Abstract
Immunity to common bacteria requires the generation of antibodies that promote opsonophagocytosis and neutralise toxins. Pooled human immunoglobulin is widely advocated as an adjunctive treatment for clinical Streptococcus pyogenes infection however, the protein targets of the reagent remain ill defined. Affinity purification of the anti-streptococcal antibodies present within pooled immunoglobulin resulted in the generation of an IgG preparation that promoted opsonophagocytic killing of S. pyogenes in vitro and provided passive immunity in vivo. Isolation of the streptococcal surface proteins recognised by pooled human immunoglobulin permitted identification and ranking of 94 protein antigens, ten of which were reproducibly identified across four contemporary invasive S. pyogenes serotypes (M1, M3, M12 and M89). The data provide novel insight into the action of pooled human immunoglobulin during invasive S. pyogenes infection, and demonstrate a potential route to enhance the efficacy of antibody based therapies.
Over the past 30 years there has been a steady rise in the global rate of severe Streptococcus pyogenes infection, which now exceeds 3 cases/100,000 in some regions1,2. The attendant morbidity and mortality of invasive S. pyogenes diseases such as necrotizing fasciitis are considerable, with a case fatality rate exceeding 40% in patients who develop streptococcal toxic shock syndrome2. Although the basis for population immunity to S. pyogenes is poorly understood, recent studies have indicated that adjunctive intravenous immunoglobulin therapy (IVIG) may confer a survival benefit during invasive S. pyogenes infection3,4,5.
IVIG is a commercially available plasmapheresis product that is purified from the blood of over one thousand healthy donors. While originally developed as a replacement therapy for hypogammaglobulinemia, the presence of specific antibodies to many human pathogens makes pooled immunoglobulin an effective prophylactic treatment for several infective conditions including hepatitis A, measles and rubella. Recent attention has focused on the protective activity of IVIG against a number of Gram positive pathogens, most notably S. pyogenes3,4,5,6,7.
Experimentally, IVIG has been shown to reduce systemic inflammation associated with streptococcal superantigen production and to promote opsonophagocytic clearance of S. pyogenes both in vitro, and in a humanised mouse model8,9. Clinically, one small trial and two population-based studies have reported benefits of IVIG administration during severe S. pyogenes infection and, although controversial, adjunct IVIG therapy is advocated by many physicians3,4,5. While some attention has focused on the varying ability of IVIG to neutralise streptococcal superantigens10, little is known about the S. pyogenes surface antigens recognised by IVIG and the mechanisms by which the reagent promotes bacterial clearance remain ill defined. One study has demonstrated the presence of anti-M1 protein antibodies within commercial preparations suggesting that IVIG may contain other antibodies that target major S. pyogenes surface proteins9. We sought to further characterise the S. pyogenes surface proteins recognised by IVIG and assess their protective efficacy using standard models of S. pyogenes infection.
Results
Purification of anti-streptococcal IgG from pooled immunoglobulin
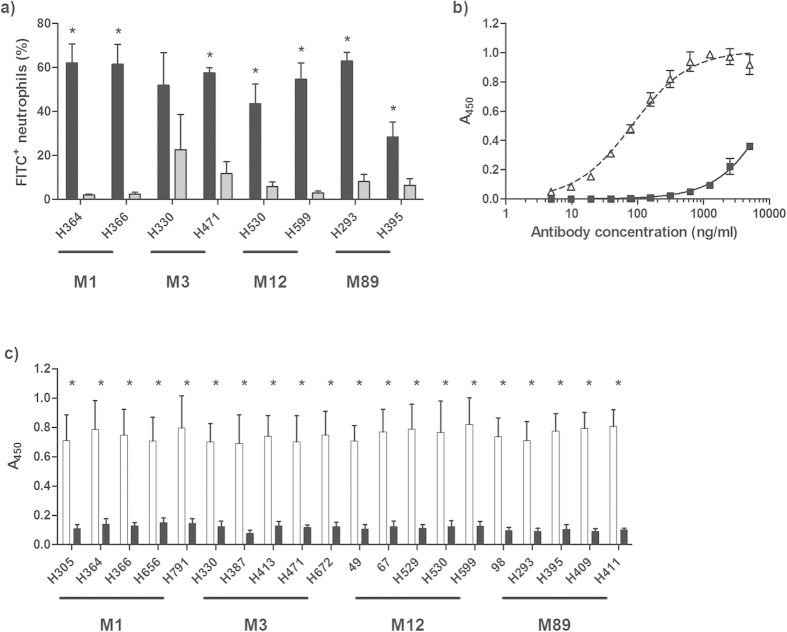
The ability of IVIG to promote phagocytic uptake of S. pyogenes was first confirmed using a purified human neutrophil opsonophagocytosis assay. S. pyogenes strains were selected to represent four of the most common contemporary serotypes associated with invasive S. pyogenes infection in Europe and North America (M1, M3, M12 and M89, Supplementary Table 1)1,2. At a fixed concentration of 5 mg/ml, IVIG was shown to promote neutrophil uptake of two representative strains from each serotype, all of which were isolated from invasive disease manifestations or toxic shock cases (Fig. 1A). Although the M89 strain H395 is hyperencapsulated, neutrophil uptake was still promoted by IVIG, albeit to a lesser degree than the other isolates examined. Interestingly, the baseline uptake of the M3 strain H330 was markedly higher than the other strains selected for study, reducing the apparent effect of IVIG despite comparable levels of overall uptake occurring in the presence of the reagent. This confirms that the anti-streptococcal antibodies present within pooled immunoglobulin are capable of opsonising a range of S. pyogenes serotypes in vitro.
Figure 1. Preparation and properties of anti-streptococcal IgG (E-IVIG).
(a) IVIG promotes neutrophil uptake of S. pyogenes. FITC labelled S. pyogenes cells were treated with 5 mg/ml of IVIG (black bars) or PBS (grey bars) and incubated with freshly isolated human neutrophils. Results from three independent experiments are expressed as percentage of FITC+ neutrophils (mean ± SD) after a 30 min co-incubation. n = 3, two-tailed t-test: p = 0.0003 (H364, H366 and H599); p = 0.0002; (H471); p = 0.0021 (H530); p < 0.0001 (H293); p = 0.0069 (H395). (b) Concentration-dependent binding of M1 S. pyogenes cell wall extract by E-IVIG (white triangles) and IVIG (black squares). (c) Serotype-independent binding of S. pyogenes cell wall extracts by E-IVIG (white bars) IVIG (black bars), at a fixed concentration (2500 ng/ml). Results from three independent experiments are expressed as triplicate A450 readings minus the background absorbance (mean ± SD). n = 3, two-tailed t-test, p < 0.0001 in all instances.
Having established the presence of opsonic anti-streptococcal antibodies within pooled immunoglobulin, serotype M1 cell wall extract was covalently conjugated to cyanogen bromide activated agarose, and used to affinity purify the anti-streptococcal IgG fraction from commercially available IVIG. The reactivity of the resulting “enhanced” (E)-IVIG was then assessed by ELISA. The apparent affinity of E-IVIG for M1 S. pyogenes cell wall extract was shown to be much higher than that of the starting IVIG preparation (Fig. 1B). Furthermore, clear differences were observed in the binding of E-IVIG or IVIG to cell wall extracts from each of the 20 isolates selected for study, suggesting that the apparent increase in affinity towards S. pyogenes was serotype independent (Fig. 1C).
Purification of S. pyogenes surface antigens by E-IVIG immunoprecipitation
In order to identify the streptococcal surface proteins targeted by IVIG, E-IVIG was covalently conjugated to cyanogen bromide activated agarose and used to purify the IVIG-reactive antigens from S. pyogenes cell wall extracts by immunoprecipitation. The resulting immunoprecipitates were visualised by immunoblot analysis; which confirmed the presence of a multitude of IVIG-reactive proteins within the purified preparations, none of which were present when a goat isotype control IgG column was used (Supplementary Figure 1). The immunoprecipitates were separated by SDS-PAGE, and the identity of the purified antigens was determined by proteomic analysis using a publically available proteomic database derived from two M1 S. pyogenes genomes (SF370 and MGAS5005).
A total of 94 E-IVIG reactive antigens were recovered from the immunoprecipitates which included several major S. pyogenes vaccine candidates previously described in the literature (Table 1 and Supplementary Table 2). As in previous surface proteome studies, a number of secreted and classically cytoplasmic proteins were clearly present within the starting cell wall extracts11,12. Remarkably many such proteins were purified by E-IVIG immunoprecipitation, suggesting that these molecules are capable of generating a strong humoral immune response during S. pyogenes infection. Of the 94 proteins identified, only ten were recovered from all 20 isolates, suggesting that these antigens are reproducibly expressed by all four invasive serotypes selected for study, and share a high degree of sequence identity between serotypes. These conserved antigens included virulence factors (C5a peptidase13, and SpyAD14), lipoproteins (maltose/maltodextrin-binding protein, oligopeptide-binding protein and nucleoside-binding protein), other cell wall-attached, LPXTG motif-containing molecules (putative pullulanase) and a single, abundant cytoplasmic protein (Chaperone protein DnaK) previously shown to stimulate in vivo antibody production15. Two of the proteins had no attributed function despite their clear degree of conservation and propensity to generate a strong humoral immune response during S. pyogenes infection (hypothetical membrane associated protein and cell surface protein).
Table 1. The S. pyogenes antigens purified by E-IVIG immunoprecipitation.
| Uniprot identifier | Protein product | SEQUEST P-values (range) | M1 | M3 | M12 | M89 | Total | |
|---|---|---|---|---|---|---|---|---|
| C5AP_STRP1 | C5a peptidase | 1 × 10−30 −7 × 10−10 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | 100% |
| DNAK_STRP1 | Chaperone protein DnaK | 8 × 10−15 −8 × 10−8 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q48Y96_STRP1 | Maltose/maltodextrin-binding protein | 2 × 10−16 −3 × 10−10 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q490V0_STRP1 | Oligopeptide-binding protein | 1 × 10−30 −2 × 10−10 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q491G2_STRP1 | Nucleoside-binding protein | 1 × 10−30 −1 × 10−15 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q99XX8_STRP1 | Putative pullulanase | 2 × 10−14 −6 × 10−8 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q99ZH4_STRP1 | Nucleoside-binding protein | 1 × 10−30 −7 × 10−11 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q99ZW9_STRP1 | Hypothetical membrane associated protein | 1 × 10−30 −2 × 10−13 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q9A0C0_STRP1 | Cell surface protein | 7 × 10−14 −3 × 10−7 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| Q9A1H3_STRP1 | SpyAD | 2 × 10−15 −4 × 10−11 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20 | |
| EFTS_STRP1 | Elongation factor Ts | 3 × 10−15 −3 × 10−8 | 5/5 | 5/5 | 4/5 | 5/5 | 19/20 | 70-95% |
| MTSA_STRP1 | Metal ABC transporter substrate-binding lipoprotein | 1 × 10−30 −9 × 10−10 | 5/5 | 5/5 | 4/5 | 5/5 | 19/20 | |
| PGK_STRP1 | Phosphoglycerate kinase | 6 × 10−14 −5 × 10−9 | 5/5 | 3/5 | 5/5 | 5/5 | 18/20 | |
| Q99YL6_STRP1 | Putative uncharacterised protein | 2 × 10−9 −2 × 10−6 | 4/5 | 4/5 | 5/5 | 5/5 | 18/20 | |
| Q9A0C5_STRP1 | Putative uncharacterised protein | 1 × 10−10 −6 × 10−5 | 5/5 | 3/5 | 5/5 | 5/5 | 18/20 | |
| TACY_STRP1 | Streptolysin O | 1 × 10−30 −5 × 10−7 | 5/5 | 5/5 | 5/5 | 3/5 | 18/20 | |
| PEPDB_STRP1 | Probable dipeptidase B | 1 × 10−30 −2 × 10−7 | 4/5 | 3/5 | 5/5 | 5/5 | 17/20 | |
| Q7DAN2_STRP1 | Nicotine adenine dinucleotide glycohydrolase | 8 × 10−15 −3 × 10−11 | 5/5 | 5/5 | 5/5 | 2/5 | 17/20 | |
| Q9A1G0_STRP1 | Penicillin-binding protein | 9 × 10−12 −1 × 10−4 | 2/5 | 5/5 | 5/5 | 5/5 | 17/20 | |
| Q490K8_STRP1 | SpyCEP | 3 × 10−15 −1 × 10−5 | 5/5 | 5/5 | 2/5 | 4/5 | 16/20 | |
| Q99XU2_STRP1 | Periplasmic component of efflux system | 3 × 10−13 −9 × 10−9 | 5/5 | 3/5 | 3/5 | 5/5 | 16/20 | |
| Q48XJ9_STRP1 | Sugar-binding protein | 3 × 10−15 −2 × 10−5 | 5/5 | 3/5 | 1/5 | 5/5 | 14/20 | |
| Q99XV0_STRP1 | M protein | 5 × 10−13 −5 × 10−6 | 5/5 | 5/5 | 4/5 | 0/5 | 14/20 |
Numbers represent the proportion of strains from which each target was identified. Targets identified from ≥70% of strains are displayed.
To ensure that recognition of these antigens was not limited to a single source of IVIG, E-IVIG was derived from two further commercial preparations (Intratect and Privigen) and used to immunoprecipitate reactive antigens from two representative strains of each of the four S. pyogenes serotypes. Proteomic analysis identified the same panel of highly conserved antigens within the resulting immunoprecipitates, further implicating these molecules as important targets of adjunct IVIG therapy for invasive S. pyogenes infection (Supplementary Table 3).
E-IVIG promotes opsonophagocytic killing of S. pyogenes in vitro
To determine if the higher affinity of E-IVIG for S. pyogenes surface antigens resulted in increased anti-streptococcal activity, the ability of E-IVIG and IVIG to promote opsonophagocytosis and killing of M1 S. pyogenes (strain H364) was compared. At IgG concentrations ranging from 40 μg/ml to 10 μg/ml, the percentage of neutrophils containing internalised S. pyogenes was significantly higher in the E-IVIG group than in the IVIG group (Fig. 2A). These results were echoed in a classical Lancefield assay of opsonophagocytic killing, where concentrations in excess of 1 mg/ml IVIG are normally required to enhance bacterial killing8. Supplementation of human whole blood with 40 μg/ml of E-IVIG was sufficient to increase opsonophagocytic killing by a factor of ten compared to supplementation with the same concentration of IVIG (Fig. 2B). Together these data suggest that purification of the S. pyogenes reactive IgG from IVIG results in a marked increase in opsonophagocytic killing through functional recognition of one or more serotype independent S. pyogenes surface antigens.
Figure 2. E-IVIG promotes opsonophagocytic killing of S. pyogenes in vitro.
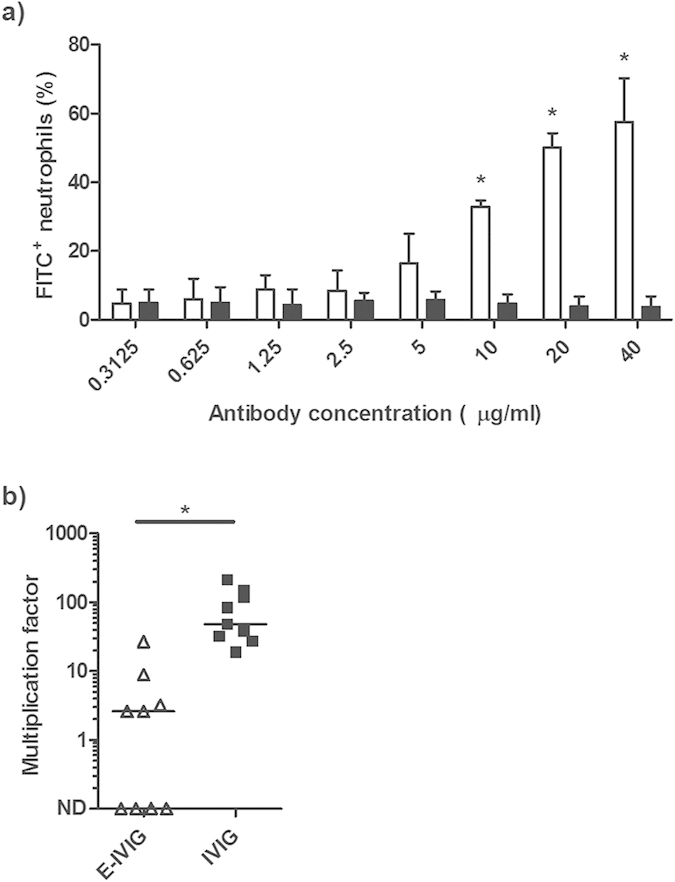
(a) FITC labelled M1 S. pyogenes cells (strain H364) were opsonised with doubling dilutions of E-IVIG (white bars) or IVIG (black bars) prior to incubation with freshly isolated human neutrophils. Results from three independent experiments are expressed as percentage of FITC+ neutrophils (mean ± SD) after a 30 min co-incubation. n = 3, two-tailed t-test: p = 0.002 (40 μg/ml); p < 0.0001 (20 μg/ml); p = 0.0001 (10 μg/ml). (b) Whole blood from healthy donors was supplemented with 40 μg/ml of E-IVIG (open triangles) or IVIG (closed squares) prior to inoculation with ~20 CFU of M1 S. pyogenes (strain H364). Results are expressed as multiplication factors (median and range). n = 9, two-tailed Mann-Whitney U: p = 0.0005. ND: Not Detected.
Passive immunisation with E-IVIG inhibits dissemination of S. pyogenes from an intramuscular focus of infection
To assess the efficacy of E-IVIG in vivo, two groups of age matched C57BL/6 mice were passively immunised with E-IVIG or IVIG and challenged intramuscularly with 5 × 106 CFU of M1 S. pyogenes (strain H305). The mice were culled 24 h post infection and the degree of bacterial dissemination was assessed by homogenisation of the organs and plating. Bacterial counts from the locally draining inguinal lymph node and blood of the E-IVIG group were significantly lower than those recovered from the IVIG group, consistent with an enhancement in bactericidal activity (Fig. 3). A similar trend was observed in the liver, although the difference was insufficient to achieve statistical significance. E-IVIG therapy had a limited effect on the bacterial burden at the site of infection, consistent with data from previous studies where treatment with antibiotics, or antibiotics plus IVIG, failed to promote bacterial clearance at the site of infection, despite promoting clearance from the blood and organs8. Together the data suggest that purification of the S. pyogenes-reactive IgG from IVIG greatly enhances the anti-streptococcal activity of the reagent both in vitro and in vivo.
Figure 3. Passive immunisation with E-IVIG inhibits dissemination of S. pyogenes from an intramuscular focus of infection.
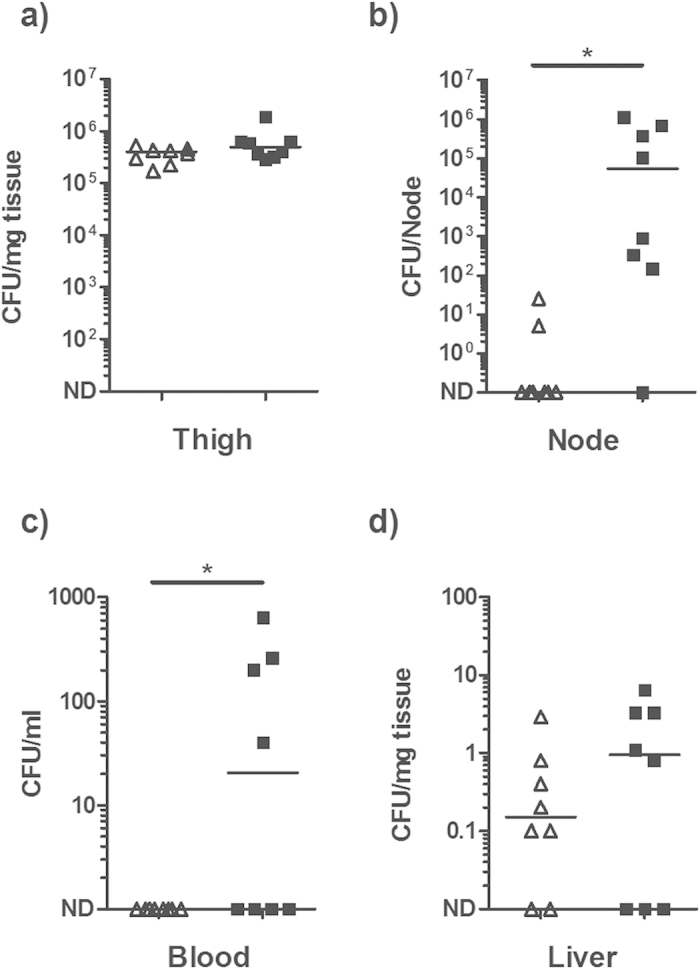
Two groups of eight age matched C57BL/6 mice were pre-treated with 800 μg of E-IVIG (open triangles) or IVIG (closed squares) and infected intramuscularly with 5 × 106 CFU of M1 S. pyogenes (strain H305). Solid lines indicate the median CFU recovered from (a) the site of original infection, (b) ipsilateral draining lymph node, (c) blood and (d) liver 24 h post-infection. n = 8, two-tailed Mann-Whitney U: p = 0.0037 (node); p = 0.0325 (blood). ND: Not Detected.
Discussion
Although IVIG is now widely advocated as an adjunctive treatment for clinical S. pyogenes infection, no studies have sought to characterise the repertoire of proteins recognised by the reagent. Our data demonstrate the full repertoire of proteinaceous surface antigens recognised by IVIG, and illustrate the capability of the anti-streptococcal antibody present within the reagent to promote opsonophagocytic killing of S. pyogenes in vitro, and provide passive protection in vivo, during invasive infection.
In Europe and North America, approximately 70% of all invasive S. pyogenes infections are caused by the ten most prominent M types1,2, four of which were represented in this study. While M1 and M3 remain the most common cause of invasive S. pyogenes infection, recent data suggest that has been an upsurge in invasive M89 infections over the past decade16,17. In this study E-IVIG was produced using serotype M1 cell wall extract, and a proteomic database derived from two M1 S. pyogenes genomes was used to identify the purified antigens. Of the 94 protein antigens isolated, only ten sequence-invariant antigens were identified from all 20 S. pyogenes strains selected for study. In addition to confirming that these antigens were expressed by four disparate S. pyogenes serotypes experimentally, the fidelity of the ten protein sequences was confirmed within all serotypes for which genome sequence data are currently available to ensure a high degree of conservation (data not shown). We considered the possibility that sequence variation and/or type-specific variation in gene content precluded proteomic identification of IVIG-reactive proteins from some isolates. However, with the exception of certain type-specific proteins such as the M protein and pilus, the core genomes of different S. pyogenes serotypes are largely conserved in sequence, and gene content18. Indeed, selective re-analysis of the purified immunoprecipitates using separate M3, M12 and M89 proteomic databases did not impact appreciably on our findings (data not shown). Therefore the ten highly conserved, invariant antigens identified herein may be of particular importance for the reported efficacy of adjunct IVIG therapy, and may warrant consideration as components of an effective anti-S. pyogenes vaccine.
Interestingly, despite the use of pooled M1 cell wall extract for affinity purification of E-IVIG, and an M1 database for proteomic analysis of the resulting immunoprecipitates, the M protein was identified in 14/20 strains evaluated. The apparent absence of the M protein from the M89 preparations could be wholly attributed to substantial differences between the M1 and M89 protein sequences; and reanalysis of the M89 immunoprecipitate data using an emm89 sequence clearly identified M protein from all five strains selected for study (data not shown). Therefore, despite considerable variation between the different emm sequences, a substantial amount of cross reactive anti-M protein antibody was clearly present within the affinity purified E-IVIG preparation. This finding supports the existence of cross protective epitopes within disparate M protein isoforms19.
Repeated exposure of the humoral immune system to a variety of common pathogens results in the accumulation of circulating antibodies with a diverse range of specificities. Affinity purification of anti-streptococcal antibodies from IVIG resulted in the generation of an antibody preparation with enhanced activity against S. pyogenes in vivo. While the breadth of protection may still be limited by serotype or strain specific differences in protein expression, E-IVIG may represent a more effective adjunct treatment for invasive S. pyogenes infection than the polyspecific IVIG that is currently employed clinically. It is possible that hyperencapsulation of S. pyogenes may impact negatively on the efficacy of therapeutic opsonic antibodies through masking of surface antigens, especially in strains that possess mutations in the global virulence regulator CovR/S20. However, IVIG was capable of promoting opsonophagocytic uptake of the hyperencapsulated covS mutant H39516, albeit at a reduced level (Fig. 1). This suggests that, while some of the epitopes targeted by IVIG may be shielded from antibody recognition by the capsule, a crucial subset remain exposed in hyperencapsulated isolates.
Although not intended to target secreted proteins, a number of extracellular toxins were purified from the cell wall extracts by E-IVIG immunoprecipitation, including streptolysin-O and the cysteine protease SpeB. Intriguingly, the repertoire of purified secreted proteins did not include any superantigens, which play a key role during invasive S. pyogenes infection and are strong targets of the human humoral immune response10,21. Thus further development of E-IVIG for S. pyogenes sepsis will have to address the possibility that the broadly acting anti-superantigen activity of IVIG may not be maintained following anti-streptococcal antibody enrichment. Furthermore, any pre-clinical evaluation of E-IVIG will require rigorous screening to ensure the purity of the antibody preparation and to rule out any unforeseen toxicity resulting from the carryover of bacterial products. Nonetheless, in light of the shortage of human immunoglobulin, it may be prudent to determine whether therapeutic antibodies against a wider range of pathogens could be sequentially isolated from a single IVIG preparation using multiple rounds of affinity purification. Such targeted reagents may prove valuable for the adjunct treatment of a wide range of common antimicrobial-resistant pathogens. Indeed while here we have applied immunoprecipitation to S. pyogenes, published reports and our own unpublished data suggest that the technique could equally be applied to other Gram positive bacteria such as Staphylococcus aureus and Enterococcus spp. where clearance is mediated through similar processes6,7,22.
As a pooled preparation of immunoglobulin from normal donor blood, IVIG is an obvious but hitherto unevaluated reagent for the systematic identification of the antigens recognised by the humoral immune response of a healthy human population. Previous studies have attempted to characterise the streptococcal antigens targeted by the human antibody response, however the results may have been influenced by the use of non-native protein preparations and/or serum of unknown protective activity, which is often employed for screening target antigens12,23,24,25. The data in this study identify for the first time a core list of highly conserved S. pyogenes antigens against which protective humoral immunity is directed. The findings enhance our understanding of population immunity to S. pyogenes as a whole, and may have implications for both passive and active vaccination.
Materials and Methods
Bacterial strains and growth conditions
The S. pyogenes isolates used are listed in Supplementary Table 1 and were routinely cultured on Columbia horse blood agar (CBA) or in Todd-Hewitt broth (THB) at 37 °C in 5% CO2.
Preparation of streptococcal cell wall extracts and immunoblotting
Streptococcal cell wall extracts were isolated as previously described26. Briefly, overnight S. pyogenes cultures were diluted 1:10 in fresh THB and grown to an A600 of 0.4–0.8. The cells were pelleted and resuspended in 1/50th volume of 10 mM Tris-HCl (pH 8.0) containing 30% raffinose, 100 U/ml mutanolysin, 1 mg/ml lysozyme and 10% Protease Inhibitor Cocktail Set III (Calbiochem), and incubated for 3 h at 37 °C with occasional inversion. The cells were pelleted and the supernatant (cell wall extract) was dialysed into PBS overnight and concentrated by centrifugation (10,000 MWCO). For immunoblotting, proteins were separated on 7% tris-acetate (TA) gels (Invitrogen) and transferred to Hybond LFP membranes (GE Healthcare). Membranes were blocked with 5% skimmed milk prior to the addition of 4 μg/ml IVIG (Endobulin®, Baxter). Bound antibodies were detected using a 1:80,000 dilution of HRP-conjugated goat anti human IgG (Sigma-Aldrich) and the ECL prime detection system (GE Healthcare).
Affinity purification of E-IVIG and immunoprecipitation of reactive streptococcal surface antigens
Pooled serotype M1 cell wall extract was dialysed into coupling buffer (0.1 M sodium bicarbonate, 0.5 M sodium chloride, pH 8.3) overnight and coupled to cyanogen bromide (CNBr) activated agarose at a concentration of ~1 mg/ml according to the manufacturer’s instructions (Sigma-Aldrich). Briefly, lyophilised CNBr-activated agarose was swollen in cold 1 mM HCl (200 ml/g dry gel), pipetted into a 5 ml column and washed sequentially with ten column volumes of distilled water and five column volumes of coupling buffer. The dialysed cell wall extract was immediately added and incubated with the swollen resin overnight at 4 °C. The resin was washed extensively with coupling buffer and incubated with two column volumes of 0.2 M glycine at RT for 2 h with gentle agitation. The resin was washed sequentially with ten column volumes of coupling buffer and ten column volumes of sodium acetate buffer (0.1 M anhydrous sodium acetate, 0.5 M sodium chloride, pH 4). This high/low pH wash cycle was repeated five times and the resulting affinity purification column was equilibrated into PBS supplemented with 0.1% sodium azide. For affinity purification, the resin was incubated with 5 ml of 5 mg/ml IVIG for 2 h at RT and washed extensively with PBS. Bound antibody was eluted into 1 ml aliquots of 1 M acetic acid and immediately neutralised using an equal volume of 3 M Tris-HCl (pH 8.8). Eluted fractions (containing the E-IVIG) were pooled, dialysed into PBS and concentrated. E-IVIG was stored in 0.1% sodium azide at 4 °C until required. For immunoprecipitation, E-IVIG or goat isotype control IgG (Abcam) was coupled to CNBr-activated agarose at a concentration of ~1 mg/ml, and incubated with 2 ml aliquots of concentrated cell wall extract. The resin was washed extensively and the bound antigens were eluted, dialysed and concentrated as outlined above. For comparative immunoprecipitation experiments, additional E-IVIG preparations were prepared from Intratect® (Biotest) and Privigen® (CSL Behring) IVIG.
Analysis of E-IVIG by ELISA
To compare binding of E-IVIG and IVIG, 96 well plates were coated with 1 μg/well of cell wall extract and incubated with 2.5 mg/ml of antibody. Bound antibodies were detected using a 1: 80,000 dilution of HRP-conjugated goat anti-human IgG and 50 μl/well tetramethylbenzidine. The detection reaction was stopped through the addition of 50 μl of 1 M H2SO4 and the absorbance at 450 nm read using a μQuant universal microplate spectrophotometer (Bio-tek instruments).
Opsonophagocytic uptake and killing assays
Human neutrophil opsonophagocytosis assays were performed using normal donor neutrophils obtained with informed consent from a subcollection of the Imperial College Tissue Bank and FITC-labelled S. pyogenes as previously described27. Briefly, bacterial cells from a 5 ml overnight culture were pelleted, washed and incubated in 500 μl of 20 μg/ml FITC in 0.1 M sodium carbonate buffer (pH 9) for 30 min at 37 °C. The cells were washed twice and resuspended to an A600 of 0.35 in PBS and incubated with an appropriate concentration of E-IVIG, IVIG or PBS at 37 °C for 30 min with agitation. Neutrophils were purified from heparinised whole blood that was combined 1:1 with 3% dextran (MW > 100,000) in 0.9% saline and allowed to separate on ice for 30 min. The leukocytes present in the supernatant layer were pelleted and resuspended in one volume of 0.9% saline. Neutrophils were then purified by Ficoll-layering as described previously28. Opsonophagocytosis assays were performed in 1 ml reaction volumes containing 100 μl of resuspended bacteria and 1 × 106 neutrophils. After a 30 min co-incubation, the S. pyogenes/neutrophil suspension was combined 1:1 with 0.02% EDTA in PBS and, immediately prior to analysis, the extracellular fluorescence was quenched through addition of one volume of trypan blue. Results are expressed as the percentage of neutrophils containing FITC+ bacteria as determined by flow cytometry. The strains selected were all isolated from invasive disease or toxic shock cases (Supplementary table 1). Whole blood opsonophagocytic killing (Lancefield) assays were performed under standard conditions using whole blood from three healthy human donors supplemented with 40 μg/ml of IVIG or E-IVIG and approximately 20 CFU of M1 S. pyogenes (strain H364) using a standard method8,29,30. Results from nine independent reactions were expressed as the bacterial multiplication factor following 3 h incubation at 37 °C with gentle agitation.
Proteomic analysis
Immunoprecipitated antigens were separated on 7% TA gels and subjected to in gel trypsin digest and subsequent liquid chromatography-tandem mass spectrometry as previously described31. Proteins were identified using a publically available proteome for serotype M1 S. pyogenes (Uniprot, CP000017)32,33; and correlation of the MS/MS peptide data with the in silico trypsin fragmentation patterns predicted using BioWorks 3.3 and the SEQUEST algorithm (Thermo Scientific). Peptides generating ions of 200–2000 amu, with a cross correlation score >1.5, 2.0, or 2.5 (where z = 1, 2 or 3 respectively) and a p < 0.01 were mapped onto the S. pyogenes proteome. Proteins that contained two or more of the identified peptides were selected for further analysis. The protein probability for each selected protein was calculated using the SEQUEST algorithm and reported as a range of up to 20 values derived independently from analysis of proteins from each of the strains examined.
Passive immunisation
5–6 week old female C57BL/6 mice were passively immunised with 800 μg of E-IVIG or IVIG in 100 μl of PBS by intraperitoneal injection. 16 h after IgG administration, the mice were challenged intramuscularly with 5 × 106 CFU of M1 S. pyogenes (Strain H305 which has been used previously in IVIG protection studies8). After 24 h, the mice were culled, the blood and organs were removed and bacterial loads at the infection site and the degree of dissemination were assessed by plating serial dilutions of each organ homogenate onto CBA. All animal procedures were approved by the local ethical review process at Imperial College London and conducted in accordance with the relevant, UK Home Office approved, project license.
Statistical analysis
Data are expressed as mean and standard deviation (bar charts and line graphs) or median and range (scatter plots) and analysed using the two-tailed student’s t-test or two-tailed Mann-Whitney U where appropriate. Statistical significance was accepted where *p < 0.05. Curve fits were calculated by non-linear regression (GraphPad Prism).
Additional Information
How to cite this article: Reglinski, M. et al. Identification of the Streptococcus pyogenes surface antigens recognised by pooled human immunoglobulin. Sci. Rep. 5, 15825; doi: 10.1038/srep15825 (2015).
Supplementary Material
Acknowledgments
The authors acknowledge Drs. Claire Turner and Lionel Tan for helpful comments during manuscript preparation. Funding: This work was supported by the UK Clinical Research Collaboration (Centre for Infection Prevention & Management G0800777), the Medical Research Council (Confidence in Concept Award) and NIHR (Imperial College Biomedical Research Centre).
Footnotes
Author Contributions M.R., R.J.E. and S.S. conceived the study, analysed the data and wrote the manuscript. M.R., M.G. and N.N.L. performed the experiments. M.R. and R.J.E. prepared the figures. R.J.E and S.S. jointly supervised the study. All authors approved the final version.
References
- O’Loughlin R. E. et al. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin. Infect. Dis. 45, 853–862 (2007). [DOI] [PubMed] [Google Scholar]
- Lamagni T. L. et al. Epidemiology of severe Streptococcus pyogenes disease in Europe. J. Clin. Microbiol. 46, 2359–2367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linner A., Darenberg J., Sjolin J., Henriques-Normark B. & Norrby-Teglund A. Clinical efficacy of polyspecific intravenous immunoglobulin therapy in patients with streptococcal toxic shock syndrome: a comparative observational study. Clin. Infect. Dis. 59, 851–857 (2014). [DOI] [PubMed] [Google Scholar]
- Carapetis J. R. et al. Effectiveness of Clindamycin and Intravenous Immunoglobulin, and Risk of Disease in Contacts, in Invasive Group A Streptococcal Infections. Clin. Infect. Dis. 59 (2014). [DOI] [PubMed] [Google Scholar]
- Darenberg J. et al. Intravenous immunoglobulin G therapy in streptococcal toxic shock syndrome: a European randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 37, 333–340 (2003). [DOI] [PubMed] [Google Scholar]
- Farag N., Mahran L., Abou-Aisha K. & El-Azizi M. Assessment of the efficacy of polyclonal intravenous immunoglobulin G (IVIG) against the infectivity of clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1149–1160 (2013). [DOI] [PubMed] [Google Scholar]
- Gaglani M. J., Baker C. J. & Edwards M. S. Contribution of antibody to neutrophil-mediated killing of Enterococcus faecalis. J. Clin. Immunol. 17, 478–484 (1997). [DOI] [PubMed] [Google Scholar]
- Sriskandan S., Ferguson M., Elliot V., Faulkner L. & Cohen J. Human intravenous immunoglobulin for experimental streptococcal toxic shock: bacterial clearance and modulation of inflammation. J. Antimicrob. Chemother. 58, 117–124 (2006). [DOI] [PubMed] [Google Scholar]
- Basma H. et al. Opsonic antibodies to the surface M protein of group A streptococci in pooled normal immunoglobulins (IVIG): potential impact on the clinical efficacy of IVIG therapy for severe invasive group A streptococcal infections. Infect. Immun. 66, 2279–2283 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrage B., Duan G. W., Yang L. P., Fraser J. D. & Proft T. Different preparations of intravenous immunoglobulin vary in their efficacy to neutralize streptococcal superantigens: Implications for treatment of streptococcal toxic shock syndrome. Clin. Infect. Dis. 43, 743–746 (2006). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ortega M. J. et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24, 191–197 (2006). [DOI] [PubMed] [Google Scholar]
- Severin A. et al. Proteomic Analysis and Identification of Streptococcus pyogenes Surface-Associated Proteins. J. Bacteriol. 189, 1514–1522 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y. D., Carlson B., Kondagunta A. & Cleary P. P. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65, 2080–2087 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallotta M. et al. SpyAD, a moonlighting protein of Group A Streptococcus contributing to bacterial division and host cell adhesion. Infect. Immun. 82, 2890–2901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A., Burne R. A. & Castro A. C. Molecular cloning, purification and immunological responses of recombinants GroEL and DnaK from Streptococcus pyogenes. FEMS Immunol. Med. Microbiol. 28, 121–128 (2000). [DOI] [PubMed] [Google Scholar]
- Turner C. E. et al. Emergence of a New Highly Successful Acapsular Group A Streptococcus Clade of Genotype emm89 in the United Kingdom. MBio 6, 10.1128/mBio.00622-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea P. R. et al. Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerg. Infect. Dis. 17, 2010–2017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres S. B. & Musser J. M. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2, 10.1371/journal.pone.0000800 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson-Smith M. et al. A Systematic and Functional Classification of Streptococcus pyogenes That Serves as a New Tool for Molecular Typing and Vaccine Development. J. Infect. Dis. 210, 1325–1338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. C. & Wessels M. R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30, 209–219 (1998). [DOI] [PubMed] [Google Scholar]
- Eriksson B. K., Andersson J., Holm S. E. & Norgren M. Invasive group A streptococcal infections: T1M1 isolates expressing pyrogenic exotoxins A and B in combination with selective lack of toxin-neutralizing antibodies are associated with increased risk of streptococcal toxic shock syndrome. J. Infect. Dis. 180, 410–418 (1999). [DOI] [PubMed] [Google Scholar]
- Miller L. S. & Cho J. S. Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 11, 505–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzer A. et al. Novel conserved group A streptococcal proteins identified by the antigenome technology as vaccine candidates for a non-M protein-based vaccine. Infect. Immun. 78, 4051–4067 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. N. et al. Surface analyses and immune reactivities of major cell wall-associated proteins of group A streptococcus. Infect. Immun. 73, 3137–3146 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombaci M. et al. Protein array profiling of tic patient sera reveals a broad range and enhanced immune response against Group A Streptococcus antigens. PLoS One 4, 10.1371/journal.pone.0006332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I., Germon P., McDade K. & Scott J. R. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69, 7029–7038 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C. E. et al. Molecular Analysis of an Outbreak of Lethal Postpartum Sepsis Caused by Streptococcus pyogenes. J. Clin. Microbiol. 51, 2089–2095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Siano B. & Diamond S. Neutrophil isolation protocol. J. Vis. Exp. 17, 10.3791/745 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield R. C. Persistence of type-specific antibodies in man following infection with group A streptococci. J. Exp. Med. 110, 271–292 (1959). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. F. & Fischetti V. A. The importance of the location of antibody binding on the M6 protein for opsonization and phagocytosis of group A M6 streptococci. J. Exp. Med. 167, 1114–1123 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Salam V. B. et al. Proteomic analysis of lung tissues from patients with pulmonary arterial hypertension. Circulation 122, 2058–2067 (2010). [DOI] [PubMed] [Google Scholar]
- Consortium T. U. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Research 42, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P. et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192, 771–782 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.