Highlight
Monomers of three developmentally regulated isoforms of glutamine synthetase holoenzyme in wheat cultivars were identified and their molecular weight was determined using modified blue native electrophoresis combined with an in-gel activity assay.
Key words: Assembly, enzyme isoform, glutamine synthetase, nitrogen, protein modification, wheat.
Abstract
Glutamine synthetase (GS; EC 6.3.1.2) plays a crucial role in the assimilation and re-assimilation of ammonia derived from a wide variety of metabolic processes during plant growth and development. Here, three developmentally regulated isoforms of GS holoenzyme in the leaf of wheat (Triticum aestivum L.) seedlings are described using native-PAGE with a transferase activity assay. The isoforms showed different mobilities in gels, with GSII>GSIII>GSI. The cytosolic GSI was composed of three subunits, GS1, GSr1, and GSr2, with the same molecular weight (39.2kDa), but different pI values. GSI appeared at leaf emergence and was active throughout the leaf lifespan. GSII and GSIII, both located in the chloroplast, were each composed of a single 42.1kDa subunit with different pI values. GSII was active mainly in green leaves, while GSIII showed brief but higher activity in green leaves grown under field conditions. LC-MS/MS experiments revealed that GSII and GSIII have the same amino acid sequence, but GSII has more modification sites. With a modified blue native electrophoresis (BNE) technique and in-gel catalytic activity analysis, only two GS isoforms were observed: one cytosolic and one chloroplastic. Mass calibrations on BNE gels showed that the cytosolic GS1 holoenzyme was ~490kDa and likely a dodecamer, and the chloroplastic GS2 holoenzyme was ~240kDa and likely a hexamer. Our experimental data suggest that the activity of GS isoforms in wheat is regulated by subcellular localization, assembly, and modification to achieve their roles during plant development.
Introduction
Glutamine synthetase (GS; EC 6.3.1.2) assimilates ammonium into glutamine, which is then used for the biosynthesis of all essential nitrogenous compounds (Miflin and Lea, 1977). All of the nitrogen within a plant, whether derived initially from nitrate, ammonium, N2 fixation, or catabolism of proteins, is channelled through reactions catalyzed by GS. Accordingly, GS plays a central role in nitrogen metabolism of vascular plants, and is a major checkpoint controlling plant growth and productivity (Brestic et al., 2014; Habash et al., 2007; Kichey et al., 2006; Lothier et al., 2011; Miflin and Habash, 2002; Simons et al., 2014; Tabuchi et al., 2005; Thomsen et al., 2014).
In vascular plants, two isoforms of GS were initially resolved by chromatography (Mann et al., 1979; McNally et al., 1983; McParland et al., 1976; O’Neal and Joy, 1973). Based on subcellular location, GS is classified as the cytosolic isoform (GS1) or the chloroplastic isoform (GS2). Electron microscopy analyses revealed that soybean (Glycine max) and common bean (Phaseolus vulgaris) GS enzymes are octamers (Llorca et al., 2006; McParland et al., 1976), whereas the crystallographic structures of GS in maize and Medicago truncatula are decamers (Torreira et al., 2014; Unno et al., 2006). GS2 is a single polypeptide (42–45kDa) encoded by one nuclear gene, whereas GS1 is composed of polypeptides with the same molecular weight (38–40kDa), but different pI values, and is encoded by three to five nuclear genes depending on the species. The GS isozymes have different metabolic roles, and their activities vary with plant development in different organs and cell types (Bernard et al., 2008; Coque et al., 2006; Finnemann and Schjoerring, 2000; Gallais et al., 2006; Habash et al., 2001; Kamachi et al., 1991; Li et al., 1993; Ohashi et al., 2015; Orsel et al., 2014; Tabuchi et al., 2007). GS2 is the predominant isozyme in leaf mesophyll cells, where it assimilates ammonia originating from nitrate reduction and photorespiration (Kumagai et al., 2011; Tobin and Yamaya, 2001). GS1 has multiple metabolic functions, involving primary ammonium assimilation in the roots, and catabolism ammonia re-assimilation for transport and distribution throughout the plant, and localizes to the vascular cells of various tissue of Arabidopsis (Guan et al., 2015), wheat (Triticum aestivum L.) (Bernard et al., 2008; Kichey et al., 2005), rice (Oryza sativa) (Tabuchi et al., 2005), tobacco (Nicotiana tabacum) (Brugiere et al., 1999), and potato (Solanum tuberosum) (Pereira et al., 1995). During leaf senescence, GS1 functions in the assimilation and recycling of the ammonia generated from catabolic processes (Avila-Ospina et al., 2014; Bernard and Habash, 2009; Kamachi et al., 1992). This role, confirmed by quantitative trait locus analysis, or gene mutation or knockout, is particularly important during grain development in cereals when nitrogen is remobilized to the reproductive sinks (Brestic et al., 2014; Guan et al., 2015; Martin et al., 2006; Tabuchi et al., 2005). To achieve these multiple non-overlapping roles, GS isozymes are regulated at the levels of transcription, translation, subcellular localization, assembly of subunits into the holoenzyme, post-translational modification of the enzyme, and protein turnover (Hirel et al., 2001; Ishiyama et al., 2004; Kamachi et al., 1991; Li et al., 1993; Lima et al., 2006; Orsel et al., 2014; Ortega et al., 1999; Riedel et al., 2001; Tabuchi et al., 2007; Tobin and Yamaya, 2001). In wheat, seven genetic loci coding for three different forms of GS1 have been identified. TaGS1a, TaGS1b, and TaGS1c code for GS1;1, TaGSr1 and TaGSr2 code for GS1;2 (also called GSr), and TaGSe1 and TaGSe2 code for GS1;3 (also called GSe). Three alleles coding for GS2 (TaGS2a, TaGS2b, and TaGS2c) are known (Bernard et al., 2008; Thomsen et al., 2014). Here, three developmentally regulated GS holoenzymes in wheat are reported that can be separated by native-PAGE in plants.
There are several methods to analyze the oligomeric active state of a native protein, including gel filtration, analytical ultracentrifugation, electron microscopy, and X-ray crystallography. However, all of these methods require a substantial amount of protein and/or investment in expensive equipment. Blue native-PAGE [blue native electrophoresis (BNE)] and clear native-PAGE [clear native electrophoresis (CNE)] are performed with smaller amounts of protein and have been widely used to study membrane protein complexes (Filoni et al., 2013; Strecker et al., 2010; Wittig et al., 2007; Wittig and Schagger, 2009). The application of these techniques to determine the native molecular weights and oligomeric states of the GS isoforms in wheat is reported here.
Materials and methods
Plant material and growth conditions
Wheat (T. aestivum L.) cvs Yumai 34, 49, and 50 were used for isolation of GS isoforms during the growth of first leaves in April in Zhengzhou, China. The other cultivars shown in Supplementary Fig. S1 (at JXB online) were grown similarly, but at different times of the year. The seeds were put in a disk covered with wet gauze at 25 °C until they germinated; they were then sown in individual pots (25cm upper diameter, 17cm lower diameter, and 25cm high) filled with vermiculite and grown outside under natural light/temperature. Each plant was sprayed with 300ml sterile water every day. The leaves were sampled three times from the onset of seedling emergence to the first leaf turning yellow, harvesting 0.5g per sample. Wheat cv. Yumai 49 was also grown under 10/14h light/dark periods at 23 °C in a growth chamber, with 800 μmol m–2 s–1 photon flux density at the top of the canopy during the light period, and watered with 200ml Hoagland solution (containing 1mM KH2PO4, 5mM KNO3, 1mM MgSO4, 0.5mM CaSO4, 4mM Ca(NO3)2, 1mM Mg(NO3)2, 0.5mM CaCl2, 1 μM H3PO3, 1 μM CuSO4.5H2O, 1 μM MnCl2.4H2O, 1 μM Na2MoO4, and 1 μM ZnSO4.7H2O) twice a week to keep the soil moist and supply sufficient nutrients. Fully expanded green leaves were collected for the preparation of intact chloroplasts and enzyme analysis when the wheat seedlings had four or five leaves.
Preparation of leaf extract
Sample (0.5g each) were ground into powder in a chilled mortar with liquid N2 and mixed with 1.5ml Extraction Buffer (100mM Tris, 1mM EDTA, 1mM MgCl2, and 10mM β-mercaptoethanol, pH 7.6). The extract was centrifuged at 13 000 g at 4 °C for 30min. The supernatant was prepared for native gel analysis.
Isolation of chloroplasts
Intact chloroplasts were isolated essentially as described by Theg et al. (1989). The washed chloroplast pellet after the Percoll step was resuspended in 350 μl Extraction Buffer and incubated on ice for 10min to break the organelles. The resuspension was used for immunoblotting or centrifuged at 13 000 g for 10min and the supernatant used for enzyme assays immediately.
In-gel detection of GS activity and molecular weight
Four gel systems (running at 4 °C) were used to separate the proteins and detect GS activities in the gel. First, a discontinuous native-PAGE system was used according to Robert and Wong (1986). The native gel system employed a 1.5 mm×170 mm×100mm gel, the analyzing gel was composed of 5% polyacrylamide (pH 8.7), and the stacking gel was 3% polyacrylamide (pH 6.7). Samples were normalized to 30 μl (~60 μg protein) from 0.5g FW leaves in each lane, and electrophoresis was carried out at 80V for the stacking gel and 120V for the resolving gel at 4 °C. Second, the BNE system was used according to Wittig et al. (2006, 2007) with the following modifications. The sample gel contained 3.5% polyacrylamide and the gradient resolving gel contained 4–13% polyacrylamide; the gel was 1.5 mm×170 mm×100mm, the sample buffer was Extraction Buffer, and before loading sample in the gel, 100 μl sample was mixed with 10 μl 50% glycerol. The current was limited to 15 mA during electrophoresis. The gel was run with cathode buffer A (0.02% Coomassie Blue G250, 50mM Tricine, and 5mM imidazole, pH 7.0) until the blue dye front was up to half of the gel length; cathode buffer A was then removed and the gel was run with cathode buffer B (0.002% Coomassie Blue G250, 50mM Tricine, and 5mM imidazole, pH 7.0) until the blue dye front moved out of the gel. Third, the BNE protocol was modified as follows. The gradient gel was prepared as the second gel system. After the gel ran for 1h with cathode buffer A, cathode buffer A was removed and the gel was run with cathode buffer C (50mM Tricine and 5mM imidazole, pH 7.0) until the blue dye front moved out of the gel. Fourth, for CNE, the gradient gel was prepared in the second gel system. Each well received 50 μl cathode buffer A before the sample was loaded. The gel was run only with cathode buffer C (50mM Tricine and 5mM imidazole, pH 7.0) until the blue dye front moved out of the gel.
After electrophoresis, GS activity was detected in-gel by the conversion of l-glutamine to γ-glutamyl hydroxamate (Barratt, 1980). The gel was immersed in 100ml reaction buffer (100mM Tricine, 1.3mM EDTA, 20mM sodium arsenate, 20mM MgSO4, 0.5mM ADP, 25mM hydroxylamine, and 50mM l-glutamate, pH 7.4) and incubated at 37 °C for 45min with slow shaking, after which the reaction buffer was removed. The reaction was terminated by adding 50 μl stop solution (370mM FeCl3, 200mM trichloroacetic acid, and 700mM HCl) for ~3min until GS activity appeared as a brownish band in the yellow background. The gel was washed twice with cool distilled H2O and scanned immediately. The GS bands were marked with a blade and then the gel was stained with Coomassie Blue R250. The molecular mass of the GS isoforms was calculated by comparison with molecular weight standards (Life Technologies) using Quantity One software.
GS recovery and GS subunit identification
After the GS activity was detected in the gel, the band of interest was excised with a scalpel, rinsed with 0.5mM EDTA, pH 7.6, and ground in a chilled mortar with this same solution. The homogenate was centrifuged at 12 000 g at 4 °C for 20min and then the extraction was mixed with an equal volume of 0.1M Tris-buffered phenol (pH 8.0). After being centrifuged (12 000 g) at 4 °C for 20min, the protein in the phenol phase was precipitated with 4 vols 0.1M ammonium acetate in methanol overnight at –20 °C. The proteins recovered by centrifugation were washed once with 1ml cold methanol and twice with 1ml cold acetone, and then resolved in SDS sample buffer for analysis. A discontinuous SDS-PAGE system was implemented according to Laemmli (1970), with a 12.5% polyacrylamide analyzing gel and a 6% polyacrylamide stacking gel, and electrophoresis was performed at room temperature. Proteins were transferred to polyvinylidene difluoride membranes for blot analysis. GS polypeptides were detected using polyclonal antisera (generously provided by Bertrand Hirel) raised against GS2 of tobacco (Bernard et al., 2008)
Protein extraction for two-dimensional immunoblots
Protein was extracted using a modification of the phenol-based method (Finnemann and Schjoerring, 2000). Wheat leaves were homogenized in an ice-cold mortar and pestle in SDS sample buffer (0.1M Tris-Cl, 2% SDS, 5% 2-mercaptoethanol, and 30% sucrose, pH 8.0) and then mixed with the same volume of Tris-buffered phenol (pH 8.0). The homogenate was centrifuged at 10 000 g for 5min at 4 °C. Protein in the upper phenol phase was precipitated with 5 vols 0.1M ammonium acetate in methanol for 30min at –20 °C. The protein recovered by centrifugation was washed twice with cold 80% acetone and then dissolved in SDS sample buffer or rehydration buffer (8M urea, 4% CHAPS, 2% IPG buffer, pH 4–7, and 20mM DTT). Protein was quantified by the Bio-Rad protein assay with BSA as standard. For two-dimensional gel electrophoresis, wheat leaf proteins (600 μg) were loaded on to pH 4–7 Immobiline Drystrips (7cm; Amersham) by passive rehydration overnight at room temperature. The rehydrated strips were resolved in a Multiphor II apparatus (Pharmacia Biotech) by isoelectric focusing for 8000 Vh at 10 °C. The resolved strips were consecutively equilibrated in DTT solution (50mM Tris, 6M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue, and 1% DTT, pH 8.8) and iodoacetamide solution (50mM Tris, 6M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue, and 2.5% iodoacetamide, pH 8.8) for 15min, and the secondary SDS–PAGE was run with 12.5% gels. After electrophoresis, immunodetection was performed as described above.
Protein identification by LC-MS/MS
After GS activity was detected in a native-PAGE gel, each band of interest was excised with a scalpel and washed with 75% ethanol. The samples were sent to the Genome Center at the University of California-Davis for identification of GS proteins and modifications by LC-MS/MS, and analyzed with Scaffold 4.0 software.
Results
Three isoforms of GS are active during wheat leaf development
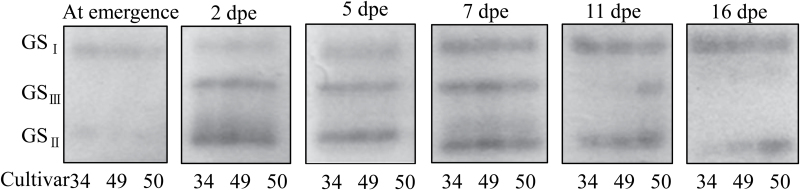
To elucidate the role of GS isoforms during wheat development, leaf extracts from three cultivars of wheat seedlings at different developmental stages were separated by native-PAGE and GS isoforms were detected using transferase activity staining. Three isoforms of GS holoenzyme were identified in the wheat leaf. GSI, GSII, and GSIII emerged sequentially with the development of the first leaf (Fig. 1). Conversely, GSIII, GSII, and GSI disappeared in turn with leaf senescence (Fig. 1). GSII had the highest mobility in native-PAGE, followed by GSIII and then GSI. GSI was present during the seed germination stage and increased progressively in activity until leaf senescence. GSII appeared with leaf expansion and maintained the highest activity in green leaves, but disappeared when the leaf turned yellow. GSIII had a shorter period of activity, albeit with higher activity, in the growing green leaf, i.e. from the stage of fast leaf expansion (Fig. 1, panels 2dpe and 5 dpe) to the full-length size (Fig. 1, panel 7dpe). It was deduced that GSI was likely cytosolic (GS1) because it was present from the onset of germination until leaf senescence. GSII was considered likely to be chloroplastic (GS2) because it was the dominant GS in green leaves. GSIII has not been described before.
Fig. 1.
GS isoforms as a function of leaf development in wheat. GS isoforms were monitored using native-PAGE (5%) in the first leaf in seedlings of wheat cvs Yumai 34, 49. and 50. Samples were taken at the indicated times; dpe, days post-emergence.
Subunit composition and subcellular localization of GS isoforms
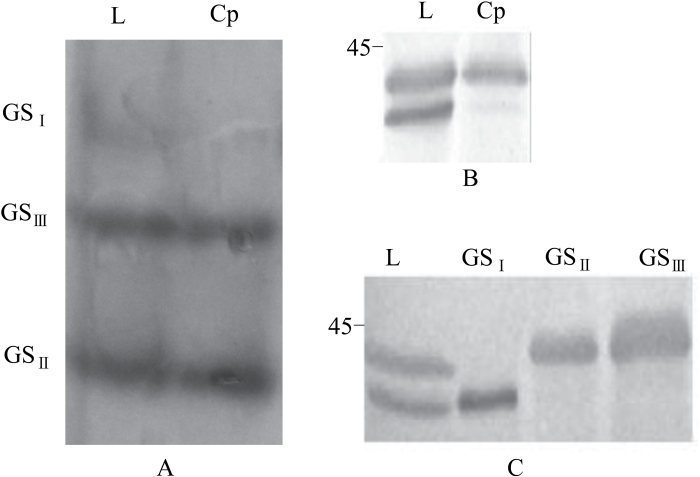
To characterize the GS isoforms further, chloroplasts were isolated and GS activity therein detected using native-PAGE. Both GSII and GSIII were found in chloroplasts (Fig. 2A) and immunoblots revealed them to be derived from only one GS polypeptide of 43.6kDa (Fig. 2B). Based on these findings, they are considered to be chloroplastic GS2-type isoforms. By contrast, GSI was not found in chloroplasts and so might be cytoplasmic. To confirm the identities of the GS isoforms, leaf proteins were separated by native-PAGE, and the bands displaying GS activity were recovered from the gel by chemical extraction, separated by SDS-PAGE, and detected by immunoblot. The band giving GSI activity was composed of a 39kDa subunit (Fig. 2C), consistent with its identification as cytosolic GS1. The bands showing GSII and GSIII activity were found to be composed of a single 43.6kDa subunit (Fig. 2C). This suggests that differential modifications of GS2-related subunits might confer different mobility to the holoenzymes (GSII and GSIII). Although the subunit of GS2 was bigger than that of GS1 (43.6 versus 39kDa), GSII and GSIII ran faster than GSI in the native-PAGE system, suggesting they may have different oligomeric states.
Fig. 2.
Electrophoretic separation and detection of cytosolic and chloroplastic GS, and GS-related subunits. (A) In-gel detection of GS activity. Protein extracts were prepared from leaves (L) and purified chloroplasts (Cp), separated by 5% native-PAGE, and GS isoforms were detected based on GS activity in the gel. (B) Detection of chloroplastic GS-related subunits. Proteins from the leaf or isolated chloroplasts were separated by 12.5% SDS-PAGE, and probed with antibodies against tobacco GS2. (C) Subunit composition of GS isozymes of wheat leaf. Protein extracts were separated using 5% native PAGE. GS isozyme bands were recovered by chemical extraction, separated by 12.5% SDS-PAGE, and probed with anti-tobacco GS2.
Identification of GS protein sequences and modifications
To determine unambiguously the proteins corresponding to GSI, GSII, and GSIII, bands containing these activities were excised from native gels and analyzed by LC-MS/MS (Supplementary Table S1 at JXB online). Protein identification revealed that the GSI band contained fragments of three previously described cytosolic GS isoforms, GS1, GSr1, and GSr2 (equivalent to GS1;1 and two forms of GS1;2), although no GSe (equivalent to GS1;3) was detected. The complete sequences of GSII and GSIII were obtained in the LC-MS/MS experiment; they were identical with a theoretical molecular weight of 42.1kDa, identical to GS2a, GS2b, and GS2c. The LC-MS/MS data (parent error <5 ppm) indicated that GSII had many more modifications than did GSIII, including acetylation, oxidation, dioxidation, and deamidation. In comparison, GSIII had fewer sites of oxidation, one site of acetylation, and more sites of deamidation (Table 1).
Table 1.
Protein modifications detected in GSII and GSIII
| Isoform | Oxidation | Acetyl | Deamidation | Dioxidation |
|---|---|---|---|---|
| GS II | ||||
| Sites | 7 | 3 | 7 | 6 |
| Amino acid | M W Q N | A L G | Q N | W M |
| GS III | ||||
| Sites | 4 | 1 | 9 | 5 |
| Amino acid | M W Q N | L | W Q N | W M |
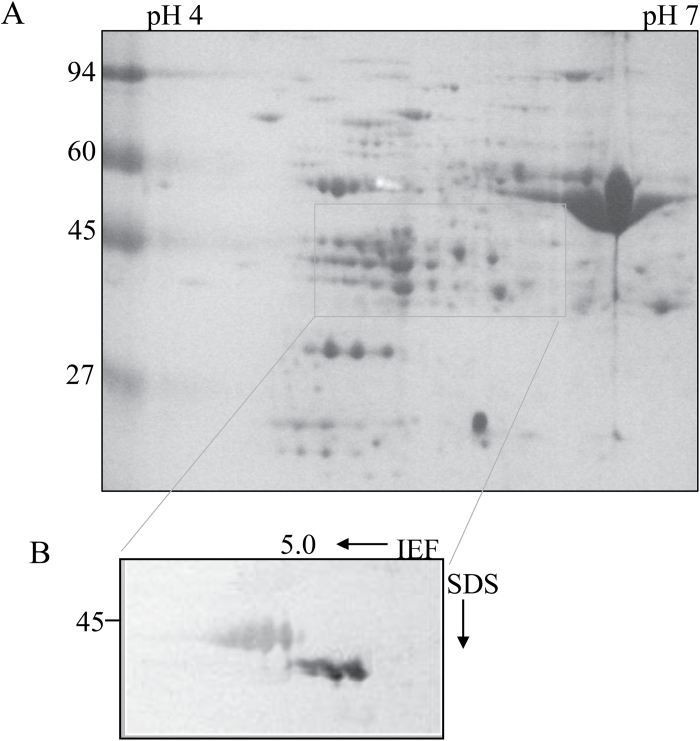
Two-dimensional separation of leaf proteins and subsequent immunoblotting revealed two groups of GS polypeptides with distinct pI values. In one, three 39kDa GS1-related polypeptides were detected with pI values of 5.08, 5.13, and 5.21 (Fig. 3). In the other, three 43.6kDa GS2-related polypeptides were detected with pI values of 4.8, 4.94, and 5.05. These combined data suggest that the GSII and GSIII activity bands seen in native-PAGE were each composed of GS2-related proteins with different pI values due to different modifications.
Fig. 3.
Two-dimensional analysis of GS isozymes in wheat leaves. (A) Two-dimensional gel of proteins extracted from the wheat leaf; stained with Coomassie Blue R250. The rectangle shows the region putatively containing GS spots. (B) Two-dimensional immunoblots of GS subunits in wheat leaf. IEF, isoelectric focussing. Leaf proteins (50 μg) were separated by two-dimensional electrophoresis, and the region thought to contain GS (larger than the rectangle) was electroblotted and probed with antibodies against tobacco GS2.
Oligomers of GS isoenzymes
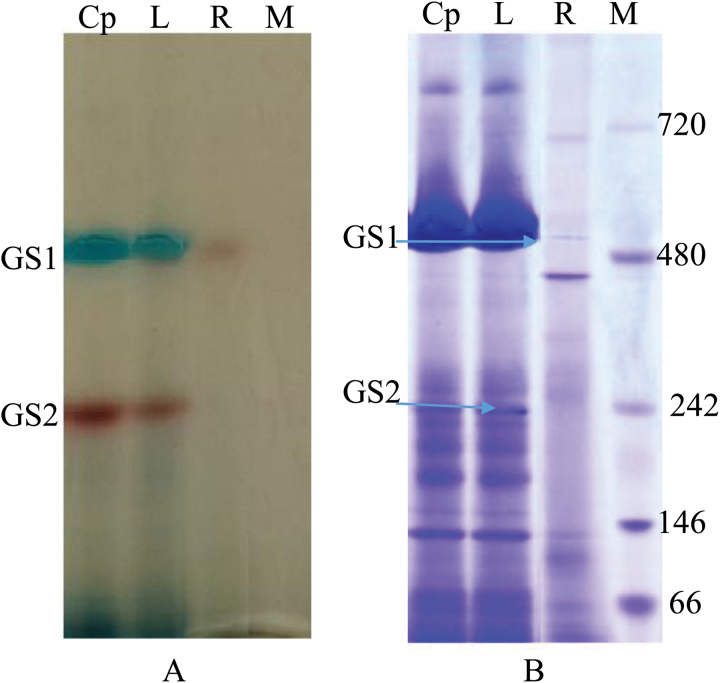
The authors next turned their attention to elucidation of the oligomeric state of the wheat GS isoforms. In BNE, protein complexes are separated according to size in acrylamide gradient gels and their sizes can be calibrated with standards. During the initial BNE experiments, the presence of the Coomassie Brilliant Blue G250 (referred to hereafter as G250) interfered with the activity stain for GS1 (Supplementary Fig. S2 at JXB online). To overcome this the BNE protocol was modified to include a 1h separation of proteins in the presence of G250 and then an additional 3h separation in which the cathode buffer was replaced with one lacking the dye. This allowed a separation of the protein complexes by molecular weight and subsequent detection by the transferase activity stain. Fig. 4 shows the results of such an analysis and reveals that the GS1 holoenzyme has a molecular weight of ~490kDa. Given the molecular weight of the monomer (39kDa), this indicates that the GS1 holoenzyme is likely a dodecamer. Interestingly, GS2, containing both GSII and GSIII activities, ran as a single band with a molecular weight of 240kDa, suggesting that the holoenzyme is most likely a hexamer. These results additionally suggest that while GS1 and GS2 have distinct migrations in native-PAGE gels, in part due to different oligomeric states of their respective holoenzymes, the different mobilities of GSII and GSIII, both GS2 isoforms, in the same native gel system must be due in part to their different modifications as described in Table 1.
Fig. 4.
Identification of the native molecular weight of GS isoforms using BNE. (A) Soluble proteins (~100 μg) from wheat seedling chloroplasts (Cp), leafs (L), and roots (R) were separated by BNE on a 4–13% polyacrylamide gradient gel, and GS isoforms were detected with a transferase activity assay. (B) The GS bands were marked and the gel stained with Coomassie Blue R250. M, high-molecular-weight markers (mass in kDa given to the right of the gel). (This figure is available in colour at JXB online.)
Discussion
In vascular plants, only two isoforms of the GS holoenzyme have been resolved by standard chromatography (Mann et al., 1979; McNally et al., 1983; McParland et al., 1976; O’Neal and Joy, 1973) and by native gel electrophoresis (Nagy et al., 2013; Péscsvéradi et al., 2009). Here, three isoforms of GS were separated using native gels in wheat seedlings (Figs 1 and 2); the third isoform, GSIII, has not been reported before. The fact that all three isoforms were observed in >20 cultivars (Supplementary Fig. S1 at JXB online) confirms they are generally present in wheat seedlings. In general, GSIII is readily observed in plants grown in the field, but is more difficult to detect in those grown in growth chambers. This might be ascribed to the lower light intensity of the growth chamber environment (<1000 μmol m–2 s–1 photon flux density), as opposed to sunlight which can provide ~2000 μmol m–2 s–1 photon flux density of photosynthetically active radiation in the field. The appearance of GSIII is also regulated with the leaf development independent of whether the wheat seedlings were grown without (Fig. 1) or with (Supplementary Fig. S3 at JXB online) nitrogen. These two factors may be responsible for it not having been identified in previous studies (Nagy et al., 2013; Péscsvéradi et al., 2009). For instance, GSIII was not found in green leaves of 14-d-old seedlings growing in chambers, whereas it was abundant in green leaves of 21-d-old seedlings from the same chamber (Fig. 2A).
The early estimates for the molecular weight of GS oligomers came from direct measurements (gel filtration, sedimentation equilibrium) with purified protein (Mann et al., 1979; McParland et al., 1976). It is difficult to obtain sufficient quantities of purified protein from plants for assembly and structure studies, and, consequently, these previous GS structural studies used proteins heterologously expressed in Escherichia coli (Llorca et al., 2006; O’Neal and Joy, 1973; Seabra et al., 2009; Torreira et al., 2014; Unno et al., 2006). BNE techniques provide an independent method for separating protein complexes with high resolution and from which their molecular weights can be determined. When soluble proteins from leaves or isolated chloroplasts were separated by BNE and GS isoform activities were detected in the gel, only one GS isoform (corresponding to chloroplastic GS2) was observed (Supplementary Fig. S2 at JXB online). This is likely because cytosolic GS1 activity appears to be sensitive to G250 and because Rubisco runs in the same part of the gel, overwhelming the transferase signal. When leaf or chloroplast soluble proteins were separated using the BNE procedure, but omitting G250 from the cathode buffer, two GS isoforms were detected in leaf extracts (GS1 and GS2) and one in the chloroplast (GS2), although chloroplastic GS in leaves and chloroplasts displayed slightly different mobility (Supplementary Fig. S4 at JXB online). Finally, the BNE protocol was modified, first running the gel with cathode buffer containing 0.02% G250 for 1h to ensure that all proteins carried same net charge and then changing to cathode buffer without G250 to reduce the influence of G250 on GS activity. Although cytosolic GS activity was weak, it was detectable. Furthermore, chloroplastic GS in leaf extracts and chloroplast extracts had the same mobility in the gel.
Based on this modified BNE procedure, the cytosolic GS, with the activity in GSI, was likely to be a dodecamer, the same as GSr1 in soybean nodules analyzed by analytical ultracentrifugation and native-PAGE (Masalkar and Roberts, 2015) and GS in prokaryotes (Eisenberg et al., 2000). Our data suggest that wheat chloroplastic GS, with activities in GSII and GSIII, was a hexamer, which differs markedly from other plants. For example, GSs from soybean (McParland et al., 1976) and common bean (Llorca et al., 2006) are octamers as determined by electron microscopy; GSs from maize and M. truncatula and GS1β from soybean are decamers (Masalkar and Roberts, 2015; Torreira et al., 2014; Unno et al., 2006) as determined by X-ray crystallography. Separated by CNE and detected by in-gel GS activity assay, GS from spinach stroma is a decamer (Kimata-Ariga and Hase, 2014). Confidence in the methodology for oligomeric state determination is strengthened by Supplementary Fig. S5 (at JXB online) in which maize GS can be seen running as a decamer. Nonetheless, the authors recognize that the oligomeric state reported here should be further evaluated by additional techniques, and future plans call for expression of recombinant TaGS1 and TaGS2 and analysis by X-ray crystallography. The authors are also working to compare GS proteins in M. truncatula, soybean, Arabidopsis, spinach, and common bean with those in wheat using the modified BNE system.
The different pI values for the wheat GS proteins have different origins. GSI, cytosolic GS, is encoded by a multigene family, GS1 and GSr, and the pI values detected here are close to those predicted by analysis of the respective gene sequences (Bernard et al., 2008). However, no GSe was identified by MS analysis, perhaps because its expression was too low to be detectable in leaves and roots during the wheat seedling stage (Bernard et al., 2008). In contrast, chloroplastic GS is encoded by three alleles (TaGS2a, TaGS2b, and TaGS2c), and the different pI values must arise from different post-translational modifications. Lima (2006) reported that phosphorylated GS2 of M. truncatula interacts with 14-3-3 proteins, which leads to selective proteolysis and thus inactivation of the plastid isoform. In E. coli, GS is reported to be inactivated by adenlylation (Liaw et al., 1993), and oxidation of soybean root GS has been reported to lead to its inactivation and increased susceptibility to degradation (Ortega et al., 1999). No evidence for phosphorylation of GS2 was found here, but numerous other modifications were detected, and they were different for GSII and GSIII (Supplementary Fig. S6 at JXB online). For instance, GSII had three acetylation sites in its N-terminal region, whereas GSIII had one such site. GSII had seven sites of oxidation, while GSIII had four, even though GSII activity was higher than that of GSIII in all but the most active stages of leaf development (Fig. 1). Whether the various modifications regulate GS2 enzyme activity or stability remains to be established.
Recently, an analysis of GS in Arabidopsis was presented in which 11 different GS1 isoforms were detected in a 7% resolving gel using a phosphate release assay and no GS2 was observed (only this group detected GS activity using this method) (Dragicevic et al., 2014). This is clearly different from the situation described herein for wheat and emphasizes the potential diversity of GS assembly configurations in different plant species.
When GS isoenzymes were originally discovered, their putative functions were deduced from their pattern of expression in different tissues during plant development and further confirmed by genetic methods (Bao et al., 2014; Gadaleta et al., 2011; Gadaleta et al., 2014; Guo et al., 2013; Habash et al., 2007; Martin et al., 2006). GS1, vascular-localized cytosolic GS, is proposed to be involved in the re-assimilation of ammonium released during leaf senescence and in transporting ammonium from source organs to sink organs, e.g. from fully expanded leaves to new leaves (Bernard et al., 2008; Kichey et al., 2006; Kichey et al., 2007). GS2, however, was found in both mitochondria and chloroplasts in Arabidopsis (Taira et al., 2004), suggesting that this isoform is active in re-assimilation of the large pool of ammonia released by photorespiration. It is noteworthy that neither GS activity nor GS subunits in mitochondria purified from wheat leaves were detected in the present report (data not shown). This, along with the detection of GSIII primarily in leaves grown under relatively high light intensity, would be consistent with a function of chloroplastic GS2 in original nitrogen assimilation, especially under conditions of abundant energy availability that would promote the conversion of nitrate to ammonium in the plastid. Although the physiological role of the newly described GSIII remains to be elucidated, findings presented here suggest that there is a complex and flexible regulation for GS isoforms in wheat that is coupled to nitrogen utilization and plant growth.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. GS isoforms in the leaf of different wheat cultivars.
Figure S2. GS isoforms in wheat chloroplasts, leaves, and roots.
Figure S3. GS isoforms as a function of leaf development in wheat.
Figure S4. GS isoforms in wheat chloroplast, leaf, and roots.
Figure S5. GS isoforms in wheat leaf and roots, and maize leaf and roots.
Figure S6. Amino acid modifications sites in GSII and GSIII.
Table S1. Identification of the composition of GSI, GSII, and GSIII by MS analysis.
Acknowledgements
The authors wish to thank Professor B. Hirel for the generous gift of anti-GS antibody. This work was supported by National Natural Science Funds of China (30771266) and open funds of the State Key Laboratory of Wheat and Maize Crop Science in China (39990004). The preparation of this manuscript was supported in part by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through grant DE-FG02-03ER15405 to SMT.
Glossary
Abbreviations:
- BNE
blue native electrophoresis
- CNE
clear native electrophoresis
- GS
glutamine synthetase.
References
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. 2014. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany 65, 3799–3811. [DOI] [PubMed] [Google Scholar]
- Bao AL, Zhao ZQ, Ding GD, Shi L, Xu FS, Cai HM. 2014. Accumulated expression level of cytosolic glutamine synthetase 1 gene (OsGS1; 1 or OsGS1; 2) alter plant development and the carbon-nitrogen metabolic status in rice. PLoS One 9, e95581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DHP. 1980. Method for the detection of glutamine synthetase activity on starch gels. Plant Science Letters 18, 249–255. [Google Scholar]
- Bernard SM, Habash DZ. 2009. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytologist 182, 608–620. [DOI] [PubMed] [Google Scholar]
- Bernard SM, Moller ALB, Dionisio G, et al. 2008. Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Molecular Biology 67, 89–105. [DOI] [PubMed] [Google Scholar]
- Brestic M, Zivcak M, Olsovska K, Shao HB, Kalaji HM, Allakhverdiev SI. 2014. Reduced glutamine synthetase activity plays a role in control of photosynthetic responses to high light in barley leaves. Plant Physiology and Biochemistry 81, 74–83. [DOI] [PubMed] [Google Scholar]
- Brugiere N, Dubois F, Limami AM, Lelandais M, Roux Y, Sangwan RS, Hirel B. 1999. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11, 1995–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque M, Bertin P, Hirel B, Gallais A. 2006. Genetic variation and QTLs for N-15 natural abundance in a set of maize recombinant inbred lines. Field Crops Research 97, 310–321. [Google Scholar]
- Dragicevic M, Todorovic S, Bogdanovic M, Filipovic B, Misic D, Simonovic A. 2014. Knockout mutants as a tool to identify the subunit composition of Arabidopsis glutamine synthetase isoforms. Plant Physiology and Biochemistry 79, 1–9. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Gill HS, Pfluege GMU, Rotstein SH. 2000. Structure-function relationships of glutamine synthetases. Biochimica et Biophysica Acta 1477, 122–145. [DOI] [PubMed] [Google Scholar]
- Filoni DN, Pesi R, Allegrini S, Camici M, Tozzi MG. 2013. A native electrophoretic technique to study oligomerization and activity of cytosolic 5′-nucleotidase II. Analytical and Bioanalytical Chemistry 405, 8951–8954. [DOI] [PubMed] [Google Scholar]
- Finnemann J, Schjoerring JK. 2000. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant Journal 24, 171–181. [DOI] [PubMed] [Google Scholar]
- Gadaleta A, Nigro D, Giancaspro A, Blanco A. 2011. The glutamine synthetase (GS2) genes in relation to grain protein content of durum wheat. Functional & Integrative Genomics 11, 665–670. [DOI] [PubMed] [Google Scholar]
- Gadaleta A, Nigro D, Marcotuli I, Giancaspro A, Giove SL, Blanco A. 2014. Isolation and characterisation of cytosolic glutamine synthetase (GSe) genes and association with grain protein content in durum wheat. Crop & Pasture Science 65, 38–45. [Google Scholar]
- Gallais A, Coque M, Quillere I, Prioul JL, Hirel B. 2006. Modelling postsilking nitrogen fluxes in maize (Zea mays) using 15N-labelling field experiments. New Phytologist 172, 696–707. [DOI] [PubMed] [Google Scholar]
- Guan M, Moller IS, Schjoerring JK. 2015. Two cytosolic glutamine synthetase isoforms play specific roles for seed germination and seed yield structure in Arabidopsis. Journal of Experimental Botany 66, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Sun JJ, Zhang GZ, Wang YY, Kong FM, Zhao Y, Li SS. 2013. Haplotype, molecular marker and phenotype effects associated with mineral nutrient and grain size traits of TaGS1 a in wheat. Field Crops Research 154, 119–125. [Google Scholar]
- Habash DZ, Bernard S, Schondelmaier J, Weyen J, Quarrie SA. 2007. The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. Theoretical and Applied Genetics 114, 403–419. [DOI] [PubMed] [Google Scholar]
- Habash DZ, Massiah AJ, Rong HL, Wallsgrove RM, Leigh RA. 2001. The role of cytosolic glutamine synthetase in wheat. Annals of Applied Botany 138, 83–89. [Google Scholar]
- Hirel B, Bertin P, Quillere I, et al. 2001. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiology 125, 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H. 2004. Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. Journal of Biological Chemistry 279, 16598–16605. [DOI] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Hayakawa T, Mae T, Ojima K. 1992. Changes in cytosolic glutamine synthetase polypeptide and its mRNA in a leaf blade of rice plants during natural senescence. Plant Physiology 98, 1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Mae T, Ojima K. 1991. A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiology 96, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichey T, Heumez E, Pocholle D, Pageau K, Vanacker H, Dubois F, Le Gouis J, Hirel B. 2006. Combined agronomic and physiological aspects of nitrogen management in wheat highlight a central role for glutamine synthetase. New Phytologist 169, 265–278. [DOI] [PubMed] [Google Scholar]
- Kichey T, Hirel B, Heumez E, Dubois F, Le Gouis J. 2007. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crops Research 102, 22–32. [Google Scholar]
- Kichey T, Le Gouis J, Sangwan B, Hirel B, Dubois F. 2005. Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.). Plant and Cell Physiology 46, 964–974. [DOI] [PubMed] [Google Scholar]
- Kimata-Ariga Y, Hase T. 2014. Multiple complexes of nitrogen assimilatory enzymes in spinach chloroplasts: possible mechanisms for the regulation of enzyme function. PLoS One 9, e108965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai E, Araki T, Hamaoka H, Ueno S. 2011. Ammonia emission from rice leaves in relation to photorespiration and genotypic differences in glutamine synthetase activity. Annals of Botany 108, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li MG, Villemur R, Hussey PJ, Silflow CD, Gantt JS, Snustad DP. 1993. Differential expression of six glutamine synthetase genes in Zea mays. Plant Molecular Biology 23, 401–407. [DOI] [PubMed] [Google Scholar]
- Liaw SH, Pan C, Eisenberg D. 1993. Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine. Proceedings of the National Academy of Sciences U S A 90, 4996–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. 2006. Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. Journal of Experimental Botany 57, 2751–2761. [DOI] [PubMed] [Google Scholar]
- Llorca O, Betti M, Gonzalez JM, Valencia A, Marquez AJ, Valpuesta JM. 2006. The three-dimensional structure of an eukaryotic glutamine synthetase: functional implications of its oligomeric structure. Journal of Structural Biology 156, 469–479. [DOI] [PubMed] [Google Scholar]
- Lothier J, Gaufichon L, Sormani R, Lemaitre T, Azzopardi M, Morin H, Chardon F, Reisdorf-Cren M, Avice JC, Masclaux-Daubresse C. 2011. The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. Journal of Experimental Botany 62, 1375–1390. [DOI] [PubMed] [Google Scholar]
- Mann AF, Fentem PA, Stewart GR. 1979. Identification of two forms of glutamine synthetase in barley (Hordeum vulgare). Biochemical and Biophysical Research Communications 88, 515–521. [DOI] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, Gerentes D, et al. 2006. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masalkar PD, Roberts DM. 2015. Glutamine synthetase isoforms in nitrogen-fixing soybean nodules: distinct oligomeric structures and thiol-based regulation. FEBS Letters 589, 215–221. [DOI] [PubMed] [Google Scholar]
- McNally SF, Hirel B, Gadal P, Mann AF, Stewart GR. 1983. Glutamine synthetases of higher plants: evidence for a specific isoform content related to their possible physiological role and their compartmentation within the leaf. Plant Physiology 72, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McParland RH, Guevara JG, Becker RR, Evans HJ. 1976. The purification and properties of the glutamine synthetase from the cytosol of soya-bean root nodules. Biochemical Journal 153, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Habash DZ. 2002. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. Journal of Experimental Botany 53, 979–987. [DOI] [PubMed] [Google Scholar]
- Miflin BJ, Lea PJ. 1977. Amino-acid metabolism. Annual Review of Plant Physiology and Plant Molecular Biology 28, 299–329. [Google Scholar]
- Nagy Z, Nemeth E, Guoth A, Bona L, Wodala B, Pecsvaradi A. 2013. Metabolic indicators of drought stress tolerance in wheat: glutamine synthetase isoenzymes and Rubisco. Plant Physiology and Biochemistry 67, 48–54. [DOI] [PubMed] [Google Scholar]
- O’Neal D, Joy KW. 1973. Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Archives of Biochemistry and Biophysics 159, 113–122. [DOI] [PubMed] [Google Scholar]
- Ohashi M, Ishiyama K, Kusano M, et al. 2015. Lack of cytosolic glutamine synthetase1;2 in vascular tissues of axillary buds causes severe reduction in their outgrowth and disorder of metabolic balance in rice seedlings. Plant Journal 81, 347–356. [DOI] [PubMed] [Google Scholar]
- Orsel M, Moison M, Clouet V, Thomas J, Leprince F, Canoy AS, Just J, Chalhoub B, Masclaux-Daubresse C. 2014. Sixteen cytosolic glutamine synthetase genes identified in the Brassica napus L. genome are differentially regulated depending on nitrogen regimes and leaf senescence. Journal of Experimental Botany 65, 3927–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Roche D, Sengupta-Gopalan C. 1999. Oxidative turnover of soybean root glutamine synthetase. In vitro and in vivo studies. Plant Physiology 119, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, Pissara J, Sunkel C, Salema R. 1995. Tissue-specific distribution of glutamine synthetase in potato tubers. Annals of Botany 77, 429–432. [Google Scholar]
- Péscsvéradi A, Nagy Z, Varga A, Vashegyi A, Labadi I, Galbacs G, Zsoldos F. 2009. Chloroplastic glutamine synthetase is activated by direct binding of aluminium. Physiologia Plantarum 135, 43–50. [DOI] [PubMed] [Google Scholar]
- Riedel J, Tischner R, Mack G. 2001. The chloroplastic glutamine synthetase (GS-2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 213, 396–401. [DOI] [PubMed] [Google Scholar]
- Robert FM, Wong PP. 1986. Isozymes of glutamine synthetase in Phaseolis vulgaris L. and Phaseolis lunatus L. root nodules. Plant Physiology 81, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra AR, Carvalho H, Pereira PJ. 2009. Crystallization and preliminary crystallographic characterization of glutamine synthetase from Medicago truncatula. Acta Crystallographia F65, 1309–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Saha R, Amiour N, et al. 2014. Assessing the metabolic impact of nitrogen availability using a compartmentalized maize leaf genome-scale model. Plant Physiology 166, 1659–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker V, Wumaier Z, Wittig I, Schagger H. 2010. Large pore gels to separate mega protein complexes larger than 10 MDa by blue native electrophoresis: isolation of putative respiratory strings or patches. Proteomics 10, 3379–3387. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T. 2007. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). Journal of Experimental Botany 58, 2319–2327. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, Takahashi H, Yamaya T. 2005. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant Journal 42, 641–651. [DOI] [PubMed] [Google Scholar]
- Taira M, Valtersson U, Burkhardt B, Ludwig RA. 2004. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16, 2048–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg SM, Bauerle C, Olsen LJ, Selman BR, Keegstra K. 1989. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. Journal of Biological Chemistry 264, 6730–6736. [PubMed] [Google Scholar]
- Thomsen HC, Eriksson D, Moller IS, Schjoerring JK. 2014. Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends in Plant Science 19, 656–663. [DOI] [PubMed] [Google Scholar]
- Tobin AK, Yamaya T. 2001. Cellular compartmentation of ammonium assimilation in rice and barley. Journal of Experimental Botany 52, 591–604. [PubMed] [Google Scholar]
- Torreira E, Seabra AR, Marriott H, Zhou M, Llorca O, Robinson CV, Carvalho HG, Fernandez-Tornero C, Pereira PJ. 2014. The structures of cytosolic and plastid-located glutamine synthetases from Medicago truncatula reveal a common and dynamic architecture. Acta Crystallographia D70, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno H, Uchida T, Sugawara H, Kurisu G, Sugiyama T, Yamaya T, Sakakibara H, Hase T, Kusunoki M. 2006. Atomic structure of plant glutamine synthetase: a key enzyme for plant productivity. Journal of Biological Chemistry 281, 29287–29296. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. 2006. Blue native PAGE. Nature Protocols 1, 418–428. [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schagger H. 2007. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Molecular and Cellular Proteomics 6, 1215–1225. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. 2009. Native electrophoretic techniques to identify protein-protein interactions. Proteomics 9, 5214–5223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.