrease the methylation of certain genes, leading to their increased transcriptional activity.
Another epigenetic mechanism involves histone modification (Figure 2). Genomic DNA is wrapped around histones that help to package and compact the extremely long DNA strand into the small volume of the nucleus. Covalent modification of histones can to some degree open or close this packaging around specific regions of DNA and alter the access of transcription factors and RNA polymerase complexes to specific genes or loci. These covalent modifications include acetylation, methylation, and phosphorylation. Acetylation of histones typically unwinds the packaging and tends to turn on the genes around that area, whereas deacetylation closes the genes and turns it off (Figure 2). Acetylation and deacetylation are catalyzed by histone acetylases (HATs) and histone deacetylases (HDACs), respectively. In addition, HATs and HDACs tend to bind coactivator or corepressor proteins and recruit them to specific regions of DNA where they bind to other transcription factors to promote and repress transcription, respectively.
Although studies of epigenetic mechanisms in mucins have been few and most have been limited to MUC2 expression, there are compelling reasons why they may be an important means of control for this family of genes. First, mucin genes clearly have cell and tissue specificity, and epigenetic mechanisms are frequently involved in the tissue-specific expression of other genes. Second, the use of epigenetic mechanisms can add an extra layer of control to help specific cells respond to the environment. For example, cytokine mediators that can activate NF-κB or MAP kinase pathways can potentially cause widespread and unwanted activation of mucin genes because these signaling pathways are active and ubiquitous in many different cell types. However, cell specificity of response can be achieved if cells primed to have mucin genes respond, e.g., airway epithelial secretory cell types, have their promoter regions relatively open by unmethylated CpGs and acetylated histones, whereas cells that are not primed to have mucin genes respond (e.g., inflammatory cells, mesenchymal cells, endothelial cells) have them turned off by methylated CpGs and deacetylated histones.
MUC2: An Example of an Epigenetically Controlled Mucin Gene
MUC2 is an example of a mucin with tissue-specific expression in the colon, small intestine, and respiratory tract. Most studies show little or no expression of MUC2 in the normal gastric mucosa. However, in carcinogenesis, there are alterations in the mucin expression pattern, with a decrease in MUC2 expression associated with de novo colon carcinogenesis (141). Studies of MUC2 in the GI tract and its expression during carcinogenesis reveal that promoter DNA methylation is a frequent mechanism that controls MUC2 expression (142, 143). Previous data indicate that the sequence of the MUC2 promoter region approximately 343 bp upstream of the transcription start site contains elements regulating MUC2 expression (21, 22). Recent results demonstrate that the methylation level of this region, comprising nine CpG sites, is related to MUC2 expression in colon cancer cells in vitro (143) and pancreatic cancer in vivo (142). The mechanisms underlying the absence of MUC2 expression in normal stomach and de novo MUC2 expression in intestinal metaplasia and gastric carcinoma have not been studied. For cases of gastric carcinogenesis, comparison was made in the levels of methylation in nine CpG sites in the −289 and +1 region of the MUC2 promoter. Methylation of this CpG region showed a significant correlation with low expression of MUC2 in normal gastric mucosa, whereas demethylation of this region was associated with high MUC2 expression in mucinous gastric carcinomas (144). Several notable CpG sites in the wider area of the MUC2 promoter region, including those in AP2 and Sp1 binding motifs, showed obvious differences in methylation level between PANC1, BxPC3, and normal colonic crypt cells (145). Similar observations showing that site-specific methylation can downregulate MUC2 gene expression have been reported in pancreatic cancer cells (142, 146). These specific sites of methylation changes in the promoters may be a road map to the critically important regulatory regions of MUC2.
Few studies have looked at the role of histone modifications in MUC2 expression. One study examined the histone deacetylase inhibitor trichostatin A (TSA) on MUC2 expression in pancreatic cancer cell lines. Treatment with TSA resulted in the upregulation of MUC2 expression, suggesting that histone acetylation can to some degree turn on the MUC2 gene (146). In the future, further studies in other epigenetic mechanisms, including histone modification and chromatin modification in MUC2 inactivation, are expected.
Other Mucin Genes and Epigenetic Mechanisms
There have been very few studies of epigenetic mechanisms in other mucin genes. MUC5B is suspected to be regulated to some degree by promoter methylation. Treatment with 5′-azacytidine can increase its expression in the KATO III and AGS gastric carcinoma cell lines. Mapping of the 5′ promoter region of MUC5B revealed, between −2044 and +3, nine putative CpG sites, some of which were methylated. Methylations of these CpGs were confirmed and were suspected to be responsible for the repression of MUC5B in these cell lines (147). For MUC5AC, a study in a pancreatic adenocarcinoma cell line showed that methylation of its 5′ promoter region showed correlation of approximately 77% with the degree of MUC5AC expression. However, when 5′-azacytidine was given, no increase of MUC5AC occurred, making the role of methylation in this cell line unclear (148). MUC1 has a CpG island in a 60-bp tandem repeat region. Comparison of the methylation of this region between breast cancer cells, noncancerous breast cells, and other non-breast tissues revealed a similar correlation between lower methylation and higher expression of the MUC1 gene (149).
CONCLUSION
The accumulated data suggest that secreted gel-forming mucins are regulated by specific pathways in surface airway epithelial cells. MAP kinase pathways, EGFR, and RA receptors signal downstream to Sp1, NF-κB, and AP-1 transcription factors that then alter the expression of various mucin genes. As our knowledge of mucins increase over time, it is becoming more and more evident that molecules of this class have an extremely complicated biology. Because of the large size and complex carbohydrate structures of mucins, mucin studies at the protein level will continue to be challenging in the future. As such, studies at the molecular genetic level on transcriptional, posttranscriptional processing, and epigenetic mechanisms should yield a greater understanding of how mucin genes are regulated in a cell- and tissue-specific manner and how their aberrant expression occurs in inflammatory lung diseases and carcinogenesis.
Acknowledgments
This review is supported in part by a California TRDRP grant (15KT-0135 to P.T.) and NIH RO1 grants (HL077902 and HL077315 to R.W.). A.L. is supported by a T32 training grant (T32 HL07013).
LITERATURE CITED
- 1.Murray JF. Text of Respiratory Medicine. 3 Philadelphia: Saunders; 2000. [Google Scholar]
- 2.Kuyper LM, Pare PD, Hogg JC, Lambert RK, Ionescu D, et al. Characterization of airway plugging in fatal asthma. Am J Med. 2003;115(1):6–11. doi: 10.1016/s0002-9343(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 3.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(26):2645–53. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 3a.Groneberg DA, Eynott PR, Lim S, Oates T, Wu R, et al. Expression of respiratory mucins in fatal status asthmaticus and mild asthma. Histopathology. 2002;40(4):367–73. doi: 10.1046/j.1365-2559.2002.01378.x. [DOI] [PubMed] [Google Scholar]
- 4.Vestbo J, Knudsen KM, Rasmussen FV. The value of mucus hypersecretion as a predictor of mortality and hospitalization. An 11-year register based follow-up study of a random population sample of 876 men. Respir Med. 1989;83(3):207–11. doi: 10.1016/s0954-6111(89)80033-2. [DOI] [PubMed] [Google Scholar]
- 5.Lange P, Nyboe J, Appleyard M, Jensen G, Schnohr P. Relation of ventilatory impairment and of chronic mucus hypersecretion to mortality from obstructive lung disease and from all causes. Thorax. 1990;45(8):579–85. doi: 10.1136/thx.45.8.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergero Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev. 2006;86(1):245–78. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 7.Caramori G, Di Gregorio C, Carlstedt I, Casolari P, Guzzinati I, et al. Mucin expression in peripheral airways of patients with chronic obstructive pulmonary disease. Histopathology. 2004;45(5):477–84. doi: 10.1111/j.1365-2559.2004.01952.x. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Vilar J. Mucin granule intraluminal organization. Am J Respir Cell Mol Biol. 2007;36(2):183–90. doi: 10.1165/rcmb.2006-0291TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergeron C, Boulet LP. Structural changes in airway diseases: characteristics, mechanisms, consequences, and pharmacologic modulation. Chest. 2006;129(4):1068–87. doi: 10.1378/chest.129.4.1068. [DOI] [PubMed] [Google Scholar]
- 10.Thornton DJ, Gray T, Nettesheim P, Howard M, Koo JS, Sheehan JK. Characterization of mucins from cultured normal human tracheobronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2000;278(6):L1118–28. doi: 10.1152/ajplung.2000.278.6.L1118. [DOI] [PubMed] [Google Scholar]
- 11.Thornton DJ, Carlstedt I, Howard M, Devine PL, Price MR, Sheehan JK. Respiratory mucins: identification of core proteins and glycoforms. Biochem J. 1996;316(Pt. 3):967–75. doi: 10.1042/bj3160967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thornton DJ, Howard M, Khan N, Sheehan JK. Identification of two glycoforms of the MUC5B mucin in human respiratory mucus. Evidence for a cysteine-rich sequence repeated within the molecule. J Biol Chem. 1997;272(14):9561–66. doi: 10.1074/jbc.272.14.9561. [DOI] [PubMed] [Google Scholar]
- 13.Koo JS, Yoon JH, Gray T, Norford D, Jetten AM, Nettesheim P. Restoration of the mucous phenotype by retinoic acid in retinoid-deficient human bronchial cell cultures: changes in mucin gene expression. Am J Respir Cell Mol Biol. 1999;20(1):43–52. doi: 10.1165/ajrcmb.20.1.3310. [DOI] [PubMed] [Google Scholar]
- 14.Buisine MP, Devisme L, Copin MC, Durand-Reville M, Gosselin B, et al. Developmental mucin gene expression in the human respiratory tract. Am J Respir Cell Mol Biol. 1999;20(2):209–18. doi: 10.1165/ajrcmb.20.2.3259. [DOI] [PubMed] [Google Scholar]
- 15.Cardoso WV, Williams MC, Mitsialis SA, Joyce-Brady M, Rishi AK, Brody JS. Retinoic acid induces changes in the pattern of airway branching and alters epithelial cell differentiation in the developing lung in vitro. Am J Respir Cell Mol Biol. 1995;12(5):464–76. doi: 10.1165/ajrcmb.12.5.7742011. [DOI] [PubMed] [Google Scholar]
- 16.Van Winkle LS, Isaac JM, Plopper CG. Distribution of epidermal growth factor receptor and ligands during bronchiolar epithelial repair from naphthalene-induced Clara cell injury in the mouse. Am J Pathol. 1997;151(2):443–59. [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HW, Ahn DH, Crawley SC, Li JD, Gum JR, Jr, et al. Phorbol 12-myristate 13-acetate up-regulates the transcription of MUC2 intestinal mucin via Ras, ERK, and NF-κB. J Biol Chem. 2002;277(36):32624–31. doi: 10.1074/jbc.M200353200. [DOI] [PubMed] [Google Scholar]
- 18.Hewson CA, Edbrooke MR, Johnston SL. PMA induces the MUC5AC respiratory mucin in human bronchial epithelial cells, via PKC, EGF/TGF-α, Ras/Raf, MEK, ERK and Sp1-dependent mechanisms. J Mol Biol. 2004;344(3):683–95. doi: 10.1016/j.jmb.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 19.Yuan-Chen Wu D, Wu R, Reddy SP, Lee YC, Chang MM. Distinctive epidermal growth factor receptor/extracellular regulated kinase-independent and -dependent signaling pathways in the induction of airway mucin 5B and mucin 5AC expression by phorbol 12-myristate 13-acetate. Am J Pathol. 2007;170(1):20–32. doi: 10.2353/ajpath.2007.060452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rousseau K, Byrne C, Kim YS, Gum JR, Swallow DM, Toribara NW. The complete genomic organization of the human MUC6 and MUC2 mucin genes. Genomics. 2004;83(5):936–39. doi: 10.1016/j.ygeno.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Li JD, Dohrman AF, Gallup MG, Miyata S, Gum JR, et al. Transcriptional activation of mucin by Pseudomonas aeruginosa lipopolysaccharide in the pathogenesis of cystic fibrosis lung disease. Proc Natl Acad Sci USA. 1997;94:967–72. doi: 10.1073/pnas.94.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen A, Hutton DA, Pearson JP. The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biol. 1998;30(7):797–801. doi: 10.1016/s1357-2725(98)00028-4. [DOI] [PubMed] [Google Scholar]
- 23.Dohrman A, Tsuda T, Escudier E, Cardone M, Jany B, et al. Distribution of lysozyme and mucin (MUC2 and MUC3) mRNA in human bronchus. Exp Lung Res. 1994;20(4):367–80. doi: 10.3109/01902149409064393. [DOI] [PubMed] [Google Scholar]
- 24.Hovenberg HW, Davies JR, Herrmann A, Linden CJ, Carlstedt I. MUC5AC, but not MUC2, is a prominent mucin in respiratory secretions. Glycoconj J. 1996;13(5):839–47. doi: 10.1007/BF00702348. [DOI] [PubMed] [Google Scholar]
- 25.Davies JR, Svitacheva N, Lannefors L, Kornfalt R, Carlstedt I. Identification of MUC5B, MUC5AC and small amounts of MUC2 mucins in cystic fibrosis airway secretions. Biochem J. 1999;344(Pt. 2):321–30. [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkham S, Sheehan JK, Knight D, Richardson PS, Thornton DJ. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem J. 2002;361(Pt. 3):537–46. doi: 10.1042/0264-6021:3610537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morcillo EJ, Cortijo J. Mucus and MUC in asthma. Curr Opin Pulm Med. 2006;12(1):1–6. doi: 10.1097/01.mcp.0000198064.27586.37. [DOI] [PubMed] [Google Scholar]
- 28.Reid CJ, Gould S, Harris A. Developmental expression of mucin genes in the human respiratory tract. Am J Respir Cell Mol Biol. 1997;17(5):592–98. doi: 10.1165/ajrcmb.17.5.2798. [DOI] [PubMed] [Google Scholar]
- 29.Buisine MP, Devisme L, Copin MC, Durand-Reville M, Gosselin B, et al. Developmental mucin gene expression in the human respiratory tract. Am J Respir Cell Mol Biol. 1999;20(2):209–18. doi: 10.1165/ajrcmb.20.2.3259. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann A, Davies JR, Lindell G, Martensson S, Packer NH, et al. Studies on the “insoluble” glycoprotein complex from human colon. Identification of reduction-insensitive MUC2 oligomers and C-terminal cleavage. J Biol Chem. 1999;274:15828–36. doi: 10.1074/jbc.274.22.15828. [DOI] [PubMed] [Google Scholar]
- 31.Perrais M, Pigny P, Copin MC, Aubert JP, Van Seuningen I. Induction of MUC2 and MUC5AC mucins by factors of the epidermal growth factor (EGF) family is mediated by EGF receptor/Ras/Raf/extracellular signal-regulated kinase cascade and Sp1. J Biol Chem. 2002;277(35):32258–67. doi: 10.1074/jbc.M204862200. [DOI] [PubMed] [Google Scholar]
- 32.Li JD, Feng W, Gallup M, Kim JH, Gum J, et al. Activation of NF-κB via a Src-dependent Ras-MAPK-pp90rsk pathway is required for Pseudomonas aeruginosa-induced mucin overproduction in epithelial cells. Proc Natl Acad Sci USA. 1998;95(10):5718–23. doi: 10.1073/pnas.95.10.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemjabbar H, Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat Med. 2002;8(1):41–46. doi: 10.1038/nm0102-41. [DOI] [PubMed] [Google Scholar]
- 34.McNamara N, Khong A, McKemy D, Caterina M, Boyer J, et al. ATP transduces signals from ASGM1, a glycolipid that functions as a bacterial receptor. Proc Natl Acad Sci USA. 2001;98(16):9086–91. doi: 10.1073/pnas.161290898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sha Q, Truong-Tran AQ, Plitt JR, Beck LA, Schleimer RP. Activation of airway epithelial cells by toll-like receptor agonists. Am J Respir Cell Mol Biol. 2004;31(3):358–64. doi: 10.1165/rcmb.2003-0388OC. [DOI] [PubMed] [Google Scholar]
- 36.Carmody RJ, Chen YH. Nuclear factor-κB: activation and regulation during Toll-like receptor signaling. Cell Mol Immunol. 2007;4(1):31–41. [PubMed] [Google Scholar]
- 37.Diamond G, Legarda D, Ryan LK. The innate immune response of the respiratory epithelium. Immunol Rev. 2000;173:27–38. doi: 10.1034/j.1600-065x.2000.917304.x. [DOI] [PubMed] [Google Scholar]
- 38.Jono H, Shuto T, Xu H, Kai H, Lim DJ, et al. Transforming growth factor-β-Smad signaling pathway cooperates with NF-κB to mediate nontypeable Haemophilus influenzae-induced MUC2 mucin transcription. J Biol Chem. 2002;277(47):45547–57. doi: 10.1074/jbc.M206883200. [DOI] [PubMed] [Google Scholar]
- 39.Chung KF. Inflammatory mediators in chronic obstructive pulmonary disease. Curr Drug Targets Inflamm Allergy. 2005;4(6):619–25. doi: 10.2174/156801005774912806. [DOI] [PubMed] [Google Scholar]
- 40.Gan WQ, Man SFP, Senthilselvan A, et al. Association between chronic obstructive pulmonary disease and systemic inflammation: a systematic review and a metaanalysis. Thorax. 2004;59:574–80. doi: 10.1136/thx.2003.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lora JM, Zhang DM, Liao SM, Burwell T, King AM, et al. Tumor necrosis factor-α triggers mucus production in airway epithelium through an IκB kinase β-dependent mechanism. J Biol Chem. 2005;280(43):36510–17. doi: 10.1074/jbc.M507977200. [DOI] [PubMed] [Google Scholar]
- 42.Lappalainen U, Whitsett JA, Wert SE, Tichelaar JW, Bry K. Interleukin-1β causes pulmonary inflammation, emphysema, and airway remodeling in the adult murine lung. Am J Respir Cell Mol Biol. 2005;32(4):311–18. doi: 10.1165/rcmb.2004-0309OC. [DOI] [PubMed] [Google Scholar]
- 43.Kao CY, Huang F, Chen Y, Thai P, Wachi S, et al. Up-regulation of CC chemokine ligand 20 expression in human airway epithelium by IL-17 through a JAK-independent but MEK/NF-κB-dependent signaling pathway. J Immunol. 2005;175(10):6676–85. doi: 10.4049/jimmunol.175.10.6676. [DOI] [PubMed] [Google Scholar]
- 44.Harper R, Wu K, Chang MM, Yoneda K, Pan R, et al. Activation of nuclear factor-κB transcriptional activity in airway epithelial cells by thioredoxin but not by N-acetyl-cysteine and glutathione. Am J Respir Cell Mol Biol. 2001;25(2):178–85. doi: 10.1165/ajrcmb.25.2.4471. [DOI] [PubMed] [Google Scholar]
- 45.Levine SJ, Larivee P, Logun C, Angus CW, Ognibene FP, Shelhamer JH. Tumor necrosis factor-α induces mucin hypersecretion and MUC-2 gene expression by human airway epithelial cells. Am J Respir Cell Mol Biol. 1995;12(2):196–204. doi: 10.1165/ajrcmb.12.2.7865217. [DOI] [PubMed] [Google Scholar]
- 46.Deleted in proof
- 47.Kim YD, Kwon EJ, Kwon TK, Baek SH, Song SY, Suh JS. Regulation of IL-1β-mediated MUC2 gene in NCI-H292 human airway epithelial cells. Biochem Biophys Res Commun. 2000;274(1):112–16. doi: 10.1006/bbrc.2000.3107. [DOI] [PubMed] [Google Scholar]
- 48.Koo JS, Kim YD, Jetten AM, Belloni P, Nettesheim P. Overexpression of mucin genes induced by interleukin-1β, tumor necrosis factor-α, lipopolysaccharide, and neutrophil elastase is inhibited by a retinoic acid receptor α antagonist. Exp Lung Res. 2002;28(4):315–32. doi: 10.1080/01902140252964393. [DOI] [PubMed] [Google Scholar]
- 49.Kim YD, Jeon JY, Woo HJ, Lee JC, Chung JH, et al. Interleukin-1β induces MUC2 gene expression and mucin secretion via activation of PKC-MEK/ERK, and PI3K in human airway epithelial cells. J Korean Med Sci. 2002;17(6):765–71. doi: 10.3346/jkms.2002.17.6.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dabbagh K, Takeyama K, Lee HM, Ueki IF, Lausier JA, Nadel JA. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J Immunol. 1999;162(10):6233–37. [PubMed] [Google Scholar]
- 51.Iwashita J, Sato Y, Sugaya H, Takahashi N, Sasaki H, Abe T. mRNA of MUC2 is stimulated by IL-4, IL-13 or TNF-α through a mitogen-activated protein kinase pathway in human colon cancer cells. Immunol Cell Biol. 2003;81(4):275–82. doi: 10.1046/j.1440-1711.2003.t01-1-01163.x. [DOI] [PubMed] [Google Scholar]
- 52.Temann UA, Prasad B, Gallup MW, Basbaum C, Ho SB, et al. A novel role for murine IL-4 in vivo: induction of MUC5AC gene expression and mucin hypersecretion. Am J Respir Cell Mol Biol. 1997;16(4):471–78. doi: 10.1165/ajrcmb.16.4.9115759. [DOI] [PubMed] [Google Scholar]
- 53.Guzman K, Randell SH, Nettesheim P. Epidermal growth factor regulates expression of the mucous phenotype of rat tracheal epithelial cells. Biochem Biophys Res Commun. 1995;217(2):412–18. doi: 10.1006/bbrc.1995.2792. [DOI] [PubMed] [Google Scholar]
- 54.Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 1996;14(1):104–12. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 55.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7(7):505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 56.Koo JS, Jetten AM, Belloni P, Yoon JH, Kim YD, Nettesheim P. Role of retinoid receptors in the regulation of mucin gene expression by retinoic acid in human tracheo-bronchial epithelial cells. Biochem J. 1999;338(Pt. 2):351–7. [PMC free article] [PubMed] [Google Scholar]
- 57.An G, Luo G, Wu R. Expression of MUC2 gene is down-regulated by vitamin A at the transcriptional level in vitro in tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10(5):546–51. doi: 10.1165/ajrcmb.10.5.8179918. [DOI] [PubMed] [Google Scholar]
- 58.Li D, Gallup M, Fan N, Szymkowski DE, Basbaum CB. Cloning of the amino-terminal and 5′-flanking region of the human MUC5AC mucin gene and transcriptional up-regulation by bacterial exoproducts. J Biol Chem. 1998;273(12):6812–20. doi: 10.1074/jbc.273.12.6812. [DOI] [PubMed] [Google Scholar]
- 59.Escande F, Aubert JP, Porchet N, Buisine MP. Human mucin gene MUC5AC: organization of its 5′-region and central repetitive region. Biochem J. 2001;358(Pt. 3):763–72. doi: 10.1042/0264-6021:3580763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buisine MP, Desseyn JL, Porchet N, Degand P, Laine A, Aubert JP. Genomic organization of the 3′-region of the human MUC5AC mucin gene: additional evidence for a common ancestral gene for the 11p15.5 mucin gene family. Biochem J. 1998;332 (Pt. 3):729–38. doi: 10.1042/bj3320729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inatomi T, Tisdale AS, Zhan Q, Spurr-Michaud S, Gipson IK. Cloning of rat Muc5AC mucin gene: comparison of its structure and tissue distribution to that of human and mouse homologues. Biochem Biophys Res Commun. 1997;236(3):789–97. doi: 10.1006/bbrc.1997.7051. [DOI] [PubMed] [Google Scholar]
- 62.Hovenberg HW, Davies JR, Carlstedt I. Different mucins are produced by the surface epithelium and the submucosa in human trachea: identification of MUC5AC as a major mucin from the goblet cells. Biochem J. 1996;318(Pt. 1):319–24. doi: 10.1042/bj3180319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuhdi Alimam M, Piazza FM, Selby DM, Letwin N, Huang L, Rose MC. Muc-5/5ac mucin messenger RNA and protein expression is a marker of goblet cell metaplasia in murine airways. Am J Respir Cell Mol Biol. 2000;22(3):253–60. doi: 10.1165/ajrcmb.22.3.3768. [DOI] [PubMed] [Google Scholar]
- 64.Chen Y, Zhao YH, Wu R. In silico cloning of mouse Muc5b gene and upregulation of its expression in mouse asthma model. Am J Respir Crit Care Med. 2001;164(6):1059–66. doi: 10.1164/ajrccm.164.6.2012114. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Zhao YH, Di YP, Wu R. Characterization of human mucin 5B gene expression in airway epithelium and the genomic clone of the amino-terminal and 5′-flanking region. Am J Respir Cell Mol Biol. 2001;25(5):542–53. doi: 10.1165/ajrcmb.25.5.4298. [DOI] [PubMed] [Google Scholar]
- 66.Ordonez CL, Khashayar R, Wong HH, Ferrando R, Wu R, et al. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am J Respir Crit Care Med. 2001;163(2):517–23. doi: 10.1164/ajrccm.163.2.2004039. [DOI] [PubMed] [Google Scholar]
- 67.Borchers MT, Carty MP, Leikauf GD. Regulation of human airway mucins by acrolein and inflammatory mediators. Am J Physiol. 1999;276(4 Pt 1):L549–55. doi: 10.1152/ajplung.1999.276.4.L549. [DOI] [PubMed] [Google Scholar]
- 68.Song KS, Lee WJ, Chung KC, Koo JS, Yang EJ, et al. Interleukin-1β and tumor necrosis factor-α induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J Biol Chem. 2003;278(26):23243–50. doi: 10.1074/jbc.M300096200. [DOI] [PubMed] [Google Scholar]
- 69.Gray T, Nettesheim P, Loftin C, Koo JS, Bonner J, et al. Interleukin-1β-induced mucin production in human airway epithelium is mediated by cyclooxygenase-2, prostaglandin E2 receptors, and cyclic AMP-protein kinase A signaling. Mol Pharmacol. 2004;66(2):337–46. doi: 10.1124/mol.66.2.337. [DOI] [PubMed] [Google Scholar]
- 70.Chen R, Lim JH, Jono H, Gu XX, Kim YS, et al. Nontypeable Haemophilus influenzae lipoprotein P6 induces MUC5AC mucin transcription via TLR2-TAK1-dependent p38 MAPK-AP1 and IKKβ-IκBα-NF-κB signaling pathways. Biochem Biophys Res Commun. 2004;324(3):1087–94. doi: 10.1016/j.bbrc.2004.09.157. [DOI] [PubMed] [Google Scholar]
- 71.Song JS, Cho KS, Yoon HK, Moon HS, Park SH. Neutrophil elastase causes MUC5AC mucin synthesis via EGF receptor, ERK and NF-kB pathways in A549 cells. Korean J Intern Med. 2005;20(4):275–83. doi: 10.3904/kjim.2005.20.4.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voynow JA, Young LR, Wang Y, Horger T, Rose MC, Fischer BM. Neutrophil elastase increases MUC5AC mRNA and protein expression in respiratory epithelial cells. Am J Physiol. 1999;276(5 Pt 1):L835–43. doi: 10.1152/ajplung.1999.276.5.L835. [DOI] [PubMed] [Google Scholar]
- 73.Kohri K, Ueki IF, Nadel JA. Neutrophil elastase induces mucin production by ligand-dependent epidermal growth factor receptor activation. Am J Physiol Lung Cell Mol Physiol. 2002;283(3):L531–40. doi: 10.1152/ajplung.00455.2001. [DOI] [PubMed] [Google Scholar]
- 74.Shao MX, Nadel JA. Neutrophil elastase induces MUC5AC mucin production in human airway epithelial cells via a cascade involving protein kinase C, reactive oxygen species, and TNF-α-converting enzyme. J Immunol. 2005;175(6):4009–16. doi: 10.4049/jimmunol.175.6.4009. [DOI] [PubMed] [Google Scholar]
- 75.Fischer BM, Voynow JA. Neutrophil elastase induces MUC5AC gene expression in airway epithelium via a pathway involving reactive oxygen species. Am J Respir Cell Mol Biol. 2002;26(4):447–52. doi: 10.1165/ajrcmb.26.4.4473. [DOI] [PubMed] [Google Scholar]
- 76.Voynow JA, Fischer BM, Malarkey DE, Burch LH, Wong T, et al. Neutrophil elastase induces mucus cell metaplasia in mouse lung. Am J Physiol Lung Cell Mol Physiol. 2004;287(6):L1293–302. doi: 10.1152/ajplung.00140.2004. [DOI] [PubMed] [Google Scholar]
- 77.Zheng S, Byrd AS, Fischer BM, Grover AR, Ghio AJ, Voynow JA. Regulation of MUC5AC expression by NAD(P)H:quinone oxidoreductase 1. Free Radic Biol Med. 2007;42(9):1398–408. doi: 10.1016/j.freeradbiomed.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shao MX, Nadel JA. Dual oxidase 1-dependent MUC5AC mucin expression in cultured human airway epithelial cells. Proc Natl Acad Sci USA. 2005;102(3):767–72. doi: 10.1073/pnas.0408932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quay JL, Reed W, Samet J, Devlin RB. Air pollution particles induce IL-6 gene expression in human airway epithelial cells via NF-κB activation. Am J Respir Cell Mol Biol. 1998;19(1):98–106. doi: 10.1165/ajrcmb.19.1.3132. [DOI] [PubMed] [Google Scholar]
- 80.Brar SS, Kennedy TP, Sturrock AB, Huecksteadt TP, Quinn MT, et al. NADPH oxidase promotes NF-κB activation and proliferation in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2002;282(4):L782–95. doi: 10.1152/ajplung.00206.2001. [DOI] [PubMed] [Google Scholar]
- 81.Krunkosky TM, Martin LD, Fischer BM, Voynow JA, Adler KB. Effects of TNFα on expression of ICAM-1 in human airway epithelial cells in vitro: oxidant-mediated pathways and transcription factors. Free Radic Biol Med. 2003;35(9):1158–67. doi: 10.1016/s0891-5849(03)00498-2. [DOI] [PubMed] [Google Scholar]
- 82.Takeyama K, Dabbagh K, Lee HM, Agusti C, Lausier JA, et al. Epidermal growth factor system regulates mucin production in airways. Proc Natl Acad Sci USA. 1999;96(6):3081–86. doi: 10.1073/pnas.96.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takeyama K, Fahy JV, Nadel JA. Relationship of epidermal growth factor receptors to goblet cell production in human bronchi. Am J Respir Crit Care Med. 2001;163(2):511–16. doi: 10.1164/ajrccm.163.2.2001038. [DOI] [PubMed] [Google Scholar]
- 84.Takeyama K, Jung B, Shim JJ, Burgel PR, Dao-Pick T, et al. Activation of epidermal growth factor receptors is responsible for mucin synthesis induced by cigarette smoke. Am J Physiol Lung Cell Mol Physiol. 2001;280(1):L165–72. doi: 10.1152/ajplung.2001.280.1.L165. [DOI] [PubMed] [Google Scholar]
- 85.Burgel PR, Lazarus SC, Tam DC, Ueki IF, Atabai K, et al. Human eosinophils induce mucin production in airway epithelial cells via epidermal growth factor receptor activation. J Immunol. 2001;167(10):5948–54. doi: 10.4049/jimmunol.167.10.5948. [DOI] [PubMed] [Google Scholar]
- 86.Kohri K, Ueki IF, Shim JJ, Burgel PR, Oh YM, et al. Pseudomonas aeruginosa induces MUC5AC production via epidermal growth factor receptor. Eur Respir J. 2002;20(5):1263–70. doi: 10.1183/09031936.02.00001402. [DOI] [PubMed] [Google Scholar]
- 87.Chow CK. Cigarette smoking and oxidative damage in the lung. Ann NY Acad Sci. 1993;686:289–98. doi: 10.1111/j.1749-6632.1993.tb39189.x. [DOI] [PubMed] [Google Scholar]
- 88.Shao MX, Nakanaga T, Nadel JA. Cigarette smoke induces MUC5AC mucin overproduction via tumor necrosis factor-α-converting enzyme in human airway epithelial (NCI-H292) cells. Am J Physiol Lung Cell Mol Physiol. 2004;287(2):L420–27. doi: 10.1152/ajplung.00019.2004. [DOI] [PubMed] [Google Scholar]
- 89.Deshmukh HS, Case LM, Wesselkamper SC, Borchers MT, Martin LD, et al. Metalloproteinases mediate mucin 5AC expression by epidermal growth factor receptor activation. Am J Respir Crit Care Med. 2005;171(4):305–14. doi: 10.1164/rccm.200408-1003OC. [DOI] [PubMed] [Google Scholar]
- 90.Casalino-Matsuda SM, Monzon ME, Forteza RM. Epidermal growth factor receptor activation by epidermal growth factor mediates oxidant-induced goblet cell metaplasia in human airway epithelium. Am J Respir Cell Mol Biol. 2006;34(5):581–91. doi: 10.1165/rcmb.2005-0386OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gensch E, Gallup M, Sucher A, Li D, Gebremichael A, et al. Tobacco smoke control of mucin production in lung cells requires oxygen radicals AP-1 and JNK. J Biol Chem. 2004;279(37):39085–93. doi: 10.1074/jbc.M406866200. [DOI] [PubMed] [Google Scholar]
- 92.Longphre M, Li D, Gallup M, Drori E, Ordonez CL, et al. Allergen-induced IL-9 directly stimulates mucin transcription in respiratory epithelial cells. J Clin Invest. 1999;104(10):1375–82. doi: 10.1172/JCI6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Louahed J, Toda M, Jen J, Hamid Q, Renauld JC, et al. Interleukin-9 upregulates mucus expression in the airways. Am J Respir Cell Mol Biol. 2000;22(6):649–56. doi: 10.1165/ajrcmb.22.6.3927. [DOI] [PubMed] [Google Scholar]
- 94.Reader JR, Hyde DM, Schelegle ES, Aldrich MC, Stoddard AM, et al. Interleukin-9 induces mucous cell metaplasia independent of inflammation. Am J Respir Cell Mol Biol. 2003;28(6):664–72. doi: 10.1165/rcmb.2002-0207OC. [DOI] [PubMed] [Google Scholar]
- 95.Temann UA, Ray P, Flavell RA. Pulmonary overexpression of IL-9 induces Th2 cytokine expression, leading to immune pathology. J Clin Invest. 2002;109(1):29–39. doi: 10.1172/JCI13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whittaker L, Niu N, Temann UA, Stoddard A, Flavell RA, et al. Interleukin-13 mediates a fundamental pathway for airway epithelial mucus induced by CD4 T cells and interleukin-9. Am J Respir Cell Mol Biol. 2002;27(5):593–602. doi: 10.1165/rcmb.4838. [DOI] [PubMed] [Google Scholar]
- 97.Chen Y, Thai P, Zhao YH, Ho YS, DeSouza MM, Wu R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J Biol Chem. 2003;278(19):17036–43. doi: 10.1074/jbc.M210429200. [DOI] [PubMed] [Google Scholar]
- 98.Vermeer PD, Harson R, Einwalter LA, Moninger T, Zabner J. Interleukin-9 induces goblet cell hyperplasia during repair of human airway epithelia. Am J Respir Cell Mol Biol. 2003;28(3):286–95. doi: 10.1165/rcmb.4887. [DOI] [PubMed] [Google Scholar]
- 99.McMillan SJ, Bishop B, Townsend MJ, McKenzie AN, Lloyd CM. The absence of interleukin 9 does not affect the development of allergen-induced pulmonary inflammation nor airway hyperreactivity. J Exp Med. 2002;195(1):51–57. doi: 10.1084/jem.20011732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Atherton HC, Jones G, Danahay H. IL-13-induced changes in the goblet cell density of human bronchial epithelial cell cultures: MAP kinase and phosphatidylinositol 3-kinase regulation. Am J Physiol Lung Cell Mol Physiol. 2003;285(3):L730–39. doi: 10.1152/ajplung.00089.2003. [DOI] [PubMed] [Google Scholar]
- 101.Kondo M, Tamaoki J, Takeyama K, Nakata J, Nagai A. Interleukin-13 induces goblet cell differentiation in primary cell culture from guinea pig tracheal epithelium. Am J Respir Cell Mol Biol. 2002;27(5):536–41. doi: 10.1165/rcmb.4682. [DOI] [PubMed] [Google Scholar]
- 102.Jayawickreme SP, Gray T, Nettesheim P, Eling T. Regulation of 15-lipoxygenase expression and mucus secretion by IL-4 in human bronchial epithelial cells. Am J Physiol. 1999;276(4 Pt 1):L596–603. doi: 10.1152/ajplung.1999.276.4.L596. [DOI] [PubMed] [Google Scholar]
- 103.Rose MC, Piazza FM, Chen YA, Alimam MZ, Bautista MV, et al. Model systems for investigating mucin gene expression in airway diseases. J Aerosol Med. 2000;13(3):245–61. doi: 10.1089/jam.2000.13.245. [DOI] [PubMed] [Google Scholar]
- 104.Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med. 2002;8(8):885–89. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 105.Thai P, Chen Y, Dolganov G, Wu R. Differential regulation of MUC5AC/Muc5ac and hCLCA-1/mGob-5 expression in airway epithelium. Am J Respir Cell Mol Biol. 2005;33(6):523–30. doi: 10.1165/rcmb.2004-0220RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Young HW, Williams OW, Chandra D, Bellinghausen LK, Perez G, et al. Central role of Muc5ac expression in mucous metaplasia and its regulation by conserved 5′ elements. Am J Respir Cell Mol Biol. 2007;37(3):273–90. doi: 10.1165/rcmb.2005-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhen G, Park SW, Nguyenvu LT, Rodriguez MW, Barbeau R, et al. IL-13 and epidermal growth factor receptor have critical but distinct roles in epithelial cell mucin production. Am J Respir Cell Mol Biol. 2007;36(2):244–53. doi: 10.1165/rcmb.2006-0180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Erjefalt JS, Persson CG. Airway epithelial repair: breathtakingly quick and multipotentially pathogenic. Thorax. 1997;52(11):1010–12. doi: 10.1136/thx.52.11.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, et al. Mucin is produced by Clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol. 2004;31(4):382–94. doi: 10.1165/rcmb.2004-0060OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, et al. Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J Clin Invest. 2006;116(2):309–21. doi: 10.1172/JCI25167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park KS, Wells JM, Zorn AM, Wert SE, Laubach VE, et al. Transdifferentiation of ciliated cells during repair of the respiratory epithelium. Am J Respir Cell Mol Biol. 2006;34(2):151–57. doi: 10.1165/rcmb.2005-0332OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shimizu T, Shimizu S, Hattori R, Gabazza EC, Majima Y. In vivo and in vitro effects of macrolide antibiotics on mucus secretion in airway epithelial cells. Am J Respir Crit Care Med. 2003;168(5):581–87. doi: 10.1164/rccm.200212-1437OC. [DOI] [PubMed] [Google Scholar]
- 113.Chen Y, Nickola TJ, DiFronzo NL, Colberg-Poley AM, Rose MC. Dexamethasone-mediated repression of MUC5AC gene expression in human lung epithelial cells. Am J Respir Cell Mol Biol. 2006;34(3):338–47. doi: 10.1165/rcmb.2005-0176OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wan H, Kaestner KH, Ang SL, Ikegami M, Finkelman FD, et al. Foxa2 regulates alveolarization and goblet cell hyperplasia. Development. 2004;131(4):953–64. doi: 10.1242/dev.00966. [DOI] [PubMed] [Google Scholar]
- 115.Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, et al. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280(14):13809–16. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y, Zhao YH, Di YP, Wu R. Characterization of human mucin 5B gene expression in airway epithelium and the genomic clone of the amino-terminal and 5′-flanking region. Am J Respir Cell Mol Biol. 2001;25(5):542–53. doi: 10.1165/ajrcmb.25.5.4298. [DOI] [PubMed] [Google Scholar]
- 117.Deleted in proof
- 118.Inoue D, Yamaya M, Kubo H, Sasaki T, Hosoda M, et al. Mechanisms of mucin production by rhinovirus infection in cultured human airway epithelial cells. Respir Physiol Neurobiol. 2006;154(3):484–99. doi: 10.1016/j.resp.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 119.Aarbiou J, Verhoosel RM, Van Wetering S, De Boer WI, Van Krieken JH, et al. Neutrophil defensins enhance lung epithelial wound closure and mucin gene expression in vitro. Am J Respir Cell Mol Biol. 2004;30(2):193–201. doi: 10.1165/rcmb.2002-0267OC. [DOI] [PubMed] [Google Scholar]
- 120.Yoshisue H, Hasegawa K. Effect of MMP/ADAM inhibitors on goblet cell hyperplasia in cultured human bronchial epithelial cells. Biosci Biotechnol Biochem. 2004;68(10):2024–31. doi: 10.1271/bbb.68.2024. [DOI] [PubMed] [Google Scholar]
- 121.Kamio K, Matsushita I, Hijikata M, Kobashi Y, Tanaka G, et al. Promoter analysis and aberrant expression of the MUC5B gene in diffuse panbronchiolitis. Am J Respir Crit Care Med. 2005;171(9):949–57. doi: 10.1164/rccm.200409-1168OC. [DOI] [PubMed] [Google Scholar]
- 122.Deleted in proof
- 123.Chen Y, Zhao YH, Wu R. Differential regulation of airway mucin gene expression and mucin secretion by extracellular nucleotide triphosphates. Am J Respir Cell Mol Biol. 2001;25(4):409–17. doi: 10.1165/ajrcmb.25.4.4413. [DOI] [PubMed] [Google Scholar]
- 124.Lillehoj EP, Hyun SW, Kim BT, Zhang XG, Lee DI, et al. Muc1 mucins on the cell surface are adhesion sites for Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol. 2001;280(1):L181–87. doi: 10.1152/ajplung.2001.280.1.L181. [DOI] [PubMed] [Google Scholar]
- 125.von Mensdorff-Pouilly S, Snijdewint FG, Verstraeten AA, Verheijen RH, Kenemans P. Human MUC1 mucin: a multifaceted glycoprotein. Int J Biol Markers. 2000;15(4):343–56. doi: 10.1177/172460080001500413. [DOI] [PubMed] [Google Scholar]
- 126.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16(9):467–76. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 127.Kuwahara I, Lillehoj EP, Hisatsune A, Lu W, Isohama Y, et al. Neutrophil elastase stimulates MUC1 gene expression through increased Sp1 binding to the MUC1 promoter. Am J Physiol Lung Cell Mol Physiol. 2005;289(2):L355–62. doi: 10.1152/ajplung.00040.2005. [DOI] [PubMed] [Google Scholar]
- 128.Griffin S, Carroll TP, Greene CM, O’Neill SJ, Taggart CC, McElvaney NG. Effect of proinflammatory stimuli on mucin expression and inhibition by secretory leucoprotease inhibitors. Cell Microbiol. 2007;9(3):670–79. doi: 10.1111/j.1462-5822.2006.00819.x. [DOI] [PubMed] [Google Scholar]
- 129.Copin MC, Buisine MP, Leteurtre E, Marquette CH, Porte H, et al. Mucinous bronchioloalveolar carcinomas display a specific pattern of mucin gene expression among primary lung adenocarcinomas. Hum Pathol. 2001;32(3):274–81. doi: 10.1053/hupa.2001.22752. [DOI] [PubMed] [Google Scholar]
- 130.Bernacki SH, Nelson AL, Abdullah L, Sheehan JK, Harris A, et al. Mucin gene expression during differentiation of human airway epithelia in vitro: MUC4 and MUC5B are strongly induced. Am J Respir Cell Mol Biol. 1999;20(4):595–604. doi: 10.1165/ajrcmb.20.4.3442. [DOI] [PubMed] [Google Scholar]
- 131.Damera G, Xia B, Ancha HR, Sachdev GP. IL-9 modulated MUC4 gene and glycoprotein expression in airway epithelial cells. Biosci Rep. 2006;26(1):55–67. doi: 10.1007/s10540-006-9000-5. [DOI] [PubMed] [Google Scholar]
- 132.Damera G, Xia B, Sachdev GP. IL-4 induced MUC4 enhancement in respiratory epithelial cells in vitro is mediated through JAK-3 selective signaling. Respir Res. 2006;7:39. doi: 10.1186/1465-9921-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fischer BM, Cuellar JG, Diehl ML, deFreytas AM, Zhang J, et al. Neutrophil elastase increases MUC4 expression in normal human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;284(4):L671–79. doi: 10.1152/ajplung.00220.2002. [DOI] [PubMed] [Google Scholar]
- 134.Sharma P, Dudus L, Nielsen PA, Clausen H, Yankaskas JR, et al. MUC5B and MUC7 are differentially expressed in mucous and serous cells of submucosal glands in human bronchial airways. Am J Respir Cell Mol Biol. 1998;19(1):30–37. doi: 10.1165/ajrcmb.19.1.3054. [DOI] [PubMed] [Google Scholar]
- 135.Li S, Intini G, Bobek LA. Modulation of MUC7 mucin expression by exogenous factors in airway cells in vitro and in vivo. Am J Respir Cell Mol Biol. 2006;35(1):95–102. doi: 10.1165/rcmb.2005-0305OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li S, Bobek LA. Functional analysis of human MUC7 mucin gene 5′-flanking region in lung epithelial cells. Am J Respir Cell Mol Biol. 2006;35(5):593–601. doi: 10.1165/rcmb.2006-0110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shankar V, Pichan P, Eddy RL, Jr, Tonk V, Nowak N, et al. Chromosomal localization of a human mucin gene (MUC8) and cloning of the cDNA corresponding to the carboxy terminus. Am J Respir Cell Mol Biol. 1997;16(3):232–41. doi: 10.1165/ajrcmb.16.3.9070607. [DOI] [PubMed] [Google Scholar]
- 138.Chen Y, Zhao YH, Kalaslavadi TB, Hamati E, Nehrke K, et al. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am J Respir Cell Mol Biol. 2004;30(2):155–65. doi: 10.1165/rcmb.2003-0103OC. [DOI] [PubMed] [Google Scholar]
- 139.Waggoner D. Related mechanisms of disease: epigenesis. Semin Pediatr Neurol. 2007;14(1):7–14. doi: 10.1016/j.spen.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 140.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19(5):269–77. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 140a.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397(6720):579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 140b.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23(1):58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 141.Mizoshita T, Tsukamoto T, Inada KI, Hirano N, Tajika M, et al. Loss of MUC2 expression correlates with progression along the adenoma-carcinoma sequence pathway as well as de novo carcinogenesis in the colon. Histol Histopathol. 2007;22(3):251–60. doi: 10.14670/HH-22.251. [DOI] [PubMed] [Google Scholar]
- 142.Hanski C, Riede E, Gratchev A, Foss HD, Bohm C, et al. MUC2 gene suppression in human colorectal carcinomas and their metastases: in vitro evidence of the modulatory role of DNA methylation. Lab Invest. 1997;77(6):685–95. [PubMed] [Google Scholar]
- 143.Siedow A, Szyf M, Gratchev A, Kobalz U, Hanski ML, et al. De novo expression of the Muc2 gene in pancreas carcinoma cells is triggered by promoter demethylation. Tumour Biol. 2002;23(1):54–60. doi: 10.1159/000048689. [DOI] [PubMed] [Google Scholar]
- 144.Mesquita P, Peixoto AJ, Seruca R, Hanski C, Almeida R, et al. Role of site-specific promoter hypomethylation in aberrant MUC2 mucin expression in mucinous gastric carcinomas. Cancer Lett. 2003;189(2):129–36. doi: 10.1016/s0304-3835(02)00549-9. [DOI] [PubMed] [Google Scholar]
- 145.Hamada T, Goto M, Tsutsumida H, Nomoto M, Higashi M, et al. Mapping of the methylation pattern of the MUC2 promoter in pancreatic cancer cell lines, using bisulfite genomic sequencing. Cancer Lett. 2005;227(2):175–84. doi: 10.1016/j.canlet.2004.11.058. [DOI] [PubMed] [Google Scholar]
- 146.Yamada N, Hamada T, Goto M, Tsutsumida H, Higashi M, et al. MUC2 expression is regulated by histone H3 modification and DNA methylation in pancreatic cancer. Int J Cancer. 2006;119(8):1850–57. doi: 10.1002/ijc.22047. [DOI] [PubMed] [Google Scholar]
- 147.Perrais M, Pigny P, Buisine MP, Porchet N, Aubert JP, Van Seuningen-Lempire I. Aberrant expression of human mucin gene MUC5B in gastric carcinoma and cancer cells. Identification and regulation of a distal promoter. J Biol Chem. 2001;276(18):15386–96. doi: 10.1074/jbc.M010534200. [DOI] [PubMed] [Google Scholar]
- 148.Ho JJ, Han SW, Pan PL, Deng G, Kuan SF, Kim YS. Methylation status of promoters and expression of MUC2 and MUC5AC mucins in pancreatic cancer cells. Int J Oncol. 2003;22(2):273–79. [PubMed] [Google Scholar]
- 149.Zrihan-Licht S, Weiss M, Keydar I, Wreschner DH. DNA methylation status of the MUC1 gene coding for a breast-cancer-associated protein. Int J Cancer. 1995;62(3):245–51. doi: 10.1002/ijc.2910620303. [DOI] [PubMed] [Google Scholar]