Highlight
A novel NAC transcription factor regulates tolerance of rice to multiple abiotic stresses through directly targeting the genes related to reactive oxygen species homeostasis.
Key words: Abiotic stress, NAC, Oryza sativa, ROS, transcription factors.
Abstract
Adverse environmental conditions such as high temperature and drought stress greatly limit the growth and production of crops worldwide. Several NAC (NAM, ATAF1/2, and CUC2) proteins have been documented as important regulators in stress responses, but the molecular mechanisms are largely unknown. Here, a stress-responsive NAC gene, SNAC3 (ONAC003, LOC_Os01g09550), conferring drought and heat tolerance in rice is reported. SNAC3 was ubiquitously expressed and its transcript level was induced by drought, high temperature, salinity stress, and abscisic acid (ABA) treatment. Overexpression (OE) of SNAC3 in rice resulted in enhanced tolerance to high temperature, drought, and oxidative stress caused by methyl viologen (MV), whereas suppression of SNAC3 by RNAi resulted in increased sensitivity to these stresses. The SNAC3-OE transgenic plants exhibited significantly lower levels of H2O2, malondiadehyde (MDA), and relative electrolyte leakage than the wild-type control under heat stress conditions, implying that SNAC3 may confer stress tolerance by modulating reactive oxygen species (ROS) homeostasis. Quantitative PCR experiments showed that the expression of a large number of ROS-scavenging genes was dramatically increased in the SNAC3-OE plants, but significantly decreased in the SNAC3-RNAi transgenic plants. Five ROS-associated genes which were up-regulated in SNAC3-OE plants showed co-expression patterns with SNAC3, and three of the co-expressed ROS-associated enzyme genes were verified to be direct target genes of SNAC3. These results suggest that SNAC3 plays important roles in stress responses, and it is likely to be useful for engineering crops with improved tolerance to heat and drought stress.
Introduction
Plants frequently encounter diverse environmental cues which restrict their growth and development. Among the adverse external stimuli, extreme temperature and water deficit are two major factors that plants frequently confront. To compensate for their sessile lifestyle, plants have evolved a series of sophisticated but efficient strategies to cope with stress conditions. Among these strategies, plants respond to the stresses through stress-specific signalling pathways, which ultimately lead to morphological, physiological, and biochemical changes to adapt to unfavourable environmental conditions. Plants perceive the external signals through sensors, and trigger changes in expression of numerous stress-responsive genes and the synthesis of diverse functional proteins to enable them to survive. Based on the biological functions, genes responsive to the stresses mainly include the functional genes, which encode functional proteins or other products to protect plant cells directly from damage, and the regulatory genes whose products can regulate signal perception and transduction, and the expression of downstream genes under stress conditions (Hirayama and Shinozaki, 2010). Various transcription factors function as central regulators and molecular switches for gene expression control in the stress signalling and adaptation networks (Zhang et al., 2011).
Increasing evidence supporting the role of NAC (NAM, ATAF1/2, and CUC2) transcription factors as key regulators in response to abiotic stresses has been reported in recent years (Puranik et al., 2012). NAC transcription factors comprise one of the largest gene families, which to date are only found in plants. NAC proteins are identified by a highly conserved DNA-binding domain, which is termed the NAC domain, in the N-terminal region, whereas the transcription regulatory region in the C-terminal of NAC proteins is usually diversified in both amino acid composition and length. NAC genes have been reported to be implicated in organ development and boundary maintenance, cell division, secondary wall synthesis, senescence, iron homeostasis, and defence agianst pathogens, and they also act as master regulators in abiotic stress responses (Takada et al., 2001; Guo and Gan, 2006; Kim et al., 2006; Uauy et al., 2006; Zhong et al., 2006; Puranik et al., 2012). The RD26 (RESPONSIVE TO DEHYDRATION 26) gene was the first NAC gene identified as a regulator in mediating cross-talk between abscisic acid (ABA) and jasmonate (JA) signalling during stress responses in Arabidopsis (Fujita et al., 2004; Shinozaki and Yamaguchi-Shinozaki, 2007). Overexpressing ANAC019, ANAC055, or ANAC072, which were induced by drought, salt, and ABA, conferred drought tolerance in transgenic Arabidopsis (Tran et al., 2004). Tran et al. (2004) identified the consensus NACRS [NAC recognition site, CGT(G/A)] and CDBS (core DNA-binding sequence, CACG) in the promoter region of the Arabidopsis ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1) gene. The recognition sequence for NAC factors might be conserved in plants, since quite a few NAC factors can bind to this NACRS (Fujita et al., 2004; Hu et al., 2006, 2008; Liu et al., 2010).
Reactive oxygen species (ROS) are byproducts mainly from aerobic respiration, and were initially considered as toxic molecules which caused oxidative damage to DNA, protein, and membrane lipids in plant cells. Recent studies have demonstrated that ROS also act as important signalling molecules in the complex networks of stress responses (Bechtold et al., 2013; Baxter et al., 2014; Shi et al., 2014). Plants have developed complicated scavenging and regulation pathways to monitor tightly the ROS redox homeostasis in order to avoid the excessive accumulation of ROS in cells. Emerging evidence supports that NAC transcription factors also participate in the regulation of ROS metabolism. An Arabidopsis NAC transcription factor, NTL4, which was induced by drought and heat, triggered ROS production by directly binding to Atrboh gene promoters, resulting in leaf senescence (Lee et al., 2012). Another NAC factor, JUB1, was described as a major longevity regulator in Arabidopsis (Wu et al., 2012). JUB1 was H2O2-responsive and could bind to the DREB2A promoter. Overexpressing JUB1 promoted leaf longevity and tolerance to salt and heat stress conditions (Shahnejat-Bushehri et al., 2012; Wu et al., 2012). A membrane-associated NAC protein, ANAC013, was shown to bind and transactivate the mitochondrial dysfunction motif (MDM) to mediate mitochondrial retrograde regulation (MRR)-induced gene expression, and conferred increased tolerance to methyl viologen (MV) and rotenone-induced oxidative stress in transgenic Arabidopsis plants (De Clerq et al., 2013).
As one of the most important crops worldwide, rice provides staple food for more than half of the world’s population. In facing climate changes and increasing global population, it is an urgent task to exploit stress resistance and yield stability of rice under adverse environmental conditions. In the past few decades, various stress-responsive transcription factors including NACs have been explored for engineering rice for improved stress tolerance through transgenic approaches (Sacar et al., 2013). SNAC1 overexpressors exhibited better performance under drought and salt stress conditions at the vegetative stage, and the transgenic plants also exhibited higher seed production under drought conditions at the reproductive stage (Hu et al., 2006). Overexpression of SNAC1, OsNAC10, and OsNAC5 driven by a root-specific promoter RCc3 enlarged the root diameter in rice, and therefore conferred increased drought resistance and grain yield under field drought conditions (Ishizaki et al., 2006; D.H. Jeong et al., 2010; J.S. Jeong et al., 2010, 2013).
In this study, another NAC transcription factor gene, SNAC3, which was responsive to various abiotic stresses, was characterized. The SNAC3 overexpression (OE) transgenic plants enhanced heat, drought, and oxidative tolerance in rice, whereas repressing SNAC3 weakened heat, osmotic, and oxidative tolerance. The data indicated that SNAC3 regulates the heat stress response by adjusting the redox homeostasis state through controlling the expression of ROS-associated enzyme genes. It was also found that the SNAC3-mediated stress responses may be ABA-independent, which is different from many previous reports of stress-responsive NAC genes in rice. The findings indicate that SNAC3 is a crucial regulator in heat and drought stress responses, and has great potential in engineering crops with enhanced heat and drought tolerance.
Materials and methods
Plant materials and stress treatments
The japonica rice (Oryza sativa) cultivar (Zhonghua 11, ZH11) was used as a transformation recipient in this study. To detect the transcript level of SNAC3 under various stresses and phytohormone treatments, ZH11 plants were planted in the greenhouse with a 14h light/10h dark cycle. Four-leaf stage seedlings were subjected to a variety of abiotic stresses including drought (stopping water supply), salt (irrigation with 200mM NaCl solution), heat (transferring the seedlings to a 42 °C growth chamber), cold (exposing the plants to 4 °C), submergence (covering the plants completely with water), wounding (cutting the leaves into pieces and floating them on water at room temperature under continuous light conditions), H2O2 treatment [irrigation with 1% (v/v) H2O2 solution], and ABA treatment (spraying 100 μM ABA on the leaves), followed by sampling at the designated times.
SNAC3-OE and RNAi transgenic plants were selected by germinating seeds on Murashige and Skoog (MS) medium containing 50mg l–1 hygromycin. Wild-type (WT) seeds were germinated on normal MS medium. For heat stress treatment at the seedling stage, transgenic plants and WT plants were grown in the same barrel in a split (half-and-half) manner under normal growth conditions. Seedlings at the four- to five-leaf stage were subjected to heat stress treatment by transferring the plants to a 42 °C growth chamber. After heat stress treatment for 1–2 d, the seedlings were allowed to recover in normal growth conditions for 1 week. For drought stress treatment at the vegetative stage, transgenic plants and WT plants were grown in the same barrel in a split manner under normal growth conditions until the four- to five-leaf stage. The water supply was withheld to cause drought stress until the WT leaves became completely wilted. After recovery by rewatering for a week, the survival rates were recorded and the plants were photographed. Drought stress treatment at the reproductive stage was executed in a refined paddy field facilitated with a movable rain-off shelter by stopping the irrigation at the booting stage followed by re-watering after flowering. To evaluate the tolerance of rice seedlings to mannitol stress, MV, and ABA, germinated seeds were transferred to MS medium containing 120mM mannitol, 2 μM MV, and 3 μM ABA, respectively. After growing on the medium for 7–10 d, the shoot lengths were measured to evaluate the tolerance.
Plasmid construction and transformation of rice
To create a SNAC3-OE construct, the coding region of SNAC3 was obtained from rice total RNA by reverse transcription–PCR (RT–PCR). The full-length cDNA of SNAC3 was inserted into the pU1301 vector downstream of the Ubiquitin promoter. To generate a dsRNAi construct of SNAC3, a 334bp SNAC3-specific fragment was introduced into the pDS1301 vector (Yuan et al., 2007). To investigate the expression pattern of SNAC3, a genomic DNA sequence from the promoter region (2536 to 303bp upstream of the predicted ATG codon of the ORF) of SNAC3 was inserted into the DX2181 vector to drive the GUS (β-glucuronidase) reporter gene. The constructs were transformed into the japonica rice cv. ZH11 using the Agrobacterium-mediated transformation method (Lin and Zhang, 2005). The primers used in this study are listed in Supplementary Table S1 available at JXB online.
RNA isolation and analysis of its expression level
Total RNA was extracted from rice leaves using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The first-strand cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative RT–PCR was conducted on a 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Premix Ex Taq (TaKaRa, Dalian, China) according to the manufacturer’s protocol. The rice Ubiquitin gene was used as the internal control. The relative expression level was determined as described previously (Livak and Schmittgen, 2001). For the promoter–GUS reporter assay, various tissues and organs of the pDX2181-SNAC3pro::GUS transgenic plants were collected for GUS histochemical staining essentially as described previously (Ning et al., 2010). Quantification of GUS activity was carried out according to the method described previously (Huang et al., 2007).
Determination of stress-associated physiological indicators
For water loss rate measurement, detached leaves from SNAC3-OE and the WT plants were weighed at the indicated times. The cell membrane permeability was evaluated by electrolyte leakage measured by the relative conductance method (Bajji et al., 2002). For estimation of the degree of membrane lipid peroxidation in cells, the MDA (malondialdehyde) content was determined as described previously (Du et al., 2010). H2O2 was visually detected by staining the leaves of the SNAC3-OE and the WT plants with 3,3′-diaminobenzidine (DAB) as described previously (Ning et al., 2010). The reddish brown coloration produced by DAB staining for H2O2 was recorded. Quantification of H2O2 was performed using a kit (Invitrogen) by following the manufacturer’s instructions. For the quantification of ABA content, leaves from the four- to five-leaf-stage WT and SNAC3-OE seedlings before and after stress treatment were sampled and prepared following a modified crude extraction procedure (Pan et al., 2008). ABA content determination was executed by an Applied Biosystems 4000Q-TRAR LC-MS system with stable isotope-labelled ABA as a standard from OlChemIm according to an ultra-fast liquid chromatography-electrospray ionization tandem mass spectrometry (UFLC-ESI-MS/MS)-based method described preciously (Ding et al., 2008; Liu et al., 2012).
Biochemical assay in yeast
The yeast one-hybrid assay was carried out using the Matchmaker one-hybrid system following the manual (Clontech, Palo Alto, CA, USA). The promoter fragments of potential SNAC3-targeted genes containing the NACRS and NDBS cis-elements were amplified and fused upstream of the HIS3 minimal promoter in the pHIS2 vector to generate reporter constructs. The ORF of SNAC3 obtained from PCR amplification were fused to the GAL4 activation domain in the pGAD7-Rec2 vector (Clontech), and subsequently co-transformed with the reporter constructs described above into the yeast strain Y187. In the meantime, the pGAD-53 and pHIS-53 plasmids were co-transformed as positive controls, while the pGAD-SNAC3 and pHIS2-53 plasmids were co-transformed into Y187 as negative controls. Growth performances of the transformants on the SD/-Leu/-Trp and SD/-Leu/-Trp/-His media containing 30mM 3-aminotriazole (3-AT) were analysed to evaluate the DNA–protein interactions.
Subcellular localization assays
The full-length cDNA of SNAC3 was amplified and fused into the pHBT-sGFP destination vector via a restriction enzyme-mediated cloning procedure, and into the pH7WGF2,0 binary vector by GATEWAY recombination reaction (Invitrogen) (Karimi et al., 2007), respectively. The SNAC3-pHBT-sGFP plasmid was transformed into rice protoplasts to perform the transient expression assay as described previously (You et al., 2013). The SNAC3-pH7WGF2,0 plasmid was introduced into Nicotiana benthamiana leaves through an Agrobacterium-mediated method. For transient expression, an Agrobacterium strain (EHA105) containing the SNAC3-pH7WGF2,0 construct was infiltrated at an OD600 of 1.0 into the 5- to 6-week-old N. benthamiana leaves. After incubation for 12–16 h, the distribution of the fusion protein was captured using a confocal fluorescence microscope (Leica TCS SP2).
Protein interaction analysis
The yeast two-hybrid assay was performed using the Matchmaker™ Gold Yeast Two-Hybrid system (Clontech). According to previous reports, two amino acids (Gly and Glu) at the N-terminal end of NAC proteins may participate in the formation of NAC protein dimers (Ernst et al., 2004; Jeong et al., 2009). To avoid the interference caused by dimer formation between SNAC3 and other NAC proteins, the coding region without the 108 amino acids at the N-terminus of SNAC3 was amplified and inserted in-frame with GALBD into the pGBKT7 vector to generate the bait construct, and the yeast two-hybrid library was constructed using the leaves from the rice seedlings exposed to heat and cold stress conditions. The two-hybrid screening was carried out using yeast mating technology according to the manufacturer’s instructions. To confirm further the proteins which interact with SNAC3 in living plant cells, bimolecular fluorescence complementation (BiFC) assay was performed in a rice protoplast system as described preciously (Tang et al., 2012).
Results
Identification and expression features of SNAC3
In the systematic analysis of the NAC family in rice, a significant portion of the family genes were found to be responsive to various abiotic stresses (Fang et al., 2008), and one of them, designated as SNAC3 or ONAC003 (LOC_Os01g09550), which belongs to subfamily II, including the closest Arabidopsis homologue At4g29230 with unknown function, was subjected to further functional characterization in this study. SNAC3 is located on chromosome 1 with four predicted differentially spliced transcripts (TIGR, http://rice.plantbiology.msu.edu/). Compared with the second longest transcript (LOC_Os01g09550.2), the longest transcript (LOC_Os01g09550.1) encoded a peptide with 52 additional amino acids at the N-terminus. However, only the full-length cDNA corresponding to the second longest transcript was isolated, and no EST was found for the extended N-terminal region for the predicted longest transcripts. To verify this, a pair of specific primers were designed to amplify the predicted extended N-terminal region by using rice cDNA (genomic DNA as the control) as the PCR template. No amplification was detected in the PCR of the cDNA template (Supplementary Fig. S1 at JXB online), indicating that LOC_Os01g09550.1 is not a transcript of SNAC3. Therefore, the following experiments were designed based on LOC_Os01g09550.2.
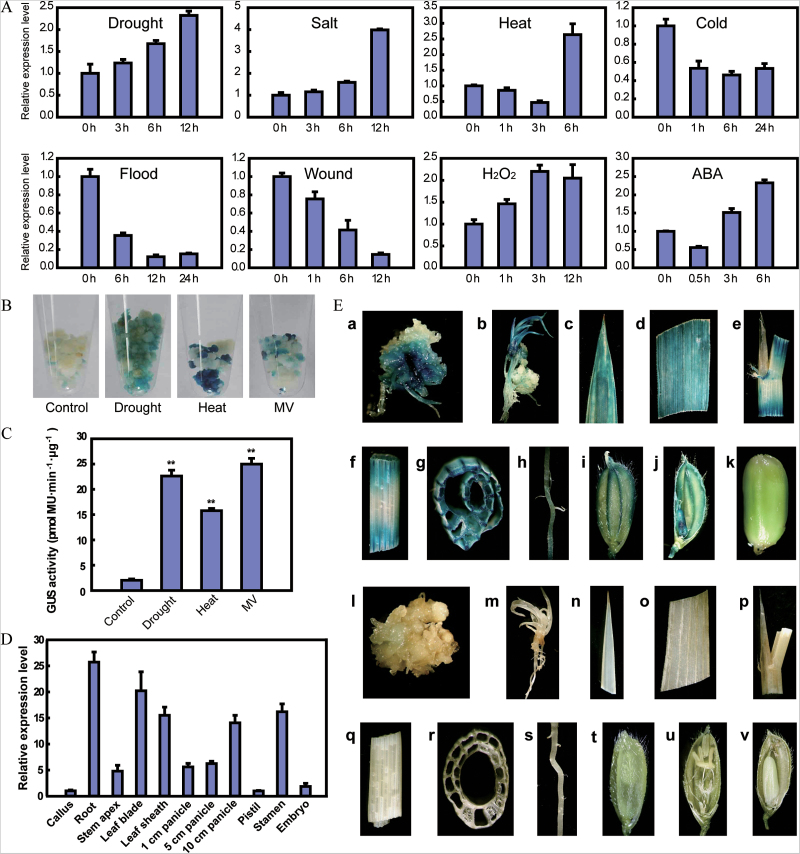
SNAC3 showed stress-inducible expression in the microarray analysis (Zhou et al., 2007). To investigate the expression profile of SNAC3 comprehensively, quantitative real-time RT–PCR (qPCR) was used to detect the transcript abundance of SNAC3 in four-leaf stage rice seedlings under a variety of abiotic stresses and hormone treatments. It was found that SNAC3 was induced by drought, high salinity, heat, oxidative stress, and ABA treatment. However, it was suppressed by cold, submergence, and wounding stresses (Fig. 1A). The induction of SNAC3 under drought, heat, and MV treatment was further confirmed in the GUS staining assay of P SNAC3:GUS transgenic callus (Fig. 1B), and the quantification of GUS activity was in agreement with the staining results (Fig. 1C).
Fig. 1.
Expression pattern analysis of SNAC3. (A) Expression level of SNAC3 under various abiotic stresses and ABA treatment. Four-leaf stage seedlings were subjected to drought, salt (200 mmol l–1 NaCl), heat (42 °C), cold (4 °C), flood, wounding, H2O2 (1% H2O2), and ABA treatment (100 mmol l–1 ABA). The relative expression level of SNAC3 was detected by qPCR at the indicated times. Error bars indicated the SE based on three replicates. (B) Expression pattern of the GUS reporter gene driven by the SNAC3 promoter in transgenic resistant callus under normal conditions (control), drought, heat, and MV stress. (C) GUS activity quantification in P SNAC3:GUS transgenic resistant callus under normal conditions, drought, heat, and MV stress. GUS activity was defined in picomoles of 4-MU generated per minute per microgram of protein. (D) Detection of SNAC3 expression in various tissues and organs using qPCR. Error bars indicate the SE based on three technical replicates. (E) Expression pattern under normal conditions. (a–k) GUS staining of tissues and organs from P SNAC3:GUS transgenic plants; (l–v) GUS staining of tissues and organs from ZH11 plants. (a and l) callus; (b and m) regenerated seedling; (c and n) leaf tip; (d and o), leaf blade; (e and p) ligule, collar, and auricle; (f and q) leaf sheath; (g and r) cross-section of the leaf sheath; (h and s) root; (i and t) hull; (j and u) stamen and pistil; (k and v) seed.
A total of 11 representative tissues/organs (callus, root, stem apex, leaf blade, leaf sheath, 1cm panicle, 5cm panicle, 10cm panicle, pistil, stamens, and embryo) were sampled for spatio-temporal expression analysis of SNAC3. As shown in Fig. 1D, SNAC3 was expressed in all of the tissues/organs tested, and the expression level was relatively lower in callus, pistil, and embryo when compared with other tissues/organs. To clarify the expression pattern of SNAC3, transgenic rice expressing the GUS reporter gene under the control of the SNAC3 promoter were generated, and GUS staining was performed to detect the expression of the reporter gene. The GUS signal was detected in almost all of the analysed tissues/organs, confirming that SNAC3 was expressed ubiquitously (Fig. 1E).
Overexpression of SNAC3 in rice enhanced heat and drought resistance
To elucidate the biological functions of SNAC3 in rice, the SNAC3-OE vector was constructed and transformed into rice ZH11. qPCR was carried out to detect the SNAC3 transcript in transgenic plants. The expression level of SNAC3 was significantly higher in the transgenic plants than in the WT (Supplementary Fig. S2 at JXB online).
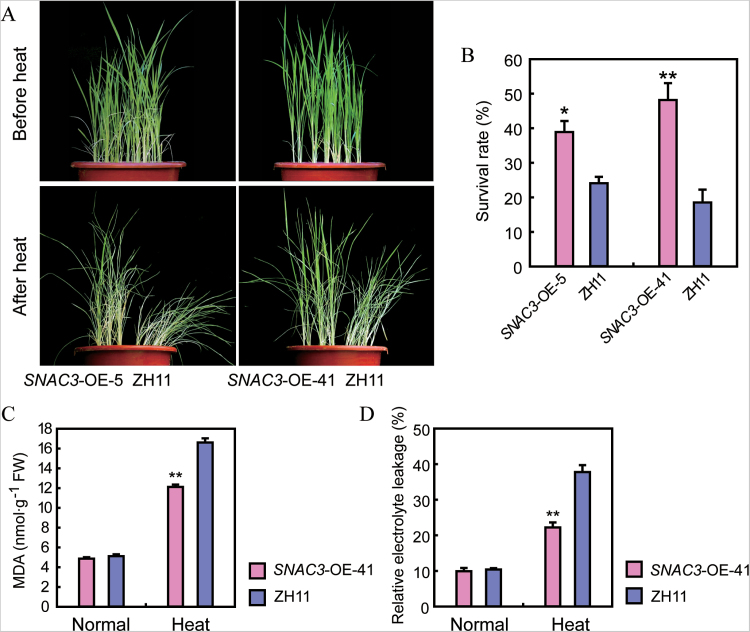
Since SNAC3 was strongly induced by high temperature stress, the performance of transgenic rice overexpressing SNAC3 was first evaluated under heat stress conditions at the vegetative stage. The seedlings at the four-leaf stage were transferred to a 42 °C growth chamber for heat stress treatment. The overexpression plants exhibited leaf rolling and wilted later than the WT during the course of heat stress treatment (Fig. 2A). About 40% of the overexpression plants survived after the heat treatment, whereas the WT only exhibited a 20% survival rate after the same stress treatment (Fig. 2B). One of the OE lines (SNAC3-OE-41) was used for further determination of the physiological parameters. Heat stress generally causes membrane lipid peroxidation of plant cells. As a major product of this process, MDA is widely used as an important indicator to evaluate the degree of membrane lipid peroxidation, which reflected the oxidative damage to lipids caused by environmental stress. In addition, cell membrane permeability increases under adverse conditions, and electrolyte leakage has been commonly considered as an indicator to assess cell membrane stability and the extent of the cell membrane damage under stress conditions (Smirnoff, 1993; Agarie et al., 1998). Therefore, the MDA content and electrolyte leakage of SNAC3-OE and the WT were measured under heat stress and normal conditions. As shown in Fig. 2, no differences were observed in the MDA content and electrolyte leakage between the SNAC3-OE and control plants under normal conditions, while both the MDA content and electrolyte leakage were obviously lower in the SNAC3-OE plants compared with the WT plants under heat stress conditions (Fig. 2C, 2D). These results suggested that overexpressing SNAC3 may cause less extensive membrane lipid peroxidation and improved cell membrane stability under the heat stress treatment, and thus contribute to attenuate the oxidative damage caused by the heat stress.
Fig. 2.
Enhanced heat tolerance of the SNAC3-OE transgenic plants at the seedling stage. (A) Phenotype of the SNAC3-OE transgenic plants under heat stress conditions. Four-leaf stage plants were subjected to 42 °C heat stress in a growth chamber (14h light/10h dark) for 1–2 d, and then transferred to normal growth conditions. (B) Survival rate of SNAC3-OE and ZH11 after heat stress treatment. Data represent the mean ±SE (n=3). *P<0.05, t-test; **P<0.01, t-test. (C) MDA content of SNAC3-OE and ZH11 seedlings under normal and heat stress conditions. Data represent the mean ±SE (n=3). *P<0.05, t-test; **P<0.01, t-test. (D) Relative electrolyte leakage of the leaves from SNAC3-OE and ZH11 seedlings under normal and heat stress conditions. Data represent the mean ±SE (n=3). *P<0.05, t-test; **P<0.01, t-test.
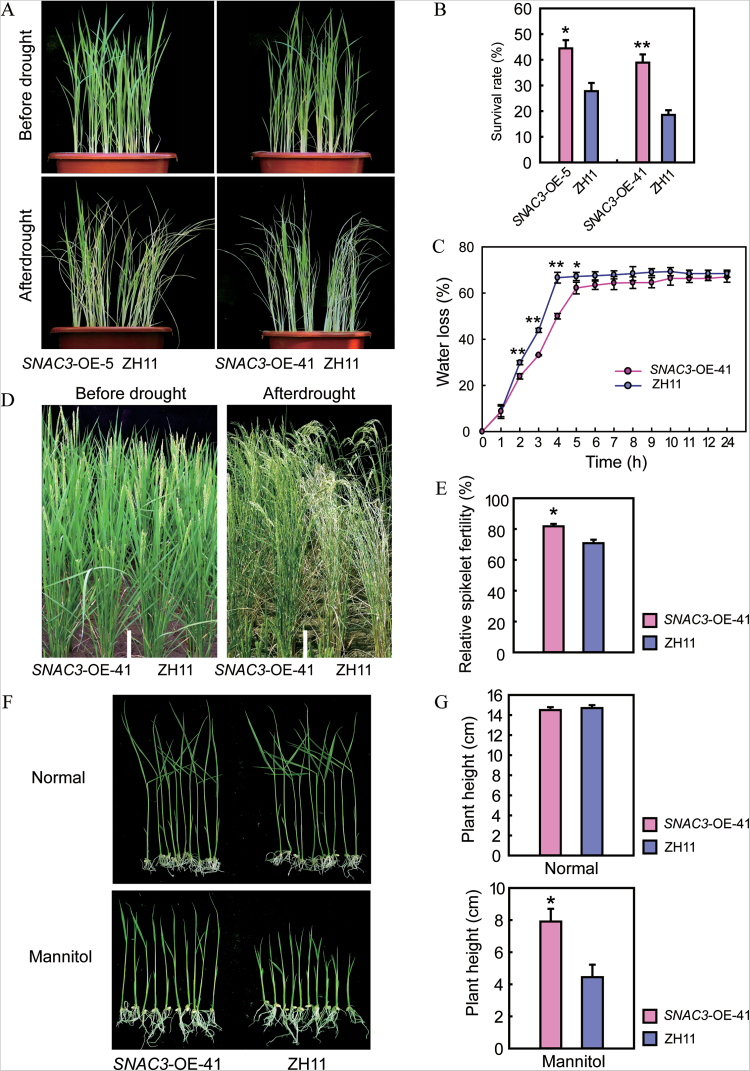
The SNAC3-OE transgenic plants were also tested for drought resistance at both the vegetative and reproductive stages. At the vegetative stage, four-leaf-stage seedlings grown in soil were subjected to drought stress treatment by withholding the water supply for 10 d. After recovery for 7 d, 40–45% of the SNAC3-OE plants had recovered, while only 20–25% of the WT plants survived (Fig. 3A, B). These results indicated that SNAC3-OE plants exhibited increased drought resistance at the vegetative growth stage. The SNAC3-OE-41 line was used for further physiological determination. The water loss rates of detached leaves from the SNAC3-OE and WT plants were gauged. In agreement with the phenotype, the detached leaves from SNAC3-OE plants lost water more slowly than the WT (Fig. 3C). For drought stress at the reproductive stage, plants were grown in a paddy field equipped with a movable rain-off shelter. Drought stress was initiated at the booting stage by stopping irrigation. SNAC3-OE transgenic plants showed later wilting than the WT during the stress treatment (Fig. 3D). After maturation, the relative spikelet fertility of the SNAC3-OE plants was significantly (P<0.05) higher than that of the WT (Fig. 3E), suggesting that overexpression of SNAC3 also enhanced drought resistance at the reproductive stage.
Fig. 3.
Enhanced drought resistance of the SNAC3-OE transgenic plants. (A) Phenotype of the SNAC3-OE plants under drought stress conditions at the seedling stage. Four-leaf stage plants were growth without water supply for 10 d, followed by rewatering for 7 d. (B) Survival rates of SNAC3-OE and ZH11 after drought stress treatment. Data represent the mean ±SE (n=3). *P<0.05, t-test; **P<0.01, t-test. (C) Water loss rates of detached leaves from the SNAC3-OE and ZH11 plants. Data represent the mean ±SE (n=3). *P<0.05, t-test; **P<0.01, t-test. (D) Enhanced drought resistance of SNAC3-OE transgenic plants at the reproductive stage. (E) Relative spikelet fertility of SNAC3-OE and ZH11 under drought stress treatment at the reproductive stage. Data represent the mean ±SE (n=12). *P<0.05, t-test. (F) Enhanced tolerance of SNAC3-OE plants to osmotic stress conditions. (G) Plant height of SNAC3-OE and ZH11 plants under normal and osmotic stress conditions. Data represent the mean ±SE (n=12). *P<0.05, t-test; **P<0.01, t-test.
The SNAC3-OE-41 plants were further tested under osmotic stress conditions. There was no obvious difference between the SNAC3-OE and WT plants grown on normal MS medium (Fig. 3F), while the shoot lengths of the SNAC3-OE plants were significantly longer than that of the WT plants grown on MS medium containing 120mM mannitol (Fig. 3F, G), indicating that overexpression of SNAC3 had a positive effect on improving osmotic stress tolerance in rice.
Suppressing SNAC3 by RNAi decreased tolerance to heat, drought, and oxidative stresses
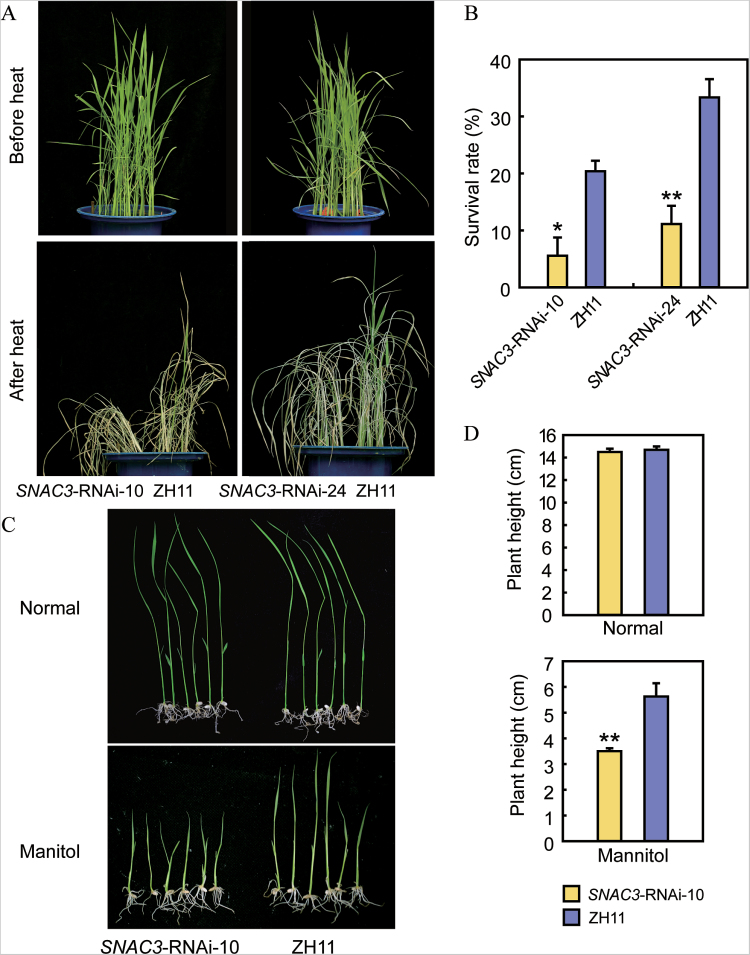
To validate further the functions of SNAC3 in rice, an RNAi construct was generated to suppress the endogenous expression of SNAC3 in transgenic plants. The transcript levels of >30 T0 SNAC3-RNAi plants were checked by qPCR, and the transcript levels of SNAC3 in >50% of the transgenic plants were significantly inhibited (Supplementary Fig. S3 at JXB online). Four-leaf stage SNAC3-RNAi and WT seedlings were subjected to heat stress treatment in a 42 °C growth chamber for 1–2 d. During the course of the stress treatment, the SNAC3-RNAi plants wilted earlier than the WT (Fig. 4A). The survival rate of the RNAi seedlings was lower than that of the WT after recovery (Fig. 4B), suggesting that suppressing SNAC3 weakened the heat tolerance in rice. The responses of the SNAC3-RNAi-10 plants to osmotic stress were also tested. In contrast to the WT plants, the RNAi plants showed increased sensitivity to osmotic stress at the post-germination stage (Fig. 4C). After treatment with 120mM mannitol for 7 d, the shoot lengths of the SNAC3-RNAi plants were significantly shorter than that of the WT (Fig. 4D). These phenotypes of the SNAC3-RNAi plants which were in contrast to the performances of the SNAC3-OE plants under the same stress conditions further supported the positive role of SNAC3 in response to heat, drought, and oxidative stress treatment.
Fig. 4.
Phenotype of SNAC3-RNAi transgenic plants under heat and osmotic stresses. (A) Four-leaf stage plants were subjected to 42 °C heat stress in a growth chamber (14h light/10h dark) for 1–2 d, and then transferred to normal growth conditions. (B) Survival rates of SNAC3-RNAi and ZH11 plants after heat stress treatment. Data represent the mean ±SE (n=3). *P<0.05, t-test; **P<0.01, t-test. (C) Enhanced sensitivity of SNAC3-RNAi plants to osmotic stress conditions. (D) Plant height of SNAC3-RNAi and ZH11 under normal and osmotic stress conditions. Data represent the mean ±SE (n=12). **P<0.01, t-test.
SNAC3 participates in the regulation of ROS metabolism and oxidative stress
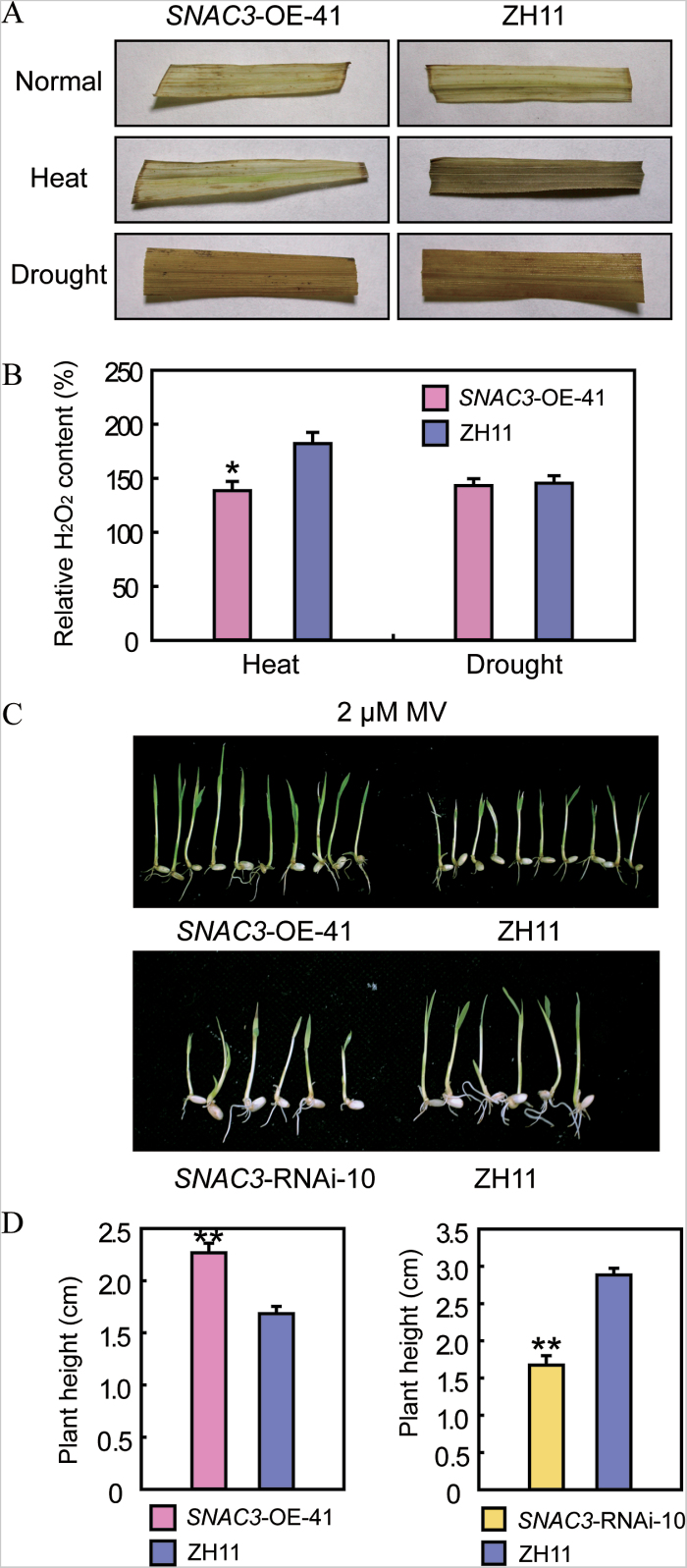
Based on the above results, it was hypothesized that SNAC3 may be involved in regulating ROS metabolism pathways. Oxidative stress usually results in excessive accumulation of H2O2. DAB staining analysis indicated that there was no significant difference in H2O2 accumulation between the SNAC3-OE and the WT plants under normal and drought stress conditions. However, the SNAC3-OE leaves showed visibly less H2O2 accumulation under heat stress conditions (Fig. 5A). The quantitative analysis also supports the DAB staining results (Fig. 5B). The potential role of SNAC3 in oxidative stress tolerance was further examined by using a well-known inducer of oxidative stress, MV (paraquat). Germinated seeds of SNAC3-OE, SNAC3-RNAi, and the WT with similar vigour were placed on MS medium containing 2 μΜ MV. After 7 d, the SNAC3-OE plants exhibited less growth inhibition and etiolation compared with the WT plants, while the RNAi plants showed more severe growth inhibition and etiolation than the WT (Fig. 5C, D). The results suggested that SNAC3 may positively regulate oxidative stress tolerance.
Fig. 5.
SNAC3 participated in regulation of ROS metabolism. (A) DAB staining of leaves from SNAC3-OE and ZH11 seedlings under normal conditions, and heat and drought stress treatments. (B) Relative H2O2 content in leaves from SNAC3-OE and ZH11 seedlings under heat, drought treatments, and normal conditions. Data are the mean ±SE (n=3). *P<0.05, t-test. (C) Enhanced tolerance of SNAC3-OE plants and enhanced sensitivity of SNAC3-RNAi plants to oxidative stress conditions. (D) Plant height of SNAC3-OE, SNAC3-RNAi, and ZH11 under oxidative stress treatments. Data are the mean ±SE (n=12). **P<0.01, t-test.
The expression of ROS scavenging-related genes was altered significantly in the SNAC3-OE and RNAi rice
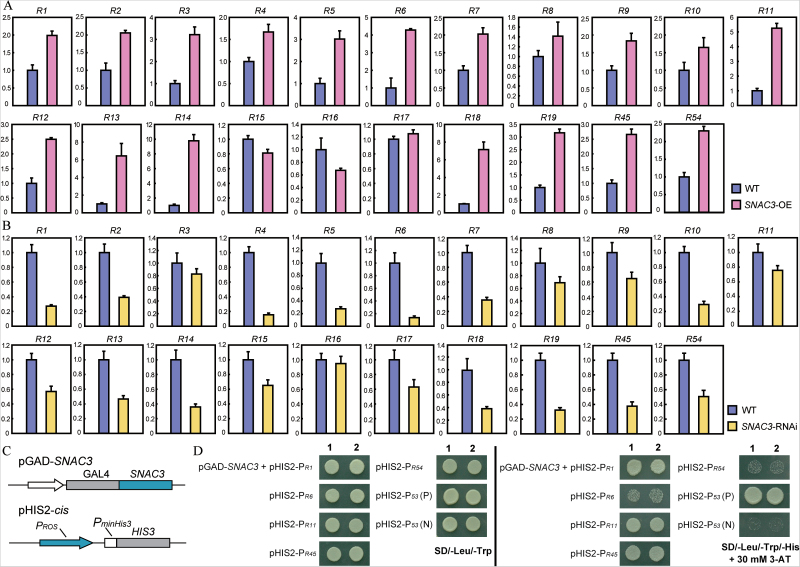
The positive effect of SNAC3-OE on oxidative stress tolerance hinted that SNAC3 may be involved in the regulation of ROS homeostasis. ROS-scavenging enzymes play crucial roles in ROS homeostasis, and ascorbate peroxidase (APX), superoxide dismutase (SOD), and catalase (CAT) are three major types of these enzymes (Apel and Hirt, 2004). Therefore, the expression levels of 19 genes (named R1–R19 in this study) encoding APX, SOD, or CAT were detected in the SNAC3-OE and RNAi transgenic plants along with the WT by using qPCR. Two other ROS-related genes, R45 (RbohF, LOC_Os08g35210) and R54 (Prx IIE2, LOC_Os02g09940), with significant co-expression with SNAC3 in the microarray database were also included in the qPCR analysis. The results indicated that 14 of the 19 tested ROS-related genes were dramatically up-regulated in the SNAC3-OE plants (Fig. 6A). In contrast, the transcript levels of 13 genes were significantly decreased in the SNAC3-RNAi plants (Fig. 6B). The above results suggested that SNAC3 may be a key regulator upstream of a large number of ROS genes, and overexpressing SNAC3 could trigger the induction of a series of ROS genes to cope positively with oxidative stress and other adverse environmental conditions.
Fig. 6.
SNAC3 directly regulates ROS-associated genes. (A) Expression of ROS-associated genes in SNAC3-overexpression materials. Error bars indicated the SE based on three technical replicates. (B) Expression of ROS-associated genes in SNAC3-repression materials. Error bars indicate the SE based on three technical replicates. (C) The schematic structure of the constructs for yeast one-hybrid analysis. (D) pGAD-SNAC3 and each of the reporter constructs were co-transformed into yeast strain Y187 and the transformants were examined by growth performance on SD/-Leu/-Trp medium and on SD/-Leu/-Trp/-His medium containing 30 mmol l–1 3-AT. pGAD-53 was co-transformed with pHIS2-P53 as a positive control (P), and pGAD-SNAC3 was co-transformed with pHIS2-P53 as a negative control (N). Labels 1 and 2 indicate two independent transformants of each transformation event. (This figure is available in colour at JXB online.)
Identification of putative targets of SNAC3
Since so many genes which were involved in ROS pathways were up-regulated in SNAC3-OE transgenic plants, and the SNAC3-OE plants accumulated less H2O2 under heat stress conditions, it was presumed that some of these genes might be directly regulated by SNAC3. Five ROS-related genes showing significant co-expression patterns with SNAC3 (Supplementary Table S2 at JXB online) were identified by using the publicly available rice expression profile databases (CREP, http://crep.ncpgr.cn/crep-cgi/home.pl; and Rice Oligonucleotide Array Database, http://www.ricearray.org/coexpression/coexpression.shtml). The five genes are R1 (LOC_Os02g02400, CATA), R6 (LOC_Os04g14680, APX3), R11 (LOC_Os02g34810, APX8), R45 (LOC_Os08g35210, NADPH oxidase, RbohF), and R54 (LOC_Os02g09940, peroxiredoxin, Prx IIE2), and the corresponding names are shown in Fig. 6. Sequence analysis indicated that the NAC-specific cis-elements including NACRS and CDBS are enriched in the promoters of these genes (Supplementary Table S3), which further supported that some of these ROS-related genes may be the direct target genes of SNAC3. Yeast one-hybrid assay was performed to examine whether SNAC3 can bind to the promoters of the five genes mentioned above. Fragments containing the NACRS and CDBS elements were amplified from the promoter regions of five ROS genes and inserted into the pHIS2 vector to generate the destination constructs (pHIS2-PR1, pHIS2-PR6, pHIS2-PR11, pHIS2-PR45, and pHIS2-PR54) which were subsequently co-transformed with the pGADT7-SNAC3 fusion construct into the yeast strain Y187 (Fig. 6C). The growth performance showed that the co-transformants of pGAD-SNAC3 along with pHIS2-PR1, pHIS2-PR11, or pHIS2-PR45 grew well on the SD/-Trp/-Leu/-His medium in the presence of 30mM 3-AT, whereas the growth of the co-transformants of pGAD-SNAC3 along with pHIS2-PR6 and pHIS2-PR54 was significantly inhibited, similar to the negative control (Fig. 6D), suggesting that SNAC3 could bind to the promoters of R1 (CATA), R11 (APX8), and R45 (RbohF) and activate the reporter gene expression in yeast.
Identification of SNAC3-interacting proteins
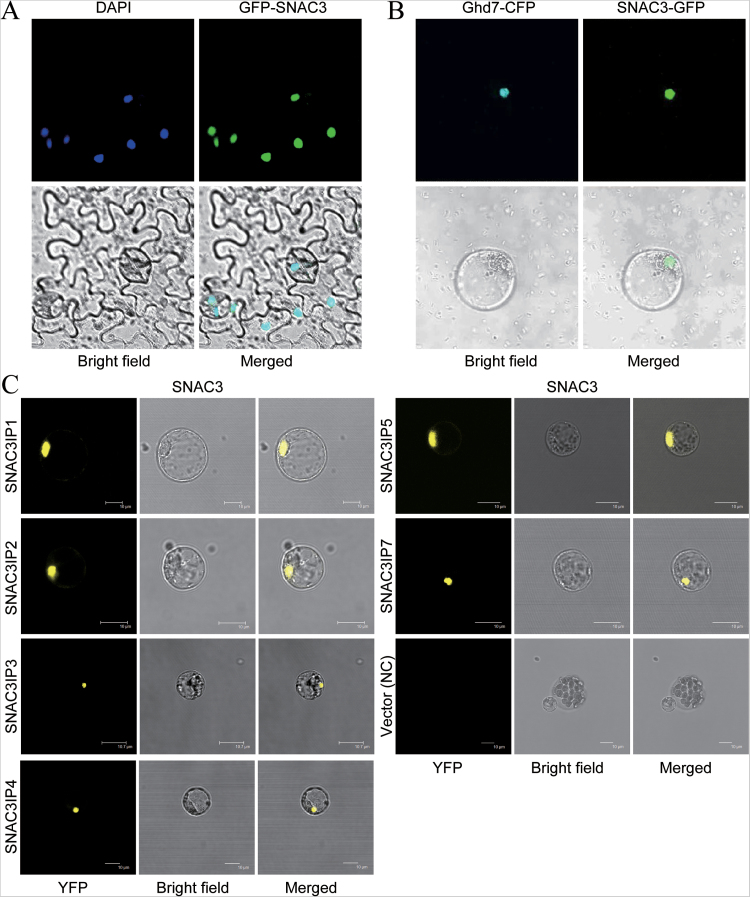
To verify SNAC3 as a transcription factor and identify the putative interacting protein of SNAC3, the subcellular location of the SNAC3 protein was first determined. A construct expressing a GFP-SNAC3 fusion gene driven by the Cauliflower mosaic virus 35S (CaMV35S) promoter was generated and introduced into tobacco using an Agrobacterium-mediated method to perform a transient expression in the leaves. As shown in Fig. 7A, a green fluorescent proteim (GFP) fluorescence signal was detected in the region which overlapped with the nucleus-specific DAPI staining, suggesting that SNAC3 is a nuclear protein. Transient expression in a rice protoplast system was applied to confirm this result. A Ghd7–cyan fluorescent protein (CFP) fusion protein was used as a nuclear localization marker, as Ghd7 is a previously reported nuclear protein in rice (Xue et al., 2008). A SNAC3–GFP fusion protein was co-expressed with Ghd7–CFP in rice protoplasts, and the fluorescence signal was observed and captured using confocal microscopy. The results revealed that the GFP fluorescence signal matched exactly with the CFP fluorescence signal (Fig. 7B), suggesting that SNAC3 is located in the nucleus.
Fig. 7.
Identification of SNAC3-interacting proteins. (A) Nuclear localization of SNAC3 in tobacco epidermal cells. Infection of tobacco leaves by Agrobacterium containing the 35S promoter-driven SNAC3–GFP fusion expression construct. DAPI staining indicates the nuclear regions. (B) Nuclear localization of SNAC3 in rice protoplasts. Ghd7–CFP and SNAC3–GFP were co-transformed into etiolated shoot protoplasts of rice. Ghd7–CFP was used as a nuclear marker. (C) Confirmation of SNAC3 and SNAC3IP1/SNAC3IP2/SNAC3IP3/SNAC3IP4/SNAC3IP5/SNAC3IP7 interaction using BiFC in rice protoplasts. NC, negative control.
By using the SNAC3 regulatory region as the bait for yeast two-hybrid screening, eight putative SNAC3-interacting proteins were obtained (Supplementary Table S4 at JXB online). The interactions between SNAC3 and six interacting proteins (SNAC3IP1, SNAC3IP2, SNAC3IP3, SNAC3IP4, SNAC3IP5, and SNAC3IP7) were confirmed by BiFC using the rice protoplast expression system (Fig. 7C). The putative SNAC3-interacting proteins, including phosphoglycerate mutase, WD domain-containing protein, cytochrome P450 72A1, protein phosphatase 2C, and oxidoreductase, have been reported to be associated with plant responses to abiotic stress (Tahtiharju and Palva, 2001; Bray, 2004; Lee et al., 2010; Kosova et al., 2011; Narsai et al., 2013).
SNAC3 function may be independent of ABA
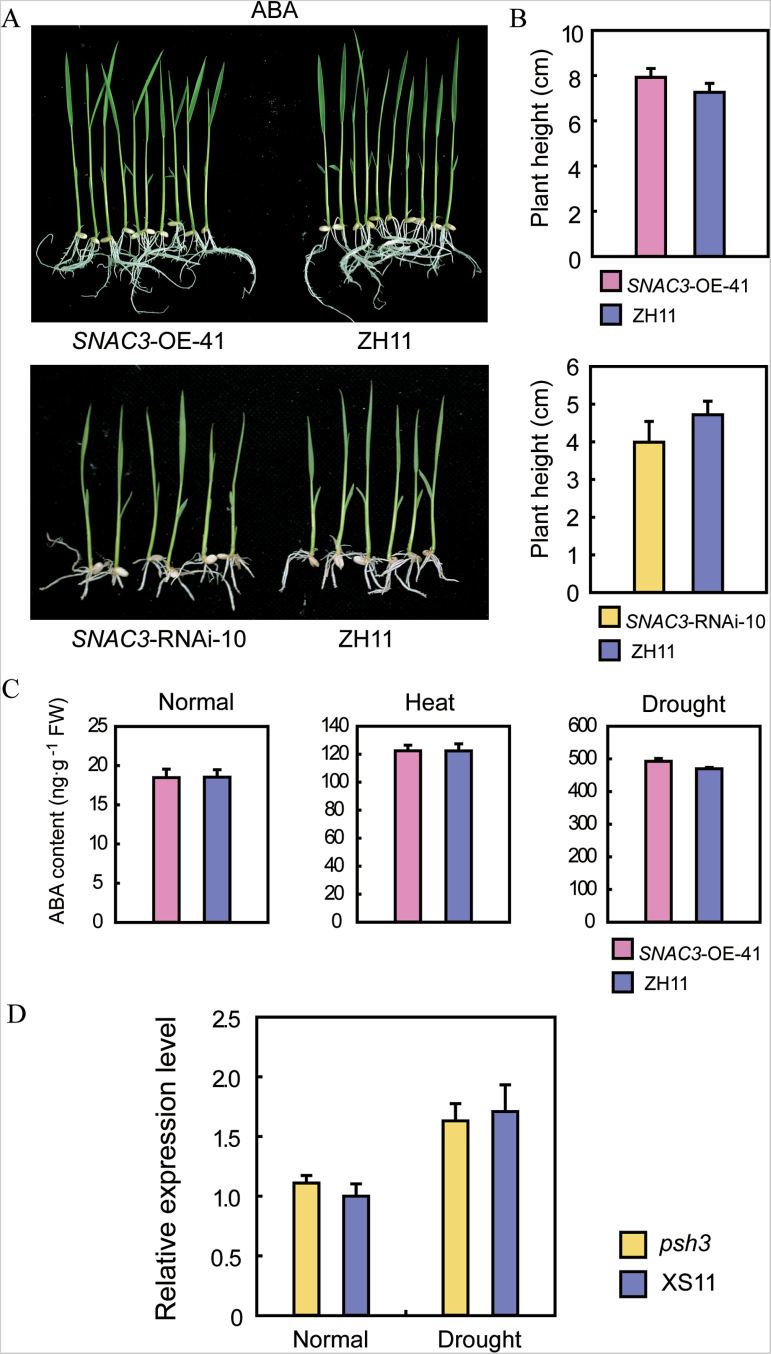
As an essential phytohormone, ABA controls various processes throughout the life cycle of plants, especially in response to external environmental stimuli (Schroeder et al., 2001). Previous studies have reported that several NAC genes are involved in stress responses in an ABA-dependent manner. To examine whether the function of SNAC3 in stress resistance relies on ABA, the sensitivity of SNAC3-OE and RNAi transgenic plants to ABA treatment was tested at the post-germination stage. Compared with the WT, neither the SNAC3-OE nor RNAi plants showed significant differences in shoot lengths under ABA treatment (Fig. 8A, B). Determination of the endogenous ABA content indicated that there was no significant difference in the ABA content between SNAC3-OE and the control plants under both normal and stress (heat or drought) conditions (Fig. 8C). The transcript levels of SNAC3 in the ABA-deficient mutant phs3 and the corresponding WT (XS11) were also analysed by qPCR. As shown in Fig. 8D, the expression levels of SNAC3 showed no obvious differences in the phs3 mutant and in the XS11 WT. Furthermore, the transcript abundance of a series of key genes for ABA biosynthesis and signal transduction showed no significant differences in the SNAC3-OE and WT plants (data not shown). These results suggest that SNAC3 may function mainly through ABA-independent pathways.
Fig. 8.
SNAC3 regulates stress responses in an ABA-independent manner. (A) No significant difference was observed between the SNAC3-OE, SNAC3-RNAi, and the control (ZH11) under ABA treatment. (B) Plant height of SNAC3-OE, SNAC3-RNAi, and ZH11 under ABA treatment. (C) Endogenous ABA content of SNAC3-OE and ZH11 WT leaves under normal conditions, drought stress, and heat stress conditions. Data represent the mean ±SE (n=3). (D) Expression analysis of SNAC3 in the ABA-deficient mutant phs3 under normal and drought stress conditions. Error bars indicate the SE based on three replicates.
Discussion
SNAC3 confers heat tolerance by modulating the expression of downstream ROS genes
Various stresses including drought, high salinity, extreme temperatures, and heavy metals may give rise to the overaccumulation of ROS, which can cause damage to plants (Mittler, 2002). Two NAC transcription factors (NTL4 and JUB1) were demonstrated to be involved in abiotic stress responses through the regulation of ROS metabolism (Lee et al., 2012; Wu et al., 2012). However, NAC transcription factors conferring stress resistance by directly regulating ROS metabolism genes have not been reported. The present data revealed that SNAC3 is a positive regulator in response to heat and oxidative stresses in rice. The H2O2 and MDA content which accumulated in the leaves of transgenic plants overexpressing SNAC3 was significantly lower than that in the control plants, and the relative ion leakage was also lower in the transgenic plants (Fig. 2C, D), suggesting that the improved heat resistance of the SNAC3-OE plants may be due to the stronger capability to scavenge ROS under heat stress conditions to maintain a lower degree of membrane lipid peroxidation. Supporting this, overexpression of SNAC3 led to the induction of many ROS-associated genes, while suppression of SNAC3 caused the down-regulation of these genes (Fig. 6A, B). Therefore, it is proposed that SNAC3 is a positive NAC regulator of heat resistance through controlling the expression of downstream genes involved in the ROS pathway.
The results indicated that SNAC3 overexpression not only enhanced heat and oxidative tolerance, but also improved the drought resistance of the transgenic rice plants. However, distinct from the up-regulated ROS-scavenging genes and reduced accumulation of H2O2 in the SNAC3-OE plants under heat stress, there were no significant differences in H2O2 accumulation between the SNAC3-OE and the control plants under drought stress conditions (Fig. 5A, B), implying that SNAC3 may confer heat and drought stress tolerance via different pathways. Measurements on the rates of water loss revealed that SNAC3-OE plants lose water more slowly than the control plants (Fig. 3C). Additionally, overexpressing SNAC3 increased the osmotic tolerance to mannitol (Fig. 3F). These results imply that SNAC3 may regulate drought resistance mainly by reducing water loss in leaves and osmotic adjustment through unknown pathways.
Inadequate water supply is often accompanied by high temperature, which severely limits the yield of crops. Therefore, it is important to pyramid the genes which can increase tolerance to drought or heat stresses or to discover genes which can enhance tolerance to both drought and heat stresses (Barnabás et al., 2002). Overexpressing SNAC3 can increase not only heat tolerance via enhancing the cell membrane stability and maintaining the redox homeostasis, but also drought tolerance by reducing water loss in rice. Therefore, this gene may be a promising candidate gene for genetic engineering in generating crops with improved drought and heat tolerance.
Target genes of SNAC3 and their potential roles in stress response
Transcription factors and cis-acting elements are central components of the regulation networks in plants. Identification of the direct downstream target genes is an effective strategy to elucidate the function of the SNAC3 transcription factor. Co-expression analysis indicated that five ROS-scavenging or metabolism-related genes, namely R1 (LOC_Os02g02400, CATA), R6 (LOC_Os04g14680, APX3), R11 (LOC_Os02g34810, APX8), R45 (LOC_Os08g35210, RbohF), and R54 (LOC_Os02g09940, Prx IIE2), shared significant correlations with the SNAC3 expression levels under normal or stress conditions. The NAC-specific NACRS and CDBS cis-elements are enriched in their promoter regions (Supplementary Table S3 at JXB online), and these genes were up-regulated in SNAC3-OE plants (Fig. 6A). Yeast one-hybrid analysis further substantiated that SNAC3 can bind to the promoter regions of three of the genes (R1, R11, and R45). R1, R11, and R45 encode OsCATA, OsAPX8, and OsRboh F, respectively, and they are all responsive to oxidative stress. Homologous genes of R1, R11, and R45 in Arabidopsis encode CAT2 (At4g35090), thylAPX (At1g77490), and NADPH oxidase F (At1g64060, AtrbohF), respectively, and their participation in the maintenance of the intracellular redox equilibrium and ROS signalling transduction has been well documented (Kalbina and Strid, 2006; Queval et al., 2007; Maruta et al., 2010; Mittler et al., 2011; Wang et al., 2013). It is speculated that R1, R11, and R45 probably have similar functions in rice, and SNAC3 confers heat resistance mainly through the control of the expression of these ROS-related target genes. CAT has been regarded as in charge of removing excess reactive oxygen intermediates (ROIs) during stress, while APX is responsible for fine-tuning of ROS signals (Mittler, 2002). As key producers of ROS under both normal and stress conditions in plants, NADPH oxidases (Rboh/Nox) are considered as the engines of ROS signalling and involved in response to stress (Suzuki et al., 2013; Wang et al., 2013). Rbohs not only regulate a multitude of vital biological processes, but also act as pivotal signalling nodes in the ROS network and integrate a great variety of signal transduction pathways with ROS signalling (Suzuki et al., 2013). OsNox6 expression was found to be slightly down-regulated by drought stress and up-regulated by high temperature (Wang et al., 2013). The interaction between SNAC3 protein and the promoter regions of three ROS-associated genes (R1-CATA, R11-APX3, and R45-RbohF) revealed that SNAC3 is likely to regulate not only ROS scavenging but also ROS metabolism.
Six SNAC3-interacting protein genes with evidence for a role in stress responses have been identified here. Further investigations are needed to illustrate the detailed mechanisms of the interactions between SNAC3 and these proteins for regulating stress responses in rice.
SNAC3 mediated stress responses in an ABA-independent manner
The phytohormone ABA co-ordinates a complicated network in response to various abiotic stress conditions in plants, and numerous transcription factors take part in stress responses in ABA-mediated pathways. Many stress-responsive genes contain an ABA-responsive promoter element (ABRE) recognition sequence with an ACGT core motif. Basic leucine zipper transcription factors called ABFs/AREBs such as ABF2 and AREB1 are documented to induce stress-responsive gene expression by interacting with the corresponding ABRE cis-acting elements (Xiong et al., 2002; Kim et al., 2004; Fujita et al., 2005). Other transcription factors including CBF4 (DREB1D), RD22BP1, and AtMYB2 are also reported to be regulated through ABA-dependent pathways (Abe et al., 1997, 2003; Haake et al., 2002). Several NAC transcription factors have been reported to play critical roles in the ABA-mediated stress response pathways in plants. In contrast, the present data indicated that SNAC3 may exert its functions in stress resistance mainly through ABA-independent pathways. Compared with the WT, neither the SNAC3-OE nor the RNAi transgenic plants exhibited obvious changes in the sensitivity to ABA treatment (Fig. 8A, B). The SNAC3-OE and the control plants had equivalent endogenous ABA levels under heat, drought, and normal conditions (Fig. 8C). The expression level of SNAC3 was not affected in the ABA-deficient mutant in rice (Fig. 8D). In addition, overexpression of SNAC3 in rice did not alter the transcript abundance of the genes which are essential for ABA biosynthesis and the signal transduction pathways (data not shown). Together, this evidence supported that SNAC3 mediates drought and heat stress resistance mainly in an ABA-independent manner.
In conclusion, SNAC3 is a stress-responsive NAC transcription factor which confers heat tolerance by means of modulating H2O2 homeostasis through controlling the expression of ROS genes. SNAC3 also contributes to drought resistance and osmotic adjustment. In contrast to some previously reported NAC genes, SNAC3 may function mainly through ABA-independent pathways in rice. The positive effect of SNAC3 in improving resistance to multiple stresses implies high potential in engineering this gene for stress resistance improvement in crops.
Supplementary data
Supplemenaty data are available at JXB online.
Figure S1. Schematic diagram of SNAC3 gene structure.
Figure S2. Overexpression of SNAC3.
Figure S3. Suppression of SNAC3 by RNAi.
Table S1. List of primers used in this study.
Table S2. Correlation between SNAC3 and the ROS genes.
Table S3. Distribution of NACRS and CDBS elements in the ROS genes.
Table S4. Information on putative SNAC3-interacting proteins.
Acknowledgements
We acknowledge the Arabidopsis Biological Resource Center (Columbus, OH, USA), Lei Wang (Huazhong Agricultural University), and Rongjian Ye (Huazhong Agricultural University) for providing pHBT-sGFP, 35S:Ghd7–CFP, and the DX2181 plasmid, respectively. We also thank Jun You for providing a portion of the qPCR primers. This work was supported by grants from the National Program on High Technology Development (2012AA10A303) and the National Program for Basic Research of China (2012CB114305).
Glossary
Abbreviations:
- DAB
3,3′-diaminobenzidine
- MDA
malondiadehyde
- MV
methyl viologen
- NAC
NAM, ATAF1/2, and CUC2
- OE
overexpression
- qPCR
quantitative real-time reverse transcription–PCR
- ROS
reactive oxygen species.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. 1997. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarie S, Hanaoka N, Ueno O, Miyazaki A, Kubota F, Agata W, Kaufman PB. 1998. Effects of silicon on tolerance to water deficit and heat stress in rice plants (Oryza sativa L.), monitored by electrolyte leakage. Plant Production Science 1, 96–103. [Google Scholar]
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Bajji M, Kinet J M, Lutts S. 2002. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regulation 36, 61–70. [Google Scholar]
- Barnabás B, Jäger K, Fehér A. 2008. The effect of drought and heat stress on reproductive processes in cereals. Plant, Cell and Environment 31, 11–38. [DOI] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Bechtold U, Albihlal WS, Lawson T, et al. 2013. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. Journal of Experimental Botany 64, 3467–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. 2004. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana . Journal of Experimental Botany 55, 2331–2341. [DOI] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, et al. 2013. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis . The Plant Cell 25, 3472–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. 2008. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. The Plant Cell 20, 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wang N, Cui F, Li X, Xiao J, Xiong L. 2010. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiology 154, 1304–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Larsen S, Lo Leggio L. 2004. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Reports 5, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L. 2008. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Molecular Genetics and Genomics 280, 547–563. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. 2004. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. The Plant Journal 39, 863–876. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. 2005. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis . The Plant Cell 17, 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612. [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. 2002. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology 130, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. 2010. Research on plant abiotic stress responses in the post-genome era: past, present and future. The Plant Journal 61, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. 2006. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA 103, 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. 2008. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology 67, 169–181. [DOI] [PubMed] [Google Scholar]
- Huang Y, Xiao B, Xiong L. 2007. Characterization of a stress responsive proteinase inhibitor gene with positive effect in improving drought resistance in rice. Planta 226, 73–85. [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ. 2006. The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. The Plant Journal 47, 751–760. [DOI] [PubMed] [Google Scholar]
- Jeong DH, German MA, Rymarquis LA, Thatcher SR, Green PJ. 2010. Abiotic stress-associated miRNAs: detection and functional analysis. Methods in Molecular Biology 592, 203–230. [DOI] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. 2010. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiology 153, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. 2013. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnology Journal 11, 101–114. [DOI] [PubMed] [Google Scholar]
- Jeong JS, Park YT, Jung H, Park SH, Kim JK. 2009. Rice NAC proteins act as homodimers and heterodimers. Plant Biotechnology Reports 3, 127–134. [Google Scholar]
- Kalbina I, Strid A. 2006. The role of NADPH oxidase and MAP kinase phosphatase in UV-B-dependent gene expression in Arabidopsis. Plant, Cell and Environment 29, 1783–1793. [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. 2007. Building blocks for plant gene assembly. Plant Physiology 145, 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY. 2004. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. The Plant Journal 40, 75–87. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. 2006. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis . The Plant Cell 18, 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova K, Vitamvas P, Prasil IT, Renaut J. 2011. Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. Journal of Proteomics 74, 1301–1322. [DOI] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee HJ, Park CM. 2012. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. The Plant Journal 70, 831–844. [DOI] [PubMed] [Google Scholar]
- Lee SC, Choi DS, Hwang IS, Hwang BK. 2010. The pepper oxidoreductase CaOXR1 interacts with the transcription factor CaRAV1 and is required for salt and osmotic stress tolerance. Plant Molecular Biology 73, 409–424. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Liu H, Li X, Xiao J, Wang S. 2012. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice–bacterium interaction. Plant Methods 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JQ, Allan DL, Vance CP. 2010. Systemic signaling and local sensing of phosphate in common bean: cross-talk between photosynthate and microRNA399. Molecular Plant 3, 428–437. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Maruta T, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S. 2010. Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant and Cell Physiology 51, 190–200. [DOI] [PubMed] [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Narsai R, Wang C, Chen J, Wu J, Shou H, Whelan J. 2013. Antagonistic, overlapping and distinct responses to biotic stress in rice (Oryza sativa) and interactions with abiotic stress. BMC Genomics 14, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Li X, Hicks LM, Xiong L. 2010. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiology 152, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. 2008. Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69, 1773–1781. [DOI] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. 2012. NAC proteins: regulation and role in stress tolerance. Trends in Plant Science 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakiere B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G. 2007. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. The Plant Journal 52, 640–657. [DOI] [PubMed] [Google Scholar]
- Sacar MD, Hamzeiy H, Allmer J. 2013. Can MiRBase provide positive data for machine learning for the detection of MiRNA hairpins? Journal of Integrative Bioinformatics 10, 215. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. 2001. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410, 327–330. [DOI] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S, Balazadeh S, Mueller-Roeber B. 2012. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signaling Behavior 7, 1518–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Ye T, Chen F, Deng J, Yang P, Zhang Y, Chan Z. 2014. The Cysteine2/Histidine2-Type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiology 165, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58, 221–227. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. 1993. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist 125, 27–58. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. 2011. Respiratory burst oxidases: the engines of ROS signaling. Current Opinion in Plant Biology 14, 691–699. [DOI] [PubMed] [Google Scholar]
- Tahtiharju S, Palva T. 2001. Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana . The Plant Journal 26, 461–470. [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. 2001. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L. 2012. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiology 158, 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. 2006. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lin A, Loake GJ, Chu C. 2013. H2O2-induced leaf cell death and the crosstalk of reactive nitric/oxygen species. Journal of Integrative Plant Biology 55, 202–208. [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, et al. 2012. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis . The Plant Cell 24, 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. 2002. Cell signaling during cold, drought, and salt stress. The Plant Cell 14 Suppl, S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- You J, Zong W, Li X, Ning J, Hu H, Xiao J, Xiong L. 2013. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. Journal of Experimental Botany 64, 569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Shen X, Li X, Xu C, Wang S. 2007. Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226, 953–960. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. 2011. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Research 39, 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. 2006. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis . The Plant Cell 18, 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, Jiao Y, et al. 2007. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Molecular Biology 63, 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.