Highlight
The wheat NDR-type AGC kinase TaAGC1 positively regulates host defence response to the necrotrophic phytopathogen Rhizoctonia cerealis through modulating expression of ROS-related and defence-associated genes.
Key words: AGC kinase, differential expression, reactive oxygen species, resistance, Rhizoctonia cerealis, Triticum aestivum, wheat.
Abstract
Considerable progress has been made in understanding the roles of AGC kinases in mammalian systems. However, very little is known about the roles of AGC kinases in wheat (Triticum aestivum). The necrotrophic fungus Rhizoctonia cerealis is the major pathogen of the destructive disease sharp eyespot of wheat. In this study, the wheat AGC kinase gene TaAGC1, responding to R. cerealis infection, was isolated, and its properties and role in wheat defence were characterized. R. cerealis-resistant wheat lines expressed TaAGC1 at higher levels than susceptible wheat lines. Sequence and phylogenetic analyses showed that the TaAGC1 protein is a serine/threonine kinase belonging to the NDR (nuclear Dbf2-related) subgroup of AGC kinases. Kinase activity assays proved that TaAGC1 is a functional kinase and the Asp-239 residue located in the conserved serine/threonine kinase domain of TaAGC1 is required for the kinase activity. Subcellular localization assays indicated that TaAGC1 localized in the cytoplasm and nucleus. Virus-induced TaAGC1 silencing revealed that the down-regulation of TaAGC1 transcripts significantly impaired wheat resistance to R. cerealis. The molecular characterization and responses of TaAGC1 overexpressing transgenic wheat plants indicated that TaAGC1 overexpression significantly enhanced resistance to sharp eyespot and reduced the accumulation of reactive oxygen species (ROS) in wheat plants challenged with R. cerealis. Furthermore, ROS-scavenging and certain defence-associated genes were up-regulated in resistant plants overexpressing TaAGC1 but down-regulated in susceptible knock-down plants. These results suggested that the kinase TaAGC1 positively contributes to wheat immunity to R. cerealis through regulating expression of ROS-related and defence-associated genes.
Introduction
Common wheat (Triticum aestivum L.) is a widely cultivated and consumed staple crop, feeding ~35% of the world’s population (Scofield et al., 2005). Sharp eyespot is a devastating disease of wheat globally (Chen et al., 2008, 2013; Hamada et al., 2011; Lemańczyk and Kwaśna, 2013) and since 2005 the epidemic has become severe across large parts of China, with 9.33 m ha of wheat affected by sharp eyespot in 2015 (http://www.agri.cn/V20/bchqb/201501/t20150121_4344729.htm). The main agent of wheat sharp eyespot is Rhizoctonia cerealis van der Hoeven, a soil-borne fungus with bi-nucleated hyphal cells (Chen et al., 2013). Sharp eyespot manifests as ‘eye’-shaped lesions on basal sheaths and stems of infected wheat plants. The disease can destroy the transport tissues in stems of plants and block transportation of substances required for nutrition, leading to yield losses of ~10–40%. Breeding resistant wheat varieties is an effective and environmentally safe approach to control disease. However, traditional resistance breeding is difficult because the resistance in wheat accessions is partial and quantitative (Cai et al., 2006; Chen et al., 2013). The wheat defence response to R. cerealis is complicated and involves a series of gene expression changes. To improve wheat resistance to sharp eyespot, it is vital to identify genes that play an important role in the defence and unravel their underlying functional mechanisms.
Plant protein kinases, including receptor-like kinases, regulate recognition and early responses to diverse signals, and frequently play critical roles in various developmental and physiological processes, including defence and symbiosis (Rentel et al., 2004; AbuQamar et al., 2008; Fu et al., 2009; Garcia et al., 2012). The AGC kinase [named after cAMP-dependent protein kinase A (PKA), cGMP-dependent protein kinase G (PKG) and phospholipid-dependent protein kinase C (PKC)] family is a group of serine/threonine (Ser/Thr) protein kinases that share sequence similarities in their catalytic kinase domains with PKA, PKG and PKC (Pearce et al., 2010), and is conserved among eukaryotic genomes. In mammalian systems, AGC kinases are important regulators of cell growth, metabolism and cell death (Pearce et al., 2010). Although the catalytic kinase domains of plant AGC kinases share sequence similarities with those of mammalian AGC kinases, little is known about their molecular functions and targets (Garcia et al., 2012). In the Arabidopsis genome, there are 39 AGC kinase members (Bogre et al., 2003). Based on a catalytic domain comparison, these AGC kinases were classified into six subfamilies: PDK1, AGC VI, AGC VII (including NDR kinases), AGC VIIIa, AGC VIIIb and ‘other’ (Bogre et al., 2003). To date, in three model plant species (Arabidopsis, tomato, and rice), several AGC kinases have been implicated in growth, morphogenesis and defence responses to pathogens (Devarenne et al., 2006; Matsui et al., 2010; Camehl et al., 2011; Garcia et al., 2012). For example, in tomato, an AvrPto-dependent Pto-interacting protein 3 (Adi3), a member of the AGC VIIIb subfamily, is phosphorylated by the resistance kinase Pto and 3-phosphoinositide- dependent protein kinase-1 (Pdk1), and negatively regulates cell death that is associated with P. syringae pv. tomato (Devarenne et al., 2006). In Arabidopsis, OXIDATIVE SIGNAL-INDUCIBLE1 (AtOXI1) is an AGC VIIIb kinase and is required for immunity against the pathogens Hyaloperonospora arabidopsidis (an oomycete) and Pseudomonas syringae pv. tomato DC3000 (a bacterial biotrophic pathogen) (Rentel et al., 2004; Petersen et al., 2009). The rice AGC kinase OsOxi1, an orthologue of AtOXI1, phosphorylates the residue Thr-233 of OsPti1a and then releases OsPti1a-mediated inhibition of disease resistance, resulting in positive contribution to basal resistance to the blast fungus Magnaporthe oryzae pv. oryzae (Matsui et al., 2010). These AGC kinases belong to the AGC VIIIb subfamily. In different plant species, AGC kinases may have different regulatory roles in response to abiotic stresses. Thus, the isolation and functional study of species-specific AGC kinase genes are important. The large genome size and polyploidy of common wheat has hampered the isolation and functional analysis of wheat genes. So far, in NCBI databases, only seven sequences of common wheat AGC kinases (GenBank accession numbers: CDM81915.1, CDM82518.1, BAF79635.1, BAD19066.1, BAD19068.1, CDM81704.1, CDM85216.1) have been identified by us based on the activation loop signature motif (Bogre et al., 2003) of AGC kinases. However, no functional study of AGC kinase in common wheat has been reported yet.
In early responses to biotic and abiotic stresses, plants often generate an excessive amount of reactive oxygen species (ROS) (Shetty et al. 2008; Dong et al., 2013; Schmidt et al., 2013). Necrotroph-induced ROS include superoxide anion (O2 −) and hydrogen peroxide (H2O2) (Shetty et al., 2008). The levels of several enzymes involved in ROS-scavenging and antioxidative defence, such as superoxide dismutase (SOD), peroxidase (POX), and catalase (CAT), were significantly altered in Alternaria-tolerant Brassica lines after infection with Alternaria brassicae and in tomato infected with Botrytis cinerea, indicating that ROS-scavenging enzymes and antioxidants may play significant roles in plant defences against these necrotrophic pathogens (Kuźniak and Skłodowska 2004; Sharma et al., 2007). How the ROS signal is perceived is currently unknown. However, a signal transduction cascade, including the activation of protein kinases and downstream transcription factors, has been proposed in Arabidopsis (Mittler et al., 2004; Schmidt et al., 2013). The Arabidopsis AGC kinase AtOXI1 is necessary for the ROS signalling pathway (Rentel et al., 2004). However, in wheat, very little is known about the roles of AGC kinases in the regulation of ROS signalling during plant defence responses to necrotrophic pathogens.
In this study, to isolate kinase genes involved in wheat defence response to R. cerealis, Agilent wheat microarray chips were used to identify wheat kinase genes expressed differentially between the sharp eyespot-resistant wheat CI12633 and the susceptible wheat Wenmai 6, following infection with R. cerealis. We identified the probe corresponding to the 3′-terminal sequence of the wheat AGC kinase gene TaAGC1, which showed higher expression in resistant wheat lines (CI12633 and Shanhongmai) than in susceptible wheat line Wenmai 6 inoculated with R. cerealis. Then, the TaAGC1 gene was cloned from resistant wheat line CI12633, and further characterized. The TaAGC1 protein was evidenced to have kinase activity. The role of TaAGC1 in host defence responses to R. cerealis was explored using both TaAGC1 silencing- and overexpressing- wheat plants. The results showed that TaAGC1, acting as a positive regulator, mediated wheat resistance responses to R. cerealis.
Materials and methods
Plant and fungal materials and treatments
Five wheat (T. aestivum) lines/cultivars—sharp eyespot-resistant CI12633 and Shanhongmai, moderately-susceptible Yangmai 158 and Yangmai 20, and susceptible Wenmai 6 (Supplementary Table S1)—were used in this research. A major Jiangsu virulent isolate of the pathogen R. cerealis, R0301, was provided by Profs Huaigu Chen and Shibin Cai of Jiangsu Academy of Agricultural Sciences, China.
Wheat plants were grown in a 14h light/10h dark (23°C/10°C) regime. At the tillering stage, the second base sheath of each wheat plant was inoculated with small toothpick fragments harbouring well-developed mycelia of R. cerealis. Inoculated plants were grown at 90% relative humidity for 4 d. The inoculated stems, leaves or roots were sampled at 0, 4, 7 and 21 d post-inoculation (dpi). Additionally, the leaves of CI12633 seedlings at the three-leaf stage were sprayed with 10mM H2O2 solution for 0, 1, 3, 6 and 12h, following the protocol of Zhu et al. (2014). The samples were harvested, frozen quickly in liquid nitrogen, and stored at −80°C prior to total RNA extraction.
DNA or RNA extraction and cDNA synthesis
Genomic DNA for each sample was isolated from the wheat leaves using the CTAB method (Saghai-Maroof et al., 1984). Total RNA was extracted using TRIzol (Invitrogen), and then subjected to RNase-free DNase I (Promega) digestion and purification. The purified RNA samples (4 µg each) were separately reverse-transcribed to cDNA.
Microarray hybridizations and data analysis
For each sample total RNA was extracted from the sheaths of 10 seedlings from sharp eyespot-resistant wheat lines (CI12633 and Shanhongmai) or the susceptible wheat cv. Wenmai 6 at 4 or 21 dpi with R. cerealis. After purification, the RNA was used for the preparation of Cy5- and Cy3-labelled complementary RNA. These probes of Cy3-labelled CI12633/Shanhongmai RNA and Cy5-labelled Wenmai 6 RNA were hybridized with Agilent Wheat Gene Expression Microarray containing 43 803 probe sets according to the manufacturer’s protocols. The hybridization signals were scanned with an Agilent G2505C Microarray Scanner System, and microarray data were extracted using Feature Extraction Software (v.10.7.1.1) available from Agilent by using the default variables. Data files were loaded into GeneSpring GX 11.5 (Agilent Technologies). The signal values between both experimental groups (CI12633 or Shanhongmai probe vs Wenmai 6 probe) were assessed for each gene based on the normalized probe signals. Differentially expressed kinase genes between CI12633 or Shanhongmai and Yangmai 12 were filtered out by a fold change threshold ≥2.0.
Isolation and characterization of the TaAGC1 sequence
The microarray analysis indicated that a probe (TIGR number: TC411796) of the Agilent wheat microarray, the 3′-terminal partial sequence of TaAGC1 cDNA, was expressed at a significantly higher level in the resistant wheat CI12633 than in the susceptible wheat Wenmai 6. TC411796 was homologous to a wheat AGC kinase gene WNDr1A (GenBank accession AB179755) in the National Center for Biotechnology Information (NCBI) database. The 3′-untranslated region (UTR) and full-length open reading frame (ORF) sequences of TaAGC1 were separately amplified using 3′-Full RACE Core Set kit v.2.0 (TaKaRa) or RT-PCR from cDNA of CI12633 stems inoculated with R. cerealis for 4 d. The deduced protein sequence was analysed using the Compute pI/Mw tool (http://web.expasy.org/compute_pi/) to determine the theoretical iso-electric point (pI) and molecular weight (MW), interPro-Scan (http://www.ebi.ac.uk/interpro/) to identify domains and Smart software (http://smart.embl-heidelberg.de/) to predict conserved motifs. A phylogenetic tree was constructed using a neighbor-joining method implemented with MEGA 5.0 software followed by an alignment with other protein kinases using ClustalX software.
Autophosphorylation and phosphorylation activity assay of TaAGC1 kinase in vitro
The residue 239 aspartate (D) at the Ser/Thr kinase active-site of TaAGC1 protein was replaced with alanine (A) to generate the D239A mutant of TaAGC1. To investigate if the TaAGC1 protein is an active kinase and the D239 is important for the kinase activity, the ORF sequence of TaAGC1 or the D239A mutant was separately subcloned in-frame to the 3′-terminus of a glutathione S-transferase (GST) gene in the pGEX-4T-1 vector (GE Healthcare). The resulting pGST-TaAGC1 and mutant pGST-D239A fusion constructs were transformed into competent cells of Escherichia coli BL21 (DE3), respectively. The recombinant GST-TaAGC1 and GST-D239A proteins were separately expressed after treatment with 0.1mM isopropyl-β-D-thiogalactopyranose at 16°C for 19h, and purified using a MicroSpin module (GE Healthcare).
Autophosphorylation and myelin basic protein (MBP) phosphorylation assays were carried out using a reaction mixture (10 μl) consisting of 40mM Hepes-NaOH (pH 8.0), 5mM Mg(CH3COO)2, 0.1mM EGTA, 2mM dithiothreitol, 100mM [γ32P]ATP and ~100ng of GST-TaAGC1 or GST-D239A. For phosphorylation experiments of protein substrates, 1 μg MBP was incubated with ~100ng of GST-TaAGC1 or GST-D239A under the phosphorylation conditions described above. The reaction was incubated at 30°C for 30min. Then, 2 μl SDS sample buffer was added to the phosphorylation mixture to stop the reaction. The autophosphorylated and phosphorylated proteins were resolved by SDS-PAGE and analysed by autoradiography. After autoradiography, the proteins were stained for 10min by Coomassie Brilliant Blue (CBB) and then photographed.
Subcellular localization of TaAGC1 in onion and wheat
The coding region of TaAGC1, lacking a stop codon, was amplified using the primers TaAGC1-GFP-F/TaAGC1-GFP-R. The amplified fragment was sub-cloned in-frame with the 5′-terminus of the green fluorescent protein’s (GFP) coding region in the p35S:GFP vector, resulting in the TaAGC1 fusion construct p35S:TaAGC1-GFP.
The resulting p35S:TaAGC1-GFP and p35S:GFP constructs were separately bombarded into white onion epidermal cells, following the protocol of Zhang et al. (2007), or were introduced into wheat mesophyll protoplasts via the PEG-mediated transfection method, following the protocol of Yoo et al. (2007). The transformed wheat mesophyll protoplasts or onion epidermal cells were incubated at 25°C for 15h. GFP signals were observed and photographed using a confocal laser scanning microscope (Zeiss LSM 700) with a Fluar 10X/0.50 M27 objective lens and SP640 filter.
Virus-induced gene silencing assay for TaAGC1 function
The virus-induced gene silencing (VIGS) assay in barley and wheat was developed using the barley stripe mosaic virus (BSMV) (Holzberg et al., 2002). BSMV-VIGS technique is an effective reverse genetic tool for rapidly investigating the functions of genes (Scofield et al., 2005; Zhou et al., 2007; Zhu et al., 2014). To generate the BSMV:TaAGC1 recombinant construct, a 302-bp sequence of TaAGC1 (from 1634 to 1935 nucleotides in TaAGC1 cDNA sequence) was sub-cloned in an antisense orientation into the NheI restriction site of the RNAγ of BSMV (Supplementary Fig. S1). Following the protocols described by Holzberg et al. (2002) and Zhou et al. (2007), the tripartite cDNA chains of BSMV:TaAGC1 or the control BSMV:GFP virus genome were separately transcribed into RNAs, mixed, and used to infect CI12633 plants at the three-leaf stage. At 10 d after infection, the fourth leaves of the inoculated seedlings were collected to monitor BSMV infection, based on the transcription of the BSMV coat protein (CP) gene, and to evaluate the transcript changes of TaAGC1. At 14 d after BSMV infection, the BSMV-infected CI12633 plants were inoculated with R. cerealis. They were scored at 14 and 40 dpi with R. cerealis, respectively.
TaAGC1-overexpressing transformation vector and wheat transformation
The TaAGC1 ORF sequence was amplified with the primers including the SpeI and SacI restriction sequences, and then subcloned into the SpeI and SacI sites of a modified monocot transformation vector pAHC25 (Christensen and Quail, 1996) with a c-myc epitope tag, resulting in the overexpression transformation vector pUBI:myc-TaAGC1. In the construct, the transcript of the myc-TaAGC1 fusion gene was driven by a maize ubiquitin (Ubi) promoter and terminated by 3′ non-transcribed region of Agrobacterium tumefaciens nopaline synthase gene (Tnos). A total of 1500 immature embryos of the wheat cultivar Yangmai 20 were transformed by biolistic bombardment using pUBI:myc-TaAGC1 following the protocol described by Chen et al. (2008).
PCR and western blot analyses on TaAGC1-overexpressing transgenic wheat
The presence of the TaAGC1-overexpressing transgene in the transformed wheat plants was monitored by PCR using the specific primers, TaAGC1-1584F (located in TaAGC1 coding sequence) and Tnos-R (in Tnos of the transformation vector). PCR was performed in a 20-μl volume containing 200ng genomic DNA, 1×PCR buffer (TaKaRa), 0.4 μM each primer, 200 μM each dNTP and 1U Taq polymerase (TaKaRa). The amplified product (386bp) specific to the introduced TaAGC1-Tnos chimera was resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
The myc-TaAGC1 fusion protein in the overexpressing transgenic wheat lines was visualized by western blotting analysis. Total proteins were extracted from 0.3g of ground leaf powder. Total soluble proteins (~12 μg) for each line were separated on 12% SDS-PAGE and transferred to polyvinyl difluoride membranes (Amersham Biosciences). The western blots were incubated with 100-fold diluted anti-c-myc antibody followed by secondary antibody conjugated to horseradish peroxidase (GE Healthcare). The myc-TaAGC1 proteins were visualized using the ECL Western Blot Detection and Analysis System (GE Healthcare).
Analysis of the transcription levels of target genes
Quantitative RT-PCR (qRT-PCR) was used to investigate the relative expression levels of TaAGC1 and the genes encoding ROS-scavenging enzymes (POX2, CAT1 and SOD3) and ROS-producing enzyme NADPH oxidase (NOX), and four defence-associated genes (defensin, nsLTP1, chitinase 2 and PR10) in wheat. qRT-PCR was performed using SYBR Green I Master Mix (TaKaRa) in a volume of 25 μl on an ABI 7300 RT-PCR system (Applied Biosystems). Reactions were set up using the following thermal cycling profile: 95°C for 5min, followed by 41 cycles of 95°C for 15 s and 60°C for 31 s. The products were examined using a melting curve analysis program. Three biological replications were run. The relative expression of the target genes was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), where the wheat Actin gene, TaActin, was used as the internal reference. RT-PCR was used to investigate the transcription of BSMV CP gene in VIGS experimental wheat plants.
The accession number of all the genes and sequences of all the primers in the study are listed in Supplementary Table S2.
Assessment on R. cerealis responses
In T1−T3 generations, 30 plants for each line of the TaAGC1-overexpressing transgenic and WT wheat were inoculated with small toothpicks harbouring the well-developed mycelia of R. cerealis following the protocol of Chen et al. (2008). The infection types (ITs) of wheat plants and disease index for each line were evaluated at 50 dpi. Based on the disease lesion squares on the base stems (Supplementary Fig. S2), ITs were categorized from 0 to 5: 0, no lesion; 1, the lesion appeared on the sheaths rather than stems; 2, the width of the lesion is <50% of the infected stem perimeter; 3, the width of the lesion is >50 and <75% of the infected stem perimeter; 4, the width of the disease lesion is >75% of the infected stem perimeter; 5, white spike or dead plant). Disease index ={(0×X0+1×Xl+2×X2+3×X3+4×X4+5×X5)/[(X0+X1+X2+X3+X4+X5)×5]}×100, where X0−X5 indicated plants with IT: 0−5.
Assay of ROS level
In situ H2O2 and O2 − accumulations were examined via histochemical staining by 3, 3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT), respectively, following the protocols of Lee et al. (2002). Briefly, leaves were harvested at the indicated time-points after R. cerealis infection, and floated in a solution of 1mg ml−1 DAB-HCl (pH 3.8) at 25°C for 8h or in NBT (pH 7.8) at 25°C for 30min, and then chlorophyll was removed with 95% ethanol in a boiling-water bath.
Results
TaAGC1 transcription is associated with defence response to R. cerealis
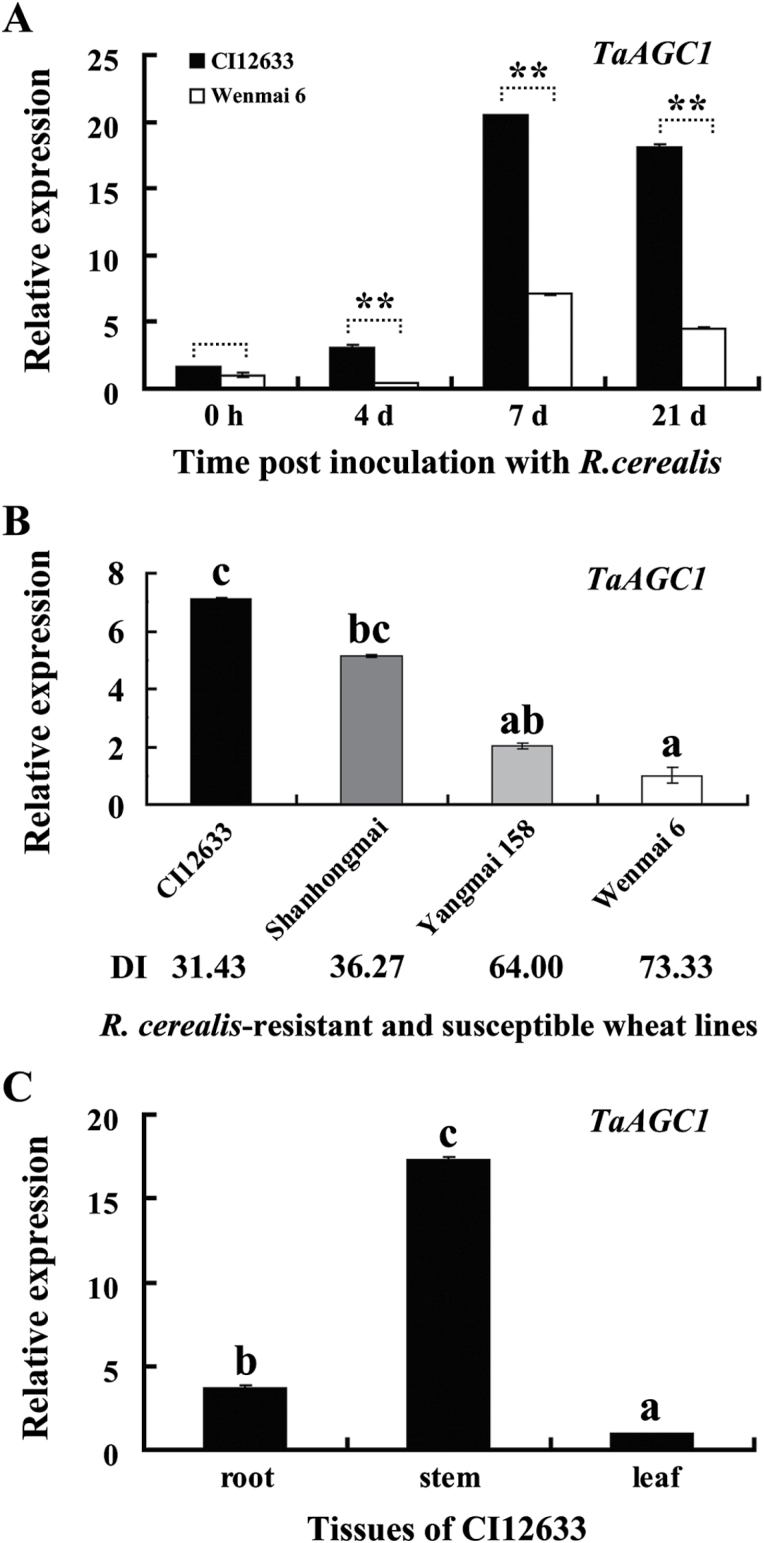
To identify kinase genes involved in the defence response to R. cerealis, the hybridization data of the Agilent wheat microarray chips were mined for wheat kinase genes that were differentially expressed between sharp eyespot-resistant wheat lines CI12633/Shanhongmai and the susceptible wheat line Wenmai 6 inoculated with R. cerealis for 4 and 21 dpi. Among differentially expressed kinase gene sequences identified from microarray data (GEO accession number GSE69245), the transcriptional level of TC411796 (probe), a cDNA 3′-terminal fragment of a wheat AGC kinase, was higher in resistant wheat lines CI12633/Shanhongmai than in susceptible wheat line Wenmai 6. For example, the probe TC411796 showed a 3.01-fold and 2.90-fold transcriptional increase in the resistant wheat line CI12633 than in the susceptible wheat line Wenmai 6 at 4 and 21 dpi, respectively. Being homologous to AGC kinases, this protein kinase gene was designated as TaAGC1. qRT-PCR analyses showed that the transcription of TaAGC1 was induced after R. cerealis inoculation, and the transcriptional induction was higher in resistant CI12633 than in susceptible Wenmai 6 at 4, 7 or 21 dpi with R. cerealis (Fig. 1A). The qRT-PCR results were in agreement with the microarray analysis trend. Furthermore, the gene transcriptional level was the highest in resistant wheat line CI12633, and the lowest in the highly-susceptible cultivar Wenmai 6 (Fig. 1B). Additionally, the transcript abundance of TaAGC1 was the highest in stems that are main disease-occurring sites, intermediate in roots, and lowest in the leaves of CI12633 (Fig. 1C). These results suggested that TaAGC1 transcription might participate in the defence response to R. cerealis in resistant wheat lines.
Fig. 1.
Transcription of the AGC kinase gene TaAGC1 in Rhizoctonia cerealis-inoculated wheat (Triticum aestivum) as measured by qRT-PCR. (A) Transcription of TaAGC1 in stems of the R. cerealis-resistant wheat line CI12633 and susceptible wheat cultivar Wenmai 6 at 0h, and 4, 7 and 21 dpi with R. cerealis. The expression level of TaAGC1 in Wenmai 6 plants at 0 dpi is set to 1. The expression levels of TaAGC1 in CI12633 are relative to those in Wenmai 6 plants at each time point (0h, 4, 7 or 21 dpi). Statistically significant differences between CI12633 and Wenmai 6 at the same time point are derived from the results of three independent replications (t-test: **, P<0.01). (B) Expression patterns of TaAGC1 in four wheat cultivars with different degrees of resistance at 21 dpi with R. cerealis. DI indicates disease index of sharp eyespot. The expression level of TaAGC1 in the Wenmai 6 plants was set to 1. (C) Transcription of TaAGC1 in roots, stems and leaves of the R. cerealis-resistant line CI12633 at 21 dpi with R. cerealis. The transcriptional levels and standard error are derived from the average of three biological replicates. The transcript levels with different letters are significantly different from each other based on statistical comparisons (t-test; P<0.01).
TaAGC1 encodes an NDR-type AGC kinase
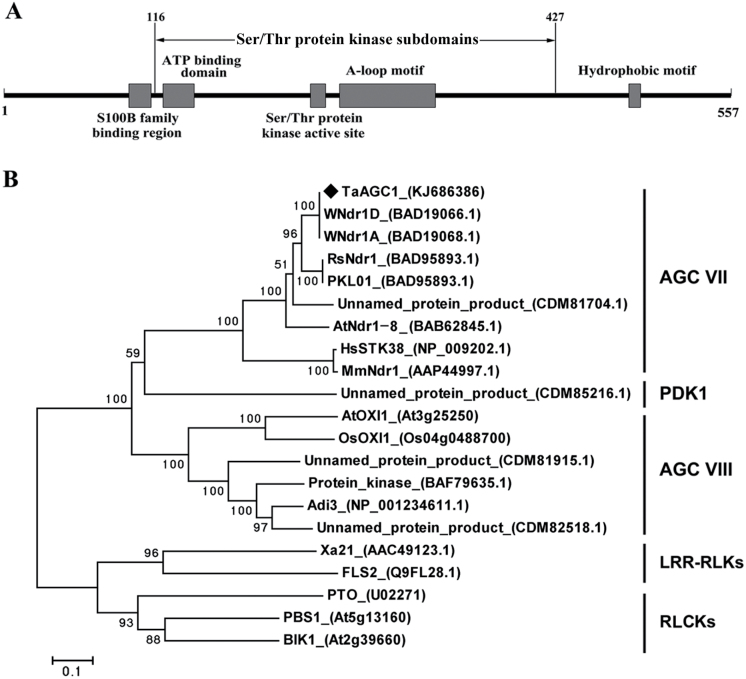
The full-length of TaAGC1 cDNA sequence (GenBank accession KJ686386) was cloned from a R. cerealis-infected stem of CI12633. A sequence analysis showed that the cDNA sequence (2077 nucleotides) of TaAGC1 includes an ORF consisting of 1674 nucleotides (from 67 to 1740) (Supplementary Fig. S3). The deduced TaAGC1 protein consists of 557 amino acid residues, with a MW of 64.14 kD and pI of 8.65. As shown in Fig. 2A and Supplementary Fig. S3, the TaAGC1 protein contains an N-terminal S100B family-binding region (amino acid 96–113), the 12 Ser/Thr protein kinase subdomains (amino acid 116–427) that harbours an ATP-binding domain (amino acid 122−146), the kinase active-site (amino acid 235−247) and an activation loop (A-loop) motif (amino acid 257−331), and a C-terminal hydrophobic motif (amino acid 479−488).
Fig. 2.
Domain and phylogenetic analyses of the wheat (Triticum aestivum) AGC kinase TaAGC1. (A) Schematic diagram of the domain structure of the TaAGC1 protein. Individual domains of TaAGC1 are represented by grey boxes, and the names of motifs are indicated on top or below. (B) Phylogenetic analysis of the deduced amino acid sequences of TaAGC1. The bootstrapped phylogenetic tree is constructed using the neighbor-joining phylogeny of MEGA 5.0. The black diamond indicates the position of TaAGC1. LRR-RLKs indicates leucine-rich repeat receptor-like kinases; RLCKs indicates receptor-like cytoplasmic kinases.
BLAST and sequence analyses indicated that the TaAGC1 protein sequence was homologous to those of plant Ser/Thr protein kinases and shared the characteristic activation loop signature motif (DFGx60STVGTPDYIAPE) of the NDR subclass of the AGC kinase family (Bogre et al., 2003; Supplementary Fig. S3). The reconstructed phylogenetic tree analysis (Fig. 2B) indicated that four wheat AGC kinases including TaAGC1, WNdr1A (GenBank accession no. BAD19068.1), WNdr1D (GenBank accession no. BAD19066.1) and an unnamed wheat AGC protein (GenBank accession no. CDM81704.1), and NDR protein kinases from other species, including the Arabidopsis thaliana NDR-type AGC AtNdr-8 (At5g09890, GenBank accession no. BAB62845), radish NDR kinase RsNdr1 (GenBank accession no. BAC76894), Lotus japonicus NDR kinase PKL01 (GenBank accession no. BAD95893), human NDR kinase STK38 (GenBank accession no. NP_009202.1) and Mus musculus NDR kinase MmNdr1 (GenBank accession no. AAP44997), were clustered on the same branch. All of them belong to AGC VII subfamily. AtOXI1, OsOXI1, Adi3 and three unnamed wheat AGC kinases (GenBank accession nos CDM81915.1, BAF79635.1, and CDM82518.1) were clustered in the other clade, which belonged to the AGC VIIIb subfamily. Another unnamed wheat AGC kinase (GenBank accession no. CDM85216.1) belonged to the PDK1 subfamily. However, the leucine-rich repeat receptor-like kinases, such as rice Xa21 (GenBank accession no. AAC49123.1) and Arabidopsis FLS2 (GenBank accession no. Q9FL28.1), and receptor-like cytoplasmic kinases, such as tomato PTO, Arabidopsis PBS1 (At5g13160) and BIK1 (At2g39660), were separately clustered into two out-groups, which do not belong to the AGC kinase family (Fig. 2B). The results suggested that the TaAGC1 protein was a member of the NDR-type AGC kinases.
TaAGC1 possesses autophosphorylation and phosphorylation capability
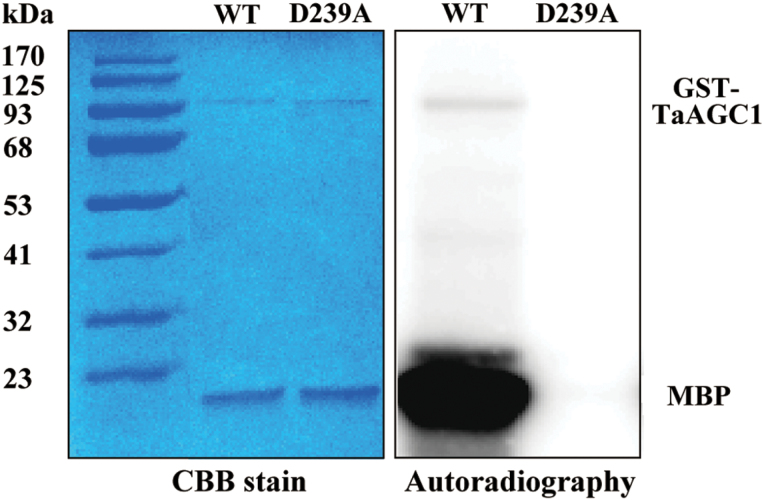
To investigate whether TaAGC1 is an active kinase, autophosphorylation and MBP phosphorylation of TaAGC1 were studied. The GST-TaAGC1 (wild-type) fusion protein with 93.2 KD and the GST-D239A (mutant) protein were separately expressed in E. coli cells and purified by affinity chromatography. In D239A mutant of TaAGC1, Asp-239 (D) at the Ser/Thr kinase domain active-site was replaced with Ala (A). The in vitro kinase assay showed that through detection by autoradiography, TaAGC1 autophosphorylated itself and phosphorylated the substrate MBP, but the D239A mutant did not display the autophosphorylation and phosphorylation activity. These results indicated that TaAGC1 is a functional kinase with autophosphorylation activity and phosphorylation activity, and that the Asp-239 residue of TaAGC1 is required for the TaAGC1 kinase activity (Fig. 3).
Fig. 3.
Analysis of the kinase activity of the TaAGC1 protein. The assays on autophosphorylation of TaAGC1 and myelin basic protein (MBP) phosphorylation by TaAGC1 were incubated for 30min in kinase buffer containing purified GST-TaAGC1 (WT) or GST-D239A mutant protein, the MBP substrate and [γ-32P] ATP. The phosphorylated proteins were separated by SDS-PAGE and visualized by autoradiography. After autoradiography, the right proteins were confirmed by Coomassie Brilliant Blue (CBB) staining. (This figure is available in colour at JXB online.)
TaAGC1 localizes in the cytoplasm and the nucleus
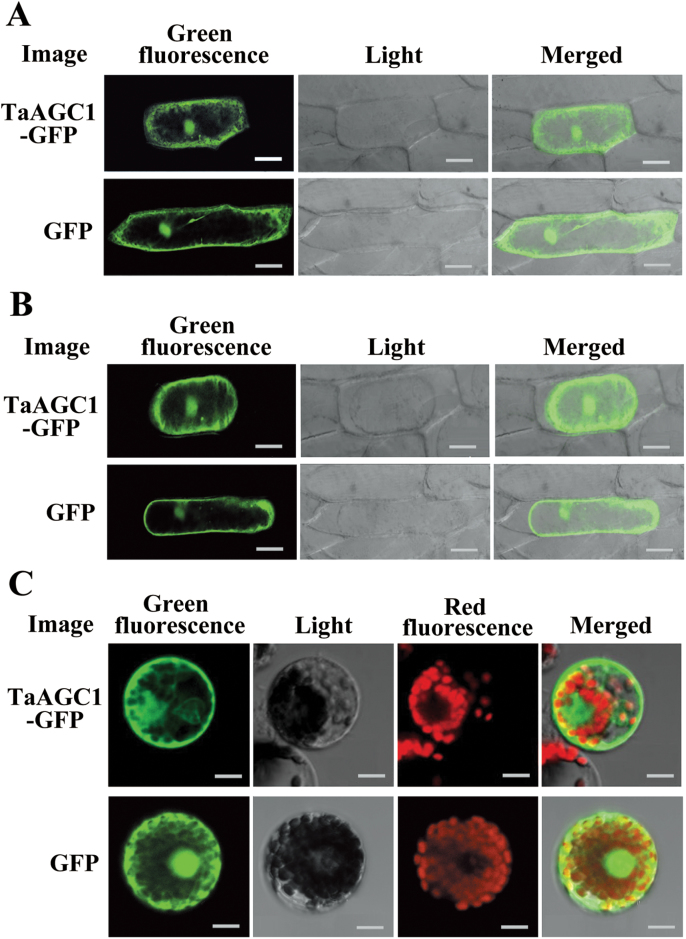
To explore the subcellular localization of TaAGC1, 35S:TaAGC1-GFP was transiently expressed either in onion epidermal cells or in wheat mesophyll protoplasts. At the same time, the transient expression of 35S:GFP was used as control. In the transient transformed onion epidermal cells, the TaAGC1-GFP protein was localized predominantly in the cytoplasm and nucleus (Fig. 4A). After plasmolysis of the bombarded onion epidermal cells, localization of TaAGC1 was more clearly observed in the cytoplasm and nucleus (Fig. 4B). Furthermore, the green fluorescent signal of the 35S:TaAGC1-GFP protein in wheat mesophyll protoplasts was detected in the cytoplasm and nucleus (Fig. 4C). The green fluorescent signal of the 35S:GFP protein was distributed in the nucleus and cytoplasm (Fig. 4A−C). Thus, the TaAGC1 protein localizes in the cytoplasm and nucleus in wheat.
Fig. 4.
Subcellular localization of the wheat (Triticum aestivum) AGC kinase TaAGC1-green fluorescent protein (GFP) fusion protein in onion and wheat. (A) The fused TaAGC1-GFP (upper panel) and control GFP (lower panel) in onion epidermal cells. (B) The fused TaAGC1-GFP (upper panel) and control GFP (lower panel) in plasmolysed onion epidermal cells. Bars, 50 μm (A, B). (C) The fused TaAGC1-GFP (upper panel) and control GFP (lower panel) in wheat protoplasts. Bar, 10 μm. (This figure is available in colour at JXB online.)
Knock-down of TaAGC1 impairs wheat resistance to R. cerealis
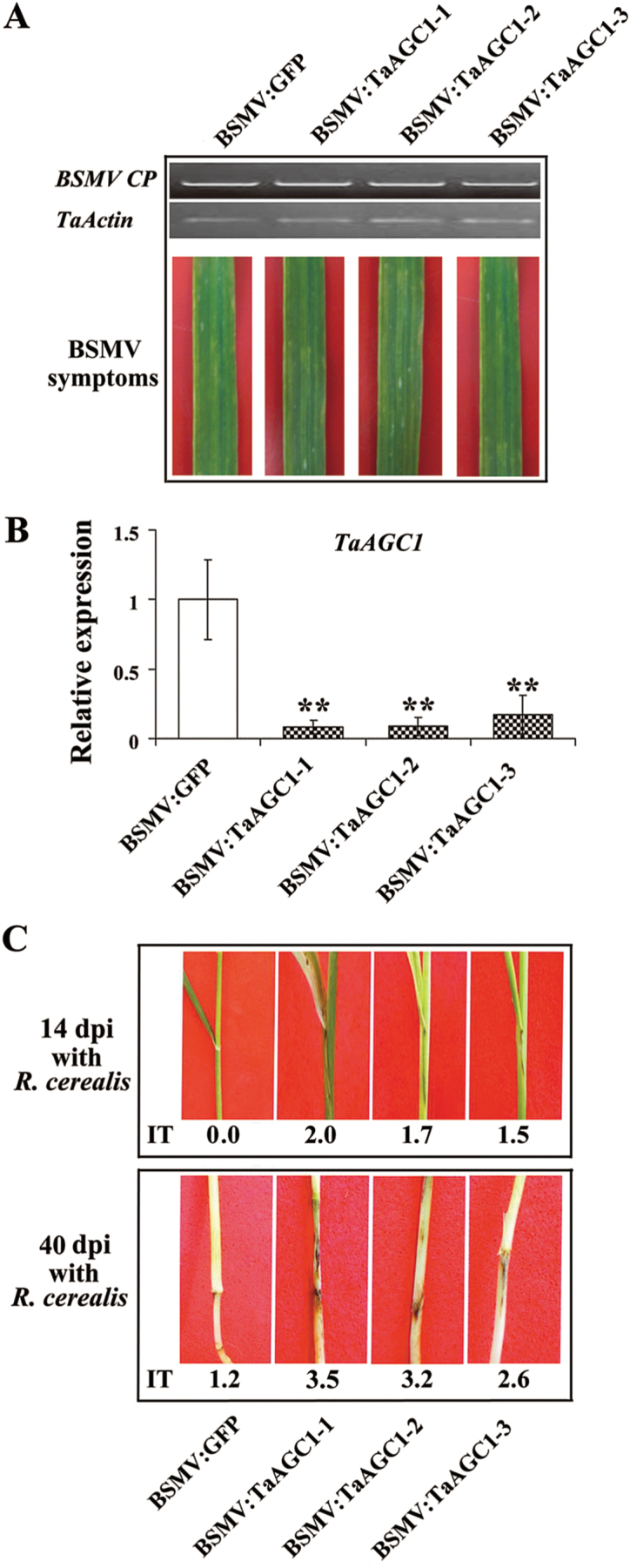
After the resistant-wheat CI12633 plants had been infected with BSMV for 10 d, symptoms and transcript analyses indicated that BSMV was present in both BSMV:GFP- and BSMV:TaAGC1-infected CI12633 plants (Fig. 5A). The TaAGC1 transcript level was substantially reduced in BSMV:TaAGC1-infected CI12633 plants, indicating that TaAGC1 was successfully silenced in BSMV:TaAGC1-infected CI12633 plants (Fig. 5B). Then, these plants were further inoculated with R. cerealis to evaluate the defence role of TaAGC1. At 14 dpi with R. cerealis, a dark brown margin, an early symptom of sharp eyespot, was present on the sheaths of BSMV:TaAGC1-infected CI12633 plants, but absent in BSMV:GFP-treated CI12633 plants. At 40 dpi with R. cerealis, obvious symptoms of sharp eyespot were present on the sheaths and stems of BSMV:TaAGC1-infected CI12633 plants, but not on the stems of BSMV:GFP-treated CI12633 plants (Fig. 5C). Compared with BSMV:GFP-treated CI12633 plants, no obvious phenotypic change was observed in BSMV:TaAGC1-infected wheat CI12633 plants except the sharp eyespot resistance alteration. These results indicated that the down-regulation of TaAGC1 transcripts in CI12633 impaired host resistance to R. cerealis, and that TaAGC1 is required for the wheat defence against R. cerealis infection.
Fig. 5.
Effect of knock-down of the wheat (Triticum aestivum) AGC kinase TaAGC1 gene on the resistance response of the resistant wheat strain CI12633 to Rhizoctonia cerealis. (A) RT-PCR analysis of the transcription levels of the barley stripe mosaic virus (BSMV) CP gene and typical symptoms of BSMV in the fourth leaves of wheat plants infected by BSMV:GFP or BSMV:TaAGC1. (B) qRT-PCR analysis of the relative transcript levels of TaAGC1 in the wheat plants infected by BSMV:GFP or BSMV:TaAGC1 at 14 dpi with R. cerealis. The relative transcript level of TaAGC1 in the wheat CI12633 plants infected by BSMV:TaAGC1 is relative to that in BSMV:GFP-infected wheat CI12633 plants. Three biological replicates per line were averaged and statistically treated (t-test; ** P<0.01). Bars indicate standard error of the mean. (C) Sharp eyespot symptoms of the BSMV:GFP- and BSMV:TaAGC1-inoculated CI12633 plants at 14 and 40 dpi with R. cerealis. Similar results were obtained from three independent replicates. (This figure is available in colour at JXB online.)
Overexpression of TaAGC1 enhances wheat resistance to sharp eyespot
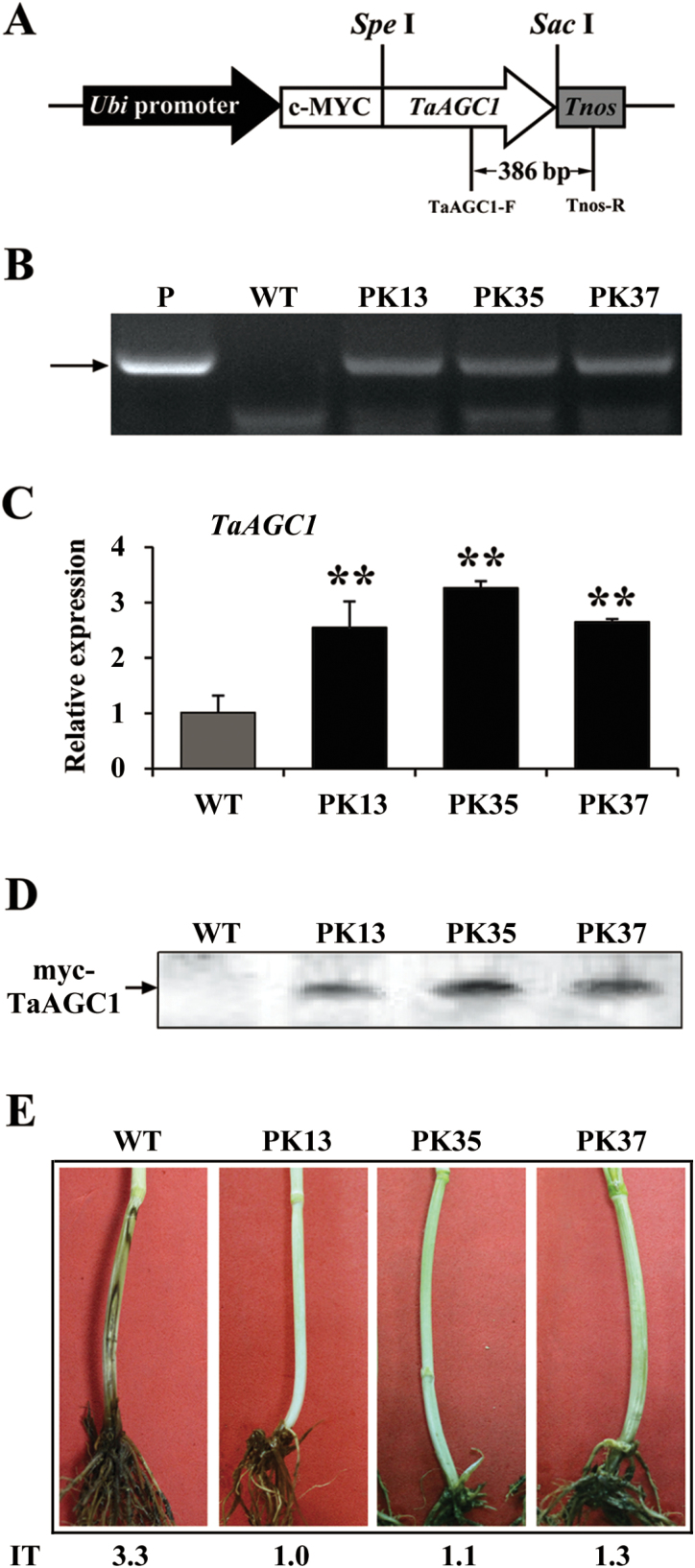
The role of TaAGC1 in the resistance to R. cerealis was further studied by developing and evaluating TaAGC1-overexpressing transgenic wheat lines. PCR and qRT-PCR assays showed that the transcript abundance of TaAGC1 in three stably-overexpressing transgenic lines (PK13, PK35 and PK37) was markedly elevated compared with untransformed recipient (wild type, WT) Yangmai 20 (Fig. 6B, C). Western blotting indicated that the introduced myc-TaAGC1 gene was translated into the fusion protein in the three TaAGC1-overexpressing transgenic lines, but not in WT Yangmai 20 (Fig. 6D). The sharp eyespot severity assessments in three successive (T1−T3) generations showed that, compared with WT Yangmai 20, all three TaAGC1-overexpressing lines (PK13, PK35 and PK37) displayed significantly enhanced resistance to R. cerealis infection (Table 1, Fig. 6E). For example, the average ITs and disease indices of the three TaAGC1-overexpressing wheat lines in T2 generation were 1.79–2.16 and 37.14–43.43%, respectively, whereas the average IT and disease index of the WT Yangmai 20 were 3.60 and 76.89%, respectively (Table 1). These results proved that TaAGC1 positively regulated wheat defence responses to R. cerealis infection.
Fig. 6.
Molecular analyses and Rhizoctonia cerealis responses of the wheat (Triticum aestivum) AGC kinase gene TaAGC1-overexpressing transgenic wheat plants. (A) TaAGC1-overexpressing transformation vector pUBI:myc-TaAGC1. The arrow indicates the fragment amplified in the PCR detection of the transgene. (B) PCR pattern of three TaAGC1-overexpressing transgenic lines (PK13, PK35 and PK37) and wild-type (WT) wheat Yangmai 20 using the TaAGC1-overexpressing transgene specific primers. P, the transformation vector pUBI:myc-TaAGC1 as a positive control. (C) qRT-PCR analyses of the relative transcript levels of TaAGC1 in three TaAGC1 transgenic lines at 7 d post R. cerealis inoculation. The relative transcript level of TaAGC1 in transgenic lines is relative to that in the WT plants. Three biological replicates per line were averaged and statistically treated (t-test; ** P<0.01). Bars indicate standard error of the mean. (D) Western blot pattern of the three TaAGC1-overexpressing transgenic lines and WT Yangmai 20 using an anti-myc antibody. Similar results were obtained from three independent replicates. (E) Typical symptoms of sharp eyespot in the three TaAGC1-overexpressing transgenic and WT Yangmai 20 plants. IT indicates infection type. (This figure is available in colour at JXB online.)
Table 1.
Rhizoctonia cerealis responses in transgenic wheat (Triticum aestivum) lines overexpressing the AGC kinase gene TaAGC1 and wild-type (WT) wheat
| Lines | Infection type | Disease index | ||||
|---|---|---|---|---|---|---|
| T 1 | T 2 | T 3 | T 1 | T 2 | T 3 | |
| PK13 | 1.38** | 2.17** | 1.63** | 27.50** | 43.43** | 32.66** |
| PK35 | 1.63** | 1.86** | 1.62** | 32.50** | 37.14** | 32.36** |
| PK37 | 1.62** | 1.98** | 1.73** | 32.42** | 39.67** | 34.61** |
| WT (Yangmai 20) | 2.28 | 3.84 | 2.72 | 45.56 | 76.89 | 54.38 |
** Significant difference between each transgenic line and untransformed (WT) wheat Yangmai 20 at P<0.01 (t-test). Infection types (ITs) were categorized from 0 to 5: 0, no lesion; 1, the lesion appeared on the sheaths rather than stems; 2, the width of the lesion is <50% of the infected stem perimeter; 3, the width of the lesion is >50 and <75% of the infected stem perimeter; 4, the width of the disease lesion is >75% of the infected stem perimeter; 5, white spike or dead plant. The infection types were the average ITs of 30 plants for each line of the TaAGC1 transgenic wheat in T1−T3 generations and WT wheat Yangmai 20 at 50 dpi with R. cerealis. Disease index={(0×X0+1×Xl+2×X2+3×X3+ 4×X4+5×X5)/[(X0+X1+X2+X3+X4+X5)×5]} ×100, where X0−X5 indicated plants with IT: 0−5. The representative symptoms of different ITs are shown in Supplementary Fig. S2.
TaAGC1 modulates ROS and related gene expression levels
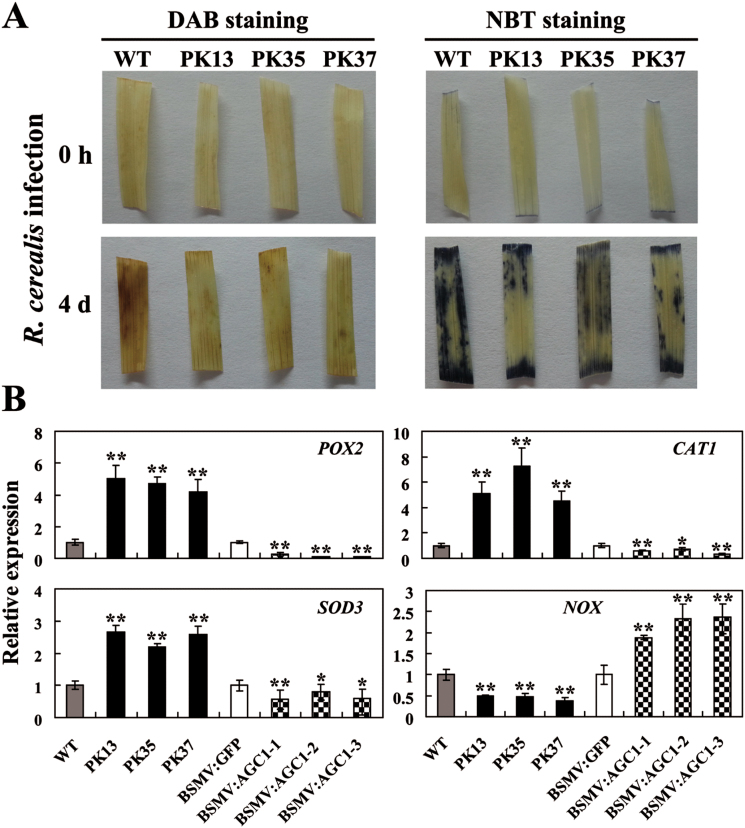
The expression of TaAGC1 was significantly induced after the H2O2 treatment from 1h to 12h and reached a peak at 3h post-treatment (Supplementary Fig. S4), suggesting that TaAGC1 might be involved in ROS signalling. To explore if the accumulation of ROS occurs in wheat challenged by R. cerealis, levels of H2O2 and O2 − in the infected-wheat Yangmai 20 were assayed using DAB and NBT staining, respectively. The results showed that levels of H2O2 and O2 − increased in the infected-wheat leaves from 1 dpi to 7 dpi (Fig. 7A, Supplementary Fig. S5), suggesting that the R. cerealis infection promoted the generation of H2O2 and O2 −. To unravel whether TaAGC1 regulates ROS homeostasis, levels of H2O2 and O2 − in TaAGC1-overexpressing and WT Yangmai 20 leaves after R. cerealis infection were further assayed. Under normal growth conditions, either WT or TaAGC1-overexpressing plants produced a small amount of H2O2 and O2 − (Fig. 7A). Following R. cerealis infection, there were less DAB- and NBT-stained spots on the leaves of TaAGC1-overexpressing plants than on WT plants (Fig. 7A), suggesting that the TaAGC1 overexpression reduced the accumulation of H2O2 and O2 − in the transgenic wheat.
Fig. 7.
Wheat (Triticum aestivum) AGC kinase TaAGC1 modulates reactive oxygen species (ROS) levels and the expression of ROS-related genes. (A) Analysis of hydrogen peroxide (H2O2) and superoxide anion (O2 −) levels in wheat. Leaves were harvested from TaAGC1-overexpressors (PK13, PK35 and PK37) and WT wheat plants at 0h and 4 dpi with Rhizoctonia cerealis, and were then stained with 3,3ʹ-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) to detect H2O2 and O2 −, respectively. Similar results were obtained from three independent replicates. (B) Transcriptional analysis of genes encoding ROS-scavenging enzymes (POX2, CAT1 and SOD3) and the ROS-producing enzyme NOX in wheat. The tested wheat samples include TaAGC1-overexpressors (PK13, PK35 and PK37), untransformed wheat Yangmai 20 (WT) and TaAGC1-knock-down plants (BSMV:TaAGC1-1, -2 and -3) and BSMV-GFP-infected controls at 7 dpi with Rhizoctonia cerealis. The reported transcript levels of the tested gene in the transgenic plants are relative to those in the WT plants. Statistically significant differences of TaAGC1-overexpression or TaAGC1-knock-down wheat plants were compared with the WT or the control, and based on three technical replications (t-test; *P<0.05, **P<0.01). Bars indicate standard error of the mean. (This figure is available in colour at JXB online.)
To explore if TaAGC1 modulated ROS-related genes, the transcription of wheat genes encoding ROS-scavenging enzymes (POX2, CAT1 and SOD3) and ROS-producing enzyme (NOX) in TaAGC1-overexpressing, TaAGC1-knock-down and control wheat plants were investigated. Under normal conditions, the transcriptional levels of POX2, CAT1 and SOD3 genes were significantly higher but the NOX transcript level was significantly lower in TaAGC1-overexpressing lines than in WT plants (Supplementary Fig. S6), suggesting that TaAGC1 positively regulated the transcription of POX2, CAT1 and SOD3 genes but negatively regulated the expression of NOX. Furthermore, following R. cerealis inoculation for 7 d and 40 d, the induced expression levels of POX2, CAT1 and SOD3 genes were significantly higher in TaAGC1 overexpressing wheat lines than in the WT plants and were lowest in the TaAGC1 knock-down plants. However, the NOX transcript level was significantly lower in TaAGC1-overexpressing lines than in WT plants and was the highest in TaAGC1-knock-down plants (Fig. 7B; Supplementary Fig. S7). The data suggested that the overexpression of TaAGC1 increased the expression of POX2, CAT1 and SOD3, but decreased the expression of NOX, resulting in a reduced ROS content in the overexpressing transgenic wheat plants infected by R. cerealis.
TaAGC1 regulates the expression of defence-associated genes
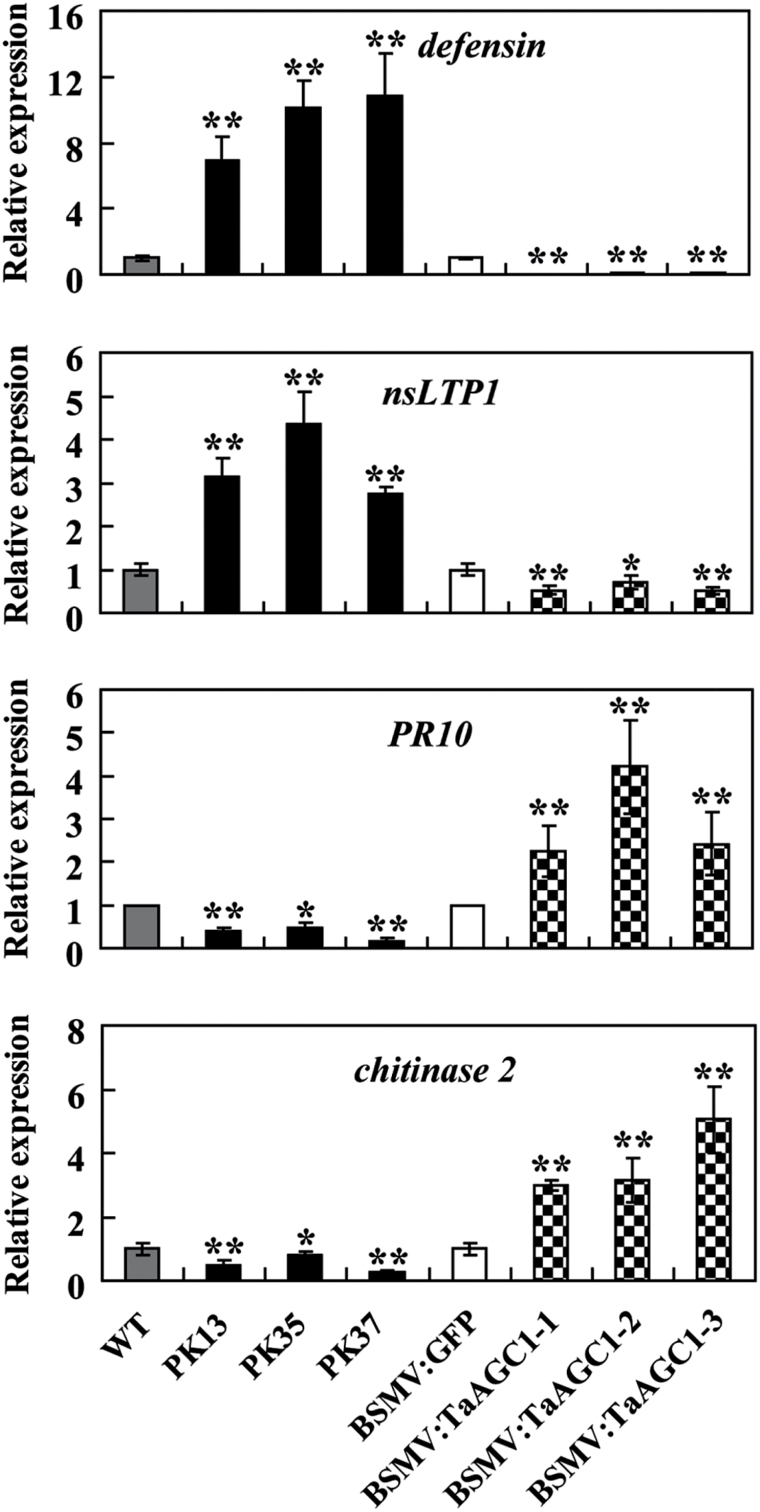
qRT-PCR analysis showed that the expression of TaAGC1 and four wheat defence-associated genes, defensin, nsLTP1, PR10 and chitinase 2, was significantly induced in WT wheat Yangmai 20 after R. cerealis inoculation for 7 d, 14 d or 40 d (Supplementary Fig. S8). To explore if TaAGC1 regulates the expression of these defence-associated genes in wheat, qRT-PCR was used to examine their transcription in TaAGC1-overexpressing, TaAGC1-knock-down, and control wheat plants. Under normal conditions, the transcriptional levels of wheat defensin and nsLTP1 genes were significantly higher but those of PR10 and chitinase 2 were significantly lower in TaAGC1-overexpressing lines than in WT plants (Supplementary Fig. S6). Furthermore, following R. cerealis inoculation for 7 d and 40 d, the transcriptional levels of wheat defensin and nsLTP1 genes elevated significantly more in TaAGC1-overexpressing lines than in WT plants and were the lowest in the TaAGC1-knock-down lines (Fig. 8; Supplementary Fig. S7). However, the transcriptional levels of PR10 and chitinase 2 were decreased in TaAGC1-overexpressing resistant lines than in WT plants and were the highest in the TaAGC1-knock-down plants (Fig. 8; Supplementary Fig. S7). The data suggested that TaAGC1 positively regulated the expression of wheat defensin and nsLTP1 genes but negatively regulated the expression of chitinase 2 and PR10.
Fig. 8.
Transcriptional analysis of four defence marker genes (defensin, nsLTP1, chitinase 2 and PR10) in wheat (Triticum aestivum). The tested wheat samples include TaAGC1-overexpressing lines (PK13, PK35 and PK37), untransformed Yangmai 20 (WT) and TaAGC1-knock-down plants (BSMV:TaAGC1-1, -2 and -3) and BSMV-GFP- infected controls at 7 dpi with Rhizoctonia cerealis. The reported transcript levels of the tested genes in the transgenic plants are relative to those in the WT plants. Statistically significant differences of TaAGC1-overexpression or TaAGC1-knock-down wheat plants were compared with WT or the controls, and based on three technical replications (t-test; *P<0.05, **P<0.01). Bars indicate standard error of the mean.
Discussion
In this study, an NDR-type AGC gene in wheat, TaAGC1, was isolated based on the microarray analysis and the qRT-PCR analyses of transcription levels in resistant and susceptible wheat genotypes. It was expressed at a higher level in R. cerealis-resistant wheat line CI12633 than in susceptible wheat line Wenmai 6. To understand the relationship between the gene transcriptional variance and the sequences in the resistant and susceptible wheat genotypes, we cloned the TaAGC1 sequence from the R. cerealis-resistant wheat line CI12633 and the homologous sequence from R. cerealis-susceptible wheat line Wenmai 6, respectively. Comparison of TaAGC1 sequences from CI12633 and Wenmai 6 showed that four single-nucleotide polymorphisms existed at their 3ʹ-terminal sequences (Supplementary Fig. S9), which may be a reason for the different expression patterns in the resistant and susceptible wheat genotypes. The protein TaAGC1 was predicted to be a Ser/Thr protein kinase and it shows the sequence characteristics (like the activation loop signature motif, DFGx60STVGTPDYIAPE) of NDR-type AGC kinases. For human NDR-type AGC kinases, three regulatory phosphorylation sites, Thr-74 in the S100B-binding domain, Ser-281 in the catalytic domain and Thr-444 in the hydrophobic motif, were previously reported to be crucial for NDR1 activity (Tamaskovic et al., 2003). All three phosphorylation sites are also found in the TaAGC1 sequence (Supplementary Fig. S3). A phylogenetic analysis provided additional evidence that the TaAGC1 protein is an NDR-type AGC kinase and is distinct from receptor-like kinases or receptor-like cytoplasmic kinases. The majority of receptor-like kinases contain transmembrane and extracellular domains and localize to the plasma membrane, which is consistent with their function in sensing extracellular signals (AbuQamar et al., 2008). The TaAGC1 protein lacks transmembrane and extracellular domains. Computational tools, such as WoLF PSORT (Horton et al., 2007), Cell-Ploc (Chou and Shen, 2010) and SCLPred (Mooney et al., 2011), predicted that TaAGC1 is likely localized to the cytoplasm and nucleus. Based on fluorescent signals of TaAGC1-GFP or GFP in the transiently transformed wheat and onion cells, TaAGC1 is localized in the cytoplasm and the nucleus in onion epidermal cell and wheat protoplast, which is consistent with the computational prediction. The protein blot analysis could help to further study TaAGC1 subcellular localization in the future. Kinase activity assays confirmed that TaAGC1 protein is a functional kinase with autophosphorylation activity and phosphorylation activity and that the residue Asp-239 (D) at the Ser/Thr kinase active-site is required for the kinase activity of TaAGC1. The biochemical properties are in agreement with the sequence traits.
Many genes are induced by environmental stresses and they offer the potential for improving biotic and abiotic stress tolerance in planta. For example, the expression of TPK1b, a tomato protein kinase gene, is induced by the necrotrophic fungus B. cinerea and wounding stimuli. Accordingly, TPK1b mediates plant responses to B. cinerea and insect herbivory (AbuQamar et al., 2008). The expression of the rice AGC kinase gene OsOxi1 was induced after the infection of the blast fungal pathogen M. oryzae pv. oryzae, and the OsOxi1-overexpression rice plants displayed enhanced resistance to blast disease (Matsui et al., 2010). However, this was not necessarily true in defence responses of distinct plant species against different lifestyle pathogens. For example, in Arabidopsis, although the expression of AtOXI1 was strongly induced after infection with B. cinerea, the Atoxi1 null mutant did not display altered susceptibility to this necrotrophic pathogen (Petersen et al., 2009). Additionally, although AtOXI1 expression was induced by H. arabidopsidis and P. syringae pv. tomato, both oxi1 null mutant and AtOXI1 overexpression lines showed increased susceptibility to virulent H. arabidopsidis and to both virulent and avirulent P. syringae pv. tomato. These data suggest that OXI1 is required for plant immunity to H. arabidopsidis and P. syringae pv. tomato, and OXI1 expression levels appear to be critical in mounting an appropriate defence response (Petersen et al., 2009). Here, the expression of TaAGC1 was significantly induced after R. cerealis infection, and the induction was higher in R. cerealis-resistant lines than in susceptible lines. Accordingly, following inoculation with R. cerealis, TaAGC1-overexpressing transgenic wheat lines (PK13, PK35 and PK37) in the T1−T3 generations displayed significantly enhanced resistance to sharp eyespot disease compared to untransformed recipient plants, while the down-regulation of TaAGC1 in resistant CI12633 significantly impaired the host resistance. These data indicated that the wheat AGC kinase TaAGC1 positively contributes to host resistance to the necrotrophic fungus R. cerealis. Interestingly, compared with host wheat plants, the TaAGC1-overexpressing and knock-down wheat plants either did not show obvious phenotypic changes in development or did not alter resistance to powdery mildew disease (Supplementary Fig. S10). It would be interesting to further study how TaAGC1-overexpressing and knock-down wheat plants perform in response to other wheat pathogens in the future. In previous studies, three AGC VIIIb kinases in other plant species were reported to participate in the immune responses to biotrophic and hemibiotrophic pathogens, including the Arabidopsis AtOXI1 to H. arabidopsidis and P. syringae pv. tomato (Rentel et al., 2004; Petersen et al., 2009), the rice OsOxi1 to Xanthomonas oryzae pv. oryzae (Matsui et al., 2010), and tomato Adi3 to P. syringae pv. tomato (Devarenne et al., 2006). The TaAGC1 protein fell into the AGC VII subfamily (namely an NDR-type AGC protein kinase), but was not in the clade containing AtOXI1, OsOXI1 and Adi3. To our knowledge, this is the first report of a defence role for a plant NDR-type AGC kinase, and the first functional characterization of a wheat AGC gene in the resistance response to necrotrophic pathogens. To date, the mechanisms of plant responses to necrotrophic pathogens have been poorly understood. These results extend the current knowledge of defence mechanisms against pathogen attacks in plants.
In early responses to pathogen invasion, plants often generate an excessive amount of ROS. During plant immunity against biotrophic pathogens, ROS not only functions in toxicity and the oxidative cross-linking of plant cell walls, but also has a signalling role in mounting a defence response (Petersen et al., 2009). A kinase cascade and downstream transcription factors represent key regulatory components of the ROS signal transduction cascade (Rentel et al., 2004; Matsui et al., 2010; Schmidt et al., 2013). For example, the Arabidopsis AGC kinase AtOxi1 is an essential part of the ROS signal transduction pathway and is required for root hair development and immunity against H. parastica and P. syringae pv. tomato, two separate H2O2-mediated processes (Rentel et al., 2004; Petersen et al., 2009). Additionally, ROS-scavenging enzymes and antioxidants play significant roles in plant defences against certain necrotrophic pathogens (Kuźniak and Skłodowska, 2004; Sharma et al., 2007). In this study, in wheat response to the infection of the necrotrophic pathogen R. cerealis, a signalling cascade in wheat is triggered, which includes the activation of TaAGC1 and the accumulation of ROS. TaAGC1 is responsive to H2O2 stimuli. Importantly, the expression of the ROS-scavenging enzyme genes (POX2, CAT1 and SOD3) is increased in uninfected wheat plants overexpressing TaAGC1, suggesting that TaAGC1 is involved in ROS signalling. Furthermore, after R. cerealis infection, the transcription levels of TaAGC1, SOD3, POX2 and CAT1 genes were more elevated in the TaAGC1-overexpressing wheat lines compared with the WT line, and were lower in TaAGC1 knock-down wheat plants than in the control plants, further indicating that TaAGC1 positively regulated the transcription of these ROS-scavenging enzyme genes but down-regulated the expression of the gene NOX encoding ROS-producing enzyme. Consequently, following R. cerealis infection, the ROS level was lower in TaAGC1-overexpressing wheat lines compared with the WT line. These data suggested that TaAGC1 maintained ROS homeostasis by regulating the expression of ROS-scavenging and -producing enzyme genes.
In plants, defence genes play vital roles in defence responses to pathogens. For example, transgenic wheat plants overexpressing TaLTP5 (a wheat LTP gene) displayed enhanced resistance to the fungal pathogens Cochliobolus sativus and Fusarium graminearum (Zhu et al., 2012). Overexpression of certain ERF or MYB transcription factors improved wheat resistance to soil-born fungal diseases through up-regulation of wheat defence marker genes (Chen et al., 2008; Zhang et al., 2012; Liu et al., 2013; Zhu et al., 2014). However, different regulatory factors, such as protein kinases and transcription factors, may regulate distinct defence-related genes in plant defence responses to pathogens. In this study, to explore if the wheat AGC kinase TaAGC1 regulates the expression of certain wheat defence-associated genes in TaAGC1-mediated resistance to R. cerealis, qRT-PCR was used to investigate expression in four of them (defensin, PR10, chitinase 2, and nsLTP1) in TaAGC1-overexpressing and knock-down wheat plants, and control wheat plants with or without R. cerealis inoculation. These four wheat defence-related genes were reported to participate in transcription factor-mediated defence responses to pathogens in previous studies (Liu et al. 2013; Zhu et al., 2014). qRT-PCR analysis results showed that TaAGC1, defensin and nsLTP1 genes were up-regulated in the resistant wheat plants overexpressing TaAGC1 but down-regulated in TaAGC1-knock-down susceptible wheat plants following R. cerealis challenge. The data suggest that TaAGC1 positively regulates the expression of defensin and nsLTP1 and that defensin and nsLTP1 might positively contribute to the TaAGC1-mediated resistance to R. cerealis infection. In contrast, PR10 and chitinase 2 genes were down-regulated in the resistant TaAGC1-overexpressing wheat plants but up-regulated in susceptible TaAGC1-knock-down wheat plants following R. cerealis inoculation, suggesting that TaAGC1 might negatively regulate the expression of PR10 and chitinase 2 genes, and that PR10 and chitinase 2 might negatively contribute to the TaAGC1-mediated resistance. A recent study showed that in pepper, the expression of PR10 triggered hypersensitive cell death that represses infection of biotrophic pathogens but might facilitate the growth and colonization of necrotrophic pathogens in plants (Choi et al., 2012), supporting our results about PR10 in wheat. The deep molecular mechanisms underlying TaAGC1 roles remain to be further studied.
In summary, the wheat NDR-type AGC kinase TaAGC1, acting as a positive regulator, mediates host resistance to R. cerealis through modulating the expression of ROS-related and certain defence-associated genes. TaAGC1 can be a candidate gene for improving sharp eyespot resistance in wheat. This study provides novel insights into the defence roles of plant NDR-type AGC kinases against necrotrophic pathogens.
Supplementary data
Supplementary data is available at JXB online.
Supplementary Fig. S1. Scheme of genomic RNAs of the barley stripe mosaic virus (BSMV) construct, BSMV with green fluorescent protein fusion construct (BSMV:GFP), and the construct of the recombinant virus expressing the wheat AGC gene TaAGC1, BSMV:TaAGC1.
Supplementary Fig. S2. The representative symptoms of different infection types of sharp eyespot disease at 40 dpi with Rhizoctonia cerealis.
Supplementary Fig. S3. Nucleotide sequence and deduced amino acid sequence of the wheat AGC kinase gene TaAGC1.
Supplementary Fig. S4. Expression patterns of the wheat AGC kinase gene TaAGC1 in wheat responding to exogenous hydrogen peroxide (H2O2) treatments.
Supplementary Fig. S5. Analysis of hydrogen peroxide (H2O2) and superoxide anion (O2 −) accumulation in wheat leaves.
Supplementary Fig. S6. Transcriptional analysis of ROS-related and defence genes in the wheat AGC kinase gene TaAGC1-overexpressing and untransformed wheat Yangmai 20 plants under normal growth conditions.
Supplementary Fig. S7. Transcription analysis of ROS-related and defence genes in the wheat AGC kinase gene TaAGC1-overexpressing and knock-down, as well as control wheat plants with Rhizoctonia cerealis inoculation for 40 d.
Supplementary Fig. S8. Transcription analysis of ROS-related and defence genes in the wild type wheat Yangmai 20 plants after Rhizoctonia cerealis inoculation for 7, 14 and 40 d.
Supplementary Fig. S9. Alignment of 3ʹ terminal sequences of TaAGC1 in resistant wheat line CI12633 and of its homologue in susceptible wheat line Wenmai 6.
Supplementary Fig. S10. The powdery mildew symptoms of the wheat AGC kinase gene TaAGC1-overexpressing and knock-down, and control wheat plants.
Supplementary Table S1. Rhizoctonia cerealis responses of four wheat lines.
Supplementary Table S2. Primers used in this study.
Acknowledgements
The authors are very grateful to Dr Yan Guo and Ms Jianfang Li (College of Biological Sciences, China Agricultural University) for help on the kinase activity assay. This work was supported by the National Natural Science Foundation program of China (Grant no. 31271799) and the National ‘Key Sci-Tech’ program of China (Grant no. 2013ZX08002-001).
References
- AbuQamar S, Chai MF, Luo H, Song F, Mengiste T. 2008. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. The Plant Cell 20, 1964–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogre L, Okresz L, Henriques R, Anthony RG. 2003. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends in Plant Science 8, 424–431. [DOI] [PubMed] [Google Scholar]
- Cai S, Ren L, Yan W, Wu J, Chen H, Wu X, Zhang X. 2006. Germplasm development and mapping of resistance to sharp eyespot (Rhizoctonia cerealis) in wheat. Scientia Agricultura Sinica 39, 928–934. [In Chinese with English abstract.] [Google Scholar]
- Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, Munnik T, Hirt H, Oelmuller R. 2011. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis . PLoS Pathogens 7, e1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li GH, Du ZY, Quan W, Zhang HY, Che MZ, Wang Z, Zhang ZJ. 2013. Mapping of QTL conferring resistance to sharp eyespot (Rhizoctonia cerealis) in bread wheat at the adult plant growth stage. Theoretical and Applied Genetics 126, 2865–2878. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang Z, Liang H, Liu H, Du L, Xu H, Xin Z. 2008. Overexpression of TiERF1 enhances resistance to sharp eyespot in transgenic wheat. Journal of Experimental Botany 59, 4195–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Hwang IS, Hwang BK. 2012. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K, Shen H. 2010. Cell-Ploc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Natural Science 2, 1090–1103. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. 1996. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Research 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Devarenne TP, Ekengren SK, Pedley KF, Martin GB. 2006. Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. The EMBO Journal 25, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Wang M, Xu F, Quan T, Peng K, Xiao L, Xia G. 2013. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiology 161, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. 2009. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AV, Al-Yousif M, Hirt H. 2012. Role of AGC kinases in plant growth and stress responses. Cellular and Molecular Life Sciences 69, 3259–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada MS, Yin Y, Chen H, Ma Z. 2011. The escalating threat of Rhizoctonia cerealis, the causal agent of sharp eyespot in wheat. Pest Management Science 67, 1411–1419. [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP. 2002. Barley stripe mosaic virus-induced gene silencing in a monocot plant. The Plant Journal 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Horton P, Rark K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35 (Web Server issue), W585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemańczyk G, Kwaśna H. 2013. Effects of sharp eyespot (Rhizoctonia cerealis) on yield and grain quality of winter wheat. European Journal of Plant Pathology 135, 187–200. [Google Scholar]
- Lee BH, Lee H, Xiong L, Zhu JK. 2002. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14, 1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Liu X, Yang L, Zhou X, Zhou M, Lu Y, Ma L, Ma H, Zhang Z. 2013. Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. Journal of Experimental Botany 64, 2243–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuźniak E, Skłodowska M. 2004. The effect of Botrytis cinerea infection on the antioxidant profile of mitochondria from tomato leaves. Journal of Experimental Botany 55, 605–612. [DOI] [PubMed] [Google Scholar]
- Matsui H, Yamazaki M, Kishi-Kaboshi M, Takahashi A, Hirochika H. 2010. AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant & Cell Physiology 51, 1731–1744. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Mooney C, Wang YH, Pollastri G. 2011. SCLpred: protein subcellular localization prediction by N-to-1 neural networks. Bioinformatics 27, 2812–2819. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR. 2010. The nuts and bolts of AGC protein kinases. Nature Reviews Molecular Cell Biology 11, 9–22. [DOI] [PubMed] [Google Scholar]
- Petersen LN, Ingle RA, Knight MR, Denby KJ. 2009. OXI1 protein kinase is required for plant immunity against Pseudomonas syringae in Arabidopsis . Journal of Experimental Botany 60, 3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, et al. 2004. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis . Nature 427, 858–861. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian in heritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences, USA 81, 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten HM, et al. 2013. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS. 2005. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiology 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Rahman MH, Strelkov S, Thiagarajah M, Bansal VK, Kav NN. 2007. Proteome-level changes in two Brassica napus lines exhibiting differential responses to the fungal pathogen Alternaria brassicae . Plant Science 172, 95–110. [Google Scholar]
- Shetty NP, Jøgersen HJL, Jensen JD, Collinge DB, Shetty HS. 2008. Roles of reactive oxygen species in interactions between plants and pathogens. European Journal of Plant Pathology 121, 267–280. [Google Scholar]
- Tamaskovic R, Bichsel SJ, Hemmings BA. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Letters 546, 73–80. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yao W, Dong N, Liang H, Liu H, Huang R. 2007. A novel ERF transcription activator in wheat and its induction kinetics after pathogen and hormone treatments. Journal of Experimental Botany 58, 2993–3003. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Liu X, Wang XD, Zhou MP, Zhou XY, Ye XG, Wei XN. 2012. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytologist 196, 1155–1170. [DOI] [PubMed] [Google Scholar]
- Zhou H, Li S, Deng Z, et al. 2007. Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. The Plant Journal 52, 420–434. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li Z, Xu H, Zhou M, Du L, Zhang Z. 2012. Overexpression of wheat lipid transfer protein gene TaLTP5 increases resistances to Cochliobolus sativus and Fusarium graminearum in transgenic wheat. Functional & Integrative Genomics 12, 481–488. [DOI] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z. 2014. The wheat ethylene response factor transcription factor PATHOGEN-INDUCED ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiology 164, 1499–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.