Highlight
PTB proteins of potato bind to the mobile RNA, StBEL5, and enhance stability and trafficking of the RNA to select organs. This protein–RNA interaction leads to enhanced tuber production.
Key words: Mobile RNA, phloem, RNA-binding protein, Solanum tuberosum, StPTB, tuberization
Abstract
Polypyrimidine tract-binding (PTB) proteins are a family of RNA-binding proteins that function in a wide range of RNA metabolic processes by binding to motifs rich in uracils and cytosines. A PTB protein of pumpkin was identified as the core protein of an RNA–protein complex that trafficks RNA. The biological function of the PTB–RNA complex, however, has not been demonstrated. In potato, six PTB proteins have been identified, and two, designated StPTB1 and StPTB6, are similar to the phloem-mobile pumpkin type. RNA binding assays confirmed the interaction of StPTB1 and StPTB6 with discrete pyrimidine-rich sequences of the 3′-untranslated regions of the phloem-mobile mRNA, StBEL5. The promoter of StPTB1 was active in companion cells of phloem in both stem and petioles. Expression of both types was evident in phloem cells of roots and in stolons during tuber formation. RNA accumulation of both PTB proteins was induced by short days in leaves in correlation with enhanced accumulation of StBEL5 RNA. StPTB suppression lines exhibited reduced tuber yields and decreased StBEL5 RNA accumulation, whereas StPTB overexpression lines displayed an increase in tuber production correlated with the enhanced production in stolons of steady-state levels of StBEL5 transcripts and RNA of key tuber identity genes. In StPTB overexpression lines, both the stability and long-distance transport of StBEL5 transcripts were enhanced, whereas in suppression lines stability and transport decreased. Using a transgenic approach, it is shown that the StPTB family of RNA-binding proteins regulate specific stages of development through an interaction with phloem-mobile transcripts of StBEL5.
Introduction
The polypyrimidine tract-binding (PTB) family of proteins represent a multifaceted group of proteins that binds numerous mRNAs and have been implicated in a wide range of RNA metabolic processes including stability (Xu and Hecht, 2007), splicing regulation (Valcarcel and Gebauer, 1997; Xue et al., 2009), intracellular localization (Kuwahata et al., 2007), translation repression (Karakasiliotis et al., 2010), and control of long-distance transport (Ham et al., 2009). PTBs have also been implicated in alternative splicing (AS) of their own pre-mRNAs (Wollerton et al., 2004). PTBs bind viral, mammalian, and plant RNAs with binding sites rich in uracils and cytosines. The structure of a PTB protein is uniquely adapted to these multiple functions. They generally contain four RNA-recognition motifs (RRMs) of ~90 amino acids connected by linker regions and designated RRM1, RRM2, RRM3, and RRM4. Each RRM is formed by 4–5 β-sheets and contains 6–8 conserved amino acids, designated RNP1 and RNP2, that interact with CU (cytosine uracil) motifs, ranging from three to five nucleotides in length (Oberstrass et al., 2005; Auweter and Allain, 2008). The four RRMs of human PTB, hnPTB1, recognize 3, 4, 5, and 3 nucleotides, respectively (Auweter and Allain, 2008). Structural analysis has revealed that RRM1 and RRM2 are linear and function independently, whereas RRM3 and RRM4 act in a tandem, compact complex that functions as an open-faced clamp on closely spaced polypyrimidine tract motifs (Oberstrass et al., 2005).
Despite their widespread function and versatility in eukaryotes, very little information is available on the role of PTB proteins in plants. There are three PTB genes in Arabidopsis, two of which are involved in pollen tube development (Wang and Okamoto, 2009). The three PTB genes from Arabidopsis auto- and cross-regulate their expression through alternative splicing coupled to nonsense-mediated decay (Stauffer et al., 2010). Clearly, some plant PTBs play an important role in regulating AS (Wachter et al., 2012). A transcriptome-wide analysis in transgenic lines that under- and overexpressed AtPTBs and a mini-exon splicing reporter system revealed that both AtPTB1 and AtPTB2 were involved in regulating AS that impacted numerous developmental processes (Rühl et al., 2012; Simpson et al., 2014). No significant AS regulatory function, however, was observed for the distantly related AtPTB3. AtPTB1 and AtPTB2 contain three RRMs, whereas AtPTB3 contains four. This latter PTB is closely related to the pumpkin PTB protein, RBP50. Ham et al. (2009) demonstrated that RBP50 was the core protein of a phloem-mobile RNA–protein complex consisting of 16 proteins and six RNAs. Included in this group were full-length transcripts for PHLOEM PROTEIN16 (PP16-1), GIBBERELLIC ACID-INSENSITIVE (GAI), a SCARECROW-LIKE protein, SHOOT MERISTEMLESS, an ETHYLENE RESPONSE FACTOR, and a MYB transcription factor. Gel mobility shift assays confirmed that specific binding of CmRBP50 to two of its target RNAs, GAI and PP16-1 of pumpkin, was facilitated by polypyrimidine tract motifs located within the transcript sequences (Ham et al., 2009). PTB proteins are ubiquitous in the plant kingdom, and as RNA chaperones some types of PTB proteins appear to play a critical role beyond a splicing repressor function in protecting and mobilizing full-length mRNAs that are transported through the phloem. Despite important information on the biochemistry of these mobile RNA-binding proteins, however, very little is known about their biological function in whole-plant systems. As an example, virtually nothing is known about the function of AtPTB3.
Through functional and genomic analyses, the PTB family of potato (Solanum tuberosum), represented by six genes, two containing four RRMs and designated StPTB1 and StPTB6, and four three-RRM types, have been characterized. Based on amino acid sequence and function, StPTB1 and StPTB6 appear to be orthologues of AtPTB3 and CmRBP50, and serve as chaperones to full-length mRNAs that are transported through the sieve element system. RNA binding assays confirmed the interaction of StPTB1 and StPTB6 with the 3′-untranslated regions (UTRs) of the mobile RNA, StBEL5. In novel analyses using transgenic whole-plant systems, it is demonstrated that PTB proteins of potato enhance stability and regulate transport of a mobile RNA and specifically control tuber development.
Materials and methods
Identification and cloning of cDNA sequences
To obtain the full-length StPTB and StGAI cDNA sequences, the DFCI Gene Index potato database (http://www.danafarber.org) was searched using the CmRBP50 and CmGAI sequence as query, respectively. From partial cDNA sequences, the rapid amplification of cDNA ends (RACE) was performed using the First Choice RLM-RACE kit (Ambion) to identify the 5′- and 3′-untranslated sequences. Full-length cDNA sequences were then obtained by PCR amplification with 5′ and 3′ gene-specific primers from RACE experiments. The amplified PCR fragments were cloned into pCRII-TOPO vector (Invitrogen) and verified by sequencing. StPTB1, StPTB6, and StPTB7 were identified in this manner. The primers used in this study (Supplementary Table S1 available at JXB online) were synthesized at the DNA Facility, Iowa State University.
RNA analysis
Transgenic and wild-type plants of S. tuberosum cv. Désirée were used for most of the studies reported here. The photoperiod-responsive potato (S. tuberosum ssp. andigena, line 7540) was used for the quantitative reverse transcription–PCR (qRT–PCR) and protein analyses. RNA extraction and qRT–PCR were performed as previously described (Lin et al., 2013). Gene-specific primers used for the quantitative qRT–PCR are listed in Supplementary Table S3 at JXB online.
Constructs
For the PromStPTB1:GUS (β-glucuronidase) and PromStPTB6:GUS plasmids, the constructs include DNA fragments containing 2519bp and 3593bp of sequence upstream of the translational start codon, respectively (Butler and Hannapel, 2012). Promoter fragments were obtained by PCR amplification using the primers listed in Supplementary Table S4 at JXB online and cloned into pBI101.1-GUS (Clontech). For transformation of the 35S:StPTB–GFP (green fluorescent protein) constructs, the pBI121-GFP vector was generated from pBI121-GUS (Clontech) by replacing the GUS gene with the GFP reporter gene in BamHI/SacI sites in the pBI121-GUS vector. The 35S:StPTB1–GFP and 35S:StPTB6–GFP constructs include the coding sequences of StPTB1 and StPTB6 without the stop codon and fused in-frame to GFP. The inserts for the constructs were obtained by PCR amplification and directly cloned into the pBI121-GFP vector (Supplementary Table S4).
RNA suppression lines for StPTB1 and StPTB6
To suppress StPTB1 and StPTB6 transcript levels, both antisense and RNAi constructs were designed. The antisense construct was made from the 611bp SacI–SpeI fragment of the StPTB6 cDNA (87% identical to the StPTB1 cDNA sequence), cloned in the antisense direction into the binary vector, pCB201 (Xiang et al., 1999), and driven by the Cauliflower mosaic virus (CaMV) 35S promoter. For generating the RNAi construct (Supplementary Table S1 at JXB online), the same conserved 611bp fragment of the StPTB6 cDNA was cloned into the pENTR/D-TOPO plasmid (Invitrogen) and inserted in opposite orientations by recombination with the LR Clonase II enzyme (Gateway Technology, Invitrogen) into the pBIN19RNAi destination vector (Navarro et al., 2011). Transformation was undertaken on leaf sections from S. tuberosum cv. Désirée as described by Banerjee et al. (2006a ). Functional transformants were screened for a reduction in the level of transcripts of both StPTB1 and StPTB6 in RNA samples from leaves and stolons.
Histological analysis of promoter activity
Select transgenic promoter lines were transferred to soil and grown for 2 weeks before tissues were harvested and stained for 16h using a 1.0mg ml–1 X-gluc (5-bromo-4-chloro-3-indolyl-β-d-GlcUA) solution and cleared using 70% ethanol. Fixation of stained materials (Fig. 2) was performed with FAA (10% formaldehyde, 50% ethanol, 5% acetic acid) using vacuum infiltration and dehydrated through an ethanol series for embedding in paraffin. Prepared paraffin sections of ~10 μm were fixed to glass slides for microscopy and photodocumented using an Axioplan compound light microscope with a colour camera mount.
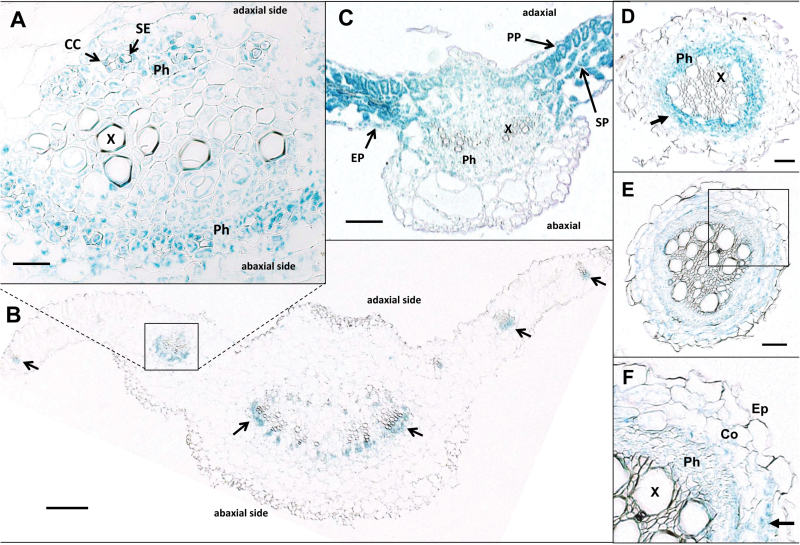
Fig. 2.
Localization of GUS activity within petioles and leaves of StPTB1 prom (A, B) and StPTB6 prom (C) transgenic lines. Petioles from 4-week-old soil-grown plants of StPTB1 prom (A, B) and leaves of 2-week-old in vitro plants of StPTB6 prom (C) were embedded in paraffin for histochemical detection of GUS activity within petiole and leaf tissues. (A) A transverse section of StPTB1 prom with a higher magnification image of a petiole vascular bundle comparable with the boxed region of (B) showing xylem (X) and phloem (Ph) tissues. The arrows in (B) indicate GUS activity of the StPTB1 prom in abaxial-side phloem cells. (C) A transverse section of StPTB6 prom showing the midvein and adjacent leaf blades with strong staining in the epidermis (EP), palisade (PP), and spongy (SP) parenchyma tissues. The scale bars represent 20 μm in (A), 200 μm in (B), and 100 μm in (C). CC, companion cell; SE, sieve element; Ph, phloem; X, xylem; EP, epidermis; PP, phloem parenchyma; SP, spongy parenchyma. GUS activity within a transverse section of primary roots of StPTB1 prom (D) and StPTB6 prom (E, F) transgenic lines. StPTB1 prom activity was observed in cells of the vascular core, mostly in phloem cells (D, arrow), whereas StPTB6 prom activity was dispersed throughout the phloem and cortex tissues (E, F). Staining was observed in the phloem (Ph) and cortex (Co), but not in the epidermis (Ep) or xylem (X). (F) A higher magnification of (E) (box). The scale bars represent 0.1mm in (D and E). The arrow in (F) designates GUS activity in cortical cells.
Preparation of polyclonal antibody
StPTB1 and StPTB6 cDNA inserts were cloned into the pET-28a (+) plasmid and transformed into Escherichia coli BL21-Codon (DE3) cells (Stratagene, La Jolla, CA, USA). After induction with 0.4mM isopropyl-β-d-thiogalactopyranoside (IPTG), the recombinant proteins were purified using Ni-NTA agarose (Qiagen, Valencia, CA, USA) and quantified. These purified proteins were used for EMSA and for preparation of polyclonal antibody. Polyclonal antibodies against StPTB6 protein were raised at the Hybridoma Facility at Iowa State University. The purified proteins (500–800 μg) were repeatedly injected into two rabbits, and standard protocols were used for bleeding and screening to obtain reliable antibodies. The sera containing specific polyclonal antibodies against StPTB6 were used for the studies without purification. Pre-immune controls are shown in Supplementary Fig. S6 at JXB online.
Electrophoretic mobility shift assays
For RNA-EMSAs, target sequences were amplified from potato leaf cDNA or genomic DNA using corresponding primers (Supplementary Table S4 at JXB online). These probes were derived from either coding sequence or the 3′-UTR, and were selected based on the yeast three-hybrid results (Supplementary Fig. S4). RNA bait was generated using in vitro transcription with the MAXIscript® T3 kit (Ambion) and biotin-labeled UTP (Bio-11-UTP, Ambion). Biotin-labelled RNA probes were purified using RNA Clean & Concentrator™-5 (Zymo Research) and quantified by NanoDrop 1000 (Thermo Scientific). Biotin-labelled RNAs (3fmol) were used for the binding assay. Binding reactions with labelled RNA and purified recombinant proteins (0–500nM) were incubated in 20 µl for 1.0h at room temperature in the presence of a binding buffer consisting of 40mM Tris (pH 8.0), 30mM KCl, 1mM MgCl2, 0.1% NP-40, and 1mM DTT. The binding reactions were resolved on a 6% (v/v) non-denaturing polyacrylamide gel, and then transferred onto a BrightStar®-Plus positively charged nylon membrane (Ambion). For detection of biotinylated RNA, the Chemiluminescent nucleic acid detection module kit (Thermo Scientific) and CL-XPosure™ film (Thermo Scientific) were used.
RNA immunoprecipitation
New tubers (~4.0–8.0mm in diameter) or petioles of S. tuberosum ssp. andigena plants grown under short-day (SD) conditions were fixed using 1.0% formaldehyde and 125mM glycine under a vacuum. The fixed samples were frozen and ground in liquid nitrogen, and total cell extracts were prepared with 0.7g of tissue sample in 2.0ml of RIP-LB buffer (Köster and Staiger, 2014), and cleared using Dynabeads® Protein A (Invitrogen). For immunoprecipitation (IP), 20 µl of Dynabeads® Protein A were coated with 2.0 µg of pre-immune serum, αStPTB6 antibody purified using the Melon Gel IgG purification kit (Thermo Scientific), or αGFP monoclonal antibody (Santa Cruz), and washed as previously described (Terzi and Simpson, 2009). For IP of StBEL5 RNA and StPTBs proteins, 600 µl of pre-cleared total cell extracts using a 0.2 µm syringe filter (VWR) were incubated with antibody-coated magnetic beads for 2h at 4 °C under rotation. After intensive washings (Terzi and Simpson, 2009), to release RNAs that were cross-linked with StPTB proteins by formaldehyde fixation, NaCl was added to 200mM final concentration, and IP reactions were incubated for 20min at 65 °C.
Diluted input (1.0%) of purified RNA from the IP was used for gel-based RT–PCR. For qRT–PCR, the qScript™ One-Step SYBR® Green qRT-PCR kit (Quanta Biosciences) with the Eco Real-Time PCR system (Illumina) was used. Each real-time PCR was performed in duplicate. For quantification and comparison of RNA levels between samples, the 2−ΔΔCt method was used (Livak and Schmittgen, 2001). To account for RNA sample preparation differences, each RIP RNA fraction’s Ct value was normalized to 1.0% of the input RNA fraction Ct value for each corresponding qPCR assay according to the RIP protocol (Sigma, Imprint® RNA RIP Kit). Primer sequences used are listed in Supplementary Table S5 at JXB online.
Potato virus X (PVX) vector system for RNA movement assay
Full-length cDNAs, including the 5′- and 3′-UTRs, of StBEL5 and StBEL14 were amplified using NEBNext High-Fidelity 2X PCR Master Mix (NEB), and cloned into MluI and EcoRV sites of the PVX/ΔCP vector (Li et al., 2009) to create PVX/B5H and PVX/B14H, respectively. The control PVX/GFP vectors were described previously by Li et al. (2009). Reverse primers for the StBEL constructs were designed to include a histidine-tag sequence for inoculum-specific detection (Supplementary Table S5 at JXB online). Transcripts for mobility assays were prepared by in vitro transcription using a T7 High Yield RNA Synthesis kit (NEB), as described in the instruction manual, and purified using LiCl precipitation. A 10 µg aliquot of in vitro transcripts was mechanically inoculated onto each leaf (2–3 leaves per plant) of wild-type S. tuberosum cv Désirée or the StPTB Désirée transgenic lines. Inoculated leaves, and non-inoculated roots and stolons were harvested at 8 d post-inoculation, and total RNA was extracted using an RNeasy Plant Mini kit (Qiagen) and followed by RNase-free DNase (Qiagen) treatment. Total RNA (150ng) was used for the gel-based RT–PCR. For qRT–PCR, the qScript One-Step SYBR Green qRT-PCR kit (Quanta Biosciences) with the Eco Real-Time PCR system (Illumina) was used. After normalization using StAct8 as an endogenous control, the relative gene quantification approach (Livak and Schmittgen, 2001) was applied to calculate the mobility level of the mechanically inoculated StBEL5, and analysed using a Student’s t-test. Levels of RNA in roots and stolons were calculated in relation to the amount of leaf inoculum.
Results
The PTB proteins of potato
Using RT–PCR and 5′ and 3′ RACE, three full-length RNAs, designated StPTB1 (1487 nucleotides), StPTB6 (1624 nucleotides), and StPTB7 (1678 nucleotides), were identified that encode for StPTB proteins. The lengths of these three proteins are 441, 444, and 467 amino acids, respectively. StPTB1 and StPTB6 closely match the amino acid sequence of the PTB protein of pumpkin, CmRBP50 (Supplementary Fig. S1 at JXB online). Each of the proteins contains conserved RRMs, the conventional set of four for StPTB1 and StPTB6 but only three for StPTB7 (a three-RRM type). StPTB7 contains 11 extra residues at the N-terminus and lacks a conserved RRM4 (Supplementary Fig. S1). The conserved motifs of the RRM (Auweter and Allain, 2008; Cléry et al., 2008), consisting of eight amino acids that interact with target RNAs, RNP1 and RNP2, are present in each of the identified RRMs (Supplementary Fig. S1, underlined sequence). One other StPTB protein has been identified from the DFCI Potato Gene Index (TC201749) and two others from the potato genome (BI920231 and TC218925; Xu et al., 2011). Each of these latter three is closely related to StPTB7 (Supplementary Fig. S2). Based on phylogenetic analysis (Supplementary Fig. S3), StPTB7 and these other three-RRM types belong to the same clade with AtPTB1 and AtPTB2 (Wang and Okamoto, 2009). Phylogenetic analysis classifies the PTBs of plants into two categories, one designated the StPTB1/PTB6 (CmRBP50-like) clade and a second designated the StPTB7 clade (Supplementary Fig. S3). Because of our interest in phloem-mobile mRNAs (Banerjee et al., 2006b ), the focus of this current study is the CmRBP50 types in potato, StPTB1 and StPTB6 (Supplementary Fig. S2).
RNA binding properties of the StPTB proteins
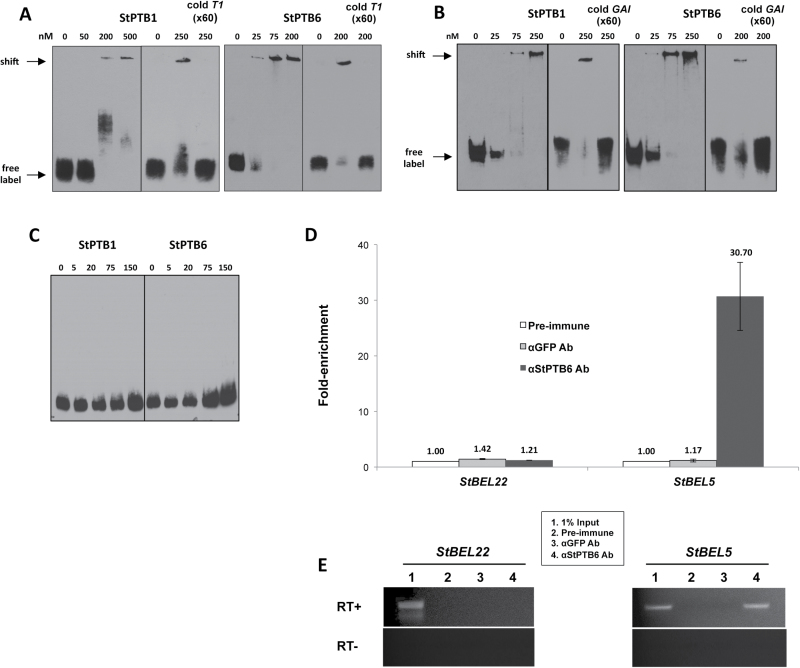
To test their putative function as chaperones to phloem-mobile RNAs, the ability of StPTB1 and StPTB6 to bind to RNA sequence was assessed. Preliminary analysis utilizing the yeast three-hybrid system suggested that StPTB1 and StPTB6 proteins interacted with specific 3′-UTR RNA sequence of both StBEL5 (T1) and StGAI (Supplementary Fig. S4 at JXB online). To verify protein–RNA interactions, RNA gel-shift assays with StPTB1 and StPTB6 were performed (Fig. 1A–C). Both proteins generated a shifted band in an interaction with both the labelled StBEL5 T1 and StGAI fragments (bait sequences for both types are shown in Supplementary Fig. S5). For the T1 bait, an interaction was observed for StPTB6 with a concentration as low as 25nM (Fig. 1A), whereas StPTB1 affected a shift at higher protein concentrations (Fig. 1A). For the StGAI bait, an interaction was observed for StPTB6 with a concentration as low as 25nM (Fig. 1B), whereas StPTB1 affected a shift at 75nM (Fig. 1B). No band shift was evident for either protein with a negative control interaction using StBEL5-N as bait (Fig. 1C) and even with protein concentrations up to 300nM (not shown). It should be noted that the negative control used in this gel shift contained no major CU motifs, whereas the StGAI and T1 baits each contains five CU motifs (Supplementary Fig. S5). Results with both the yeast three-hybrid and the RNA gel-shift assays suggest that the interaction of StPTB6 with both T1 and the StGAI UTR fragments was stronger than the interaction with StPTB1 under the current experimental conditions. RNA IP confirms that StPTB proteins bind to StBEL5 RNA in planta (Fig. 1D, E). Crude protein–RNA extracts from new tubers were fixed with formaldehyde and incubated with αStPTB6 antibody (Supplementary Figs S6, S7). Immunoprecipitation of the new tuber extract with the StPTB6 antibody yielded a 30-fold enrichment of StBEL5 RNA relative to the control sample (Fig. 1D). No transcript enrichment was observed for the negative control, full-length StBEL22. StBEL22 was used here because its 74 nucleotide 3′-UTR contained no CU motifs longer than 3 nucleotides (Supplementary Fig. S5). These results were reproducible in both replicate tuber- and petiole-derived fractions.
Fig. 1.
RNA binding assays using RNA gel shifts (A–C) and RNA immunoprecipitation (D, E). For gel-shift assays, select RNA with the PTB1 and PTB6 proteins of potato were utilized (A–C). Labelled RNA included the 137 nucleotide T1 subset sequence of the StBEL5 3′-UTR (A), a 77 nucleotide 3′-UTR sequence from StGAI (B), and a 101 nucleotide coding sequence, StBEL5-N, as a negative control (C). The complete sequence of these three RNA baits is shown in Supplementary Fig. S8 at JXB online. Unlabelled T1 or GAI probe to 60-fold excess was used with 200nM (StPTB6) or 250nM (StPTB1) of protein in competition assays for each interaction shown in the last three lanes of each StPTB panel (A, B). For these three lanes: 0 lane is label without protein, the 200 or 250 lane is label plus protein, and cold T1 or GAI (×60) lanes are the competition assays containing both unlabelled and labelled probe. RNA bait was labelled with biotin using in vitro transcription and then incubated with purified StPTB proteins in concentrations ranging from 5nM to 500nM protein. Approximately 3.0fmol of labelled bait RNA was used in each binding reaction. RNA immunoprecipitation of StBEL5 RNA was performed from new tubers using αStPTB6 antibody. New tubers of S. tuberosum ssp. andigena plants grown under short-day conditions for 14 d were used as the input sample. For quantification and comparison of RNA levels between samples (D), the 2–∆∆Ct method was used (Livak and Schmittgen, 2001). Each real-time PCR was performed in duplicate. To account for RNA sample preparation differences, each RIP RNA fraction’s Ct value was normalized to 1.0% of the input RNA fraction Ct value for the same qPCR assay. RNA for StBEL22 was quantified in parallel with StBEL5 and used as a negative control. αGFP monoclonal antibody was used as a negative immunoprecipitation control. Quantitative RT–PCR values (D) were normalized to the levels in the pre-immune sample. RNAs from the immunoprecipitations and 1.0% diluted input were used for gel-based RT–PCR with (+) and without (–) reverse transcriptase (E).
Expression patterns of StPTB genes
To determine if StPTB promoter activity coincides with StBEL5 expression, StPTB1 and StPTB6 activity was assayed in transgenic potato lines (cv. Désirée) by fusing ~3.0kb of upstream sequence of both types to a GUS marker. StBEL5 promoter activity was observed in leaf veins, petioles, and in phloem cells of the petiole (Banerjee et al., 2006b ). StPTB1 and StPTB6 exhibited promoter activity in stems, petioles, and leaf veins (Butler and Hannapel, 2012). For a more detailed look, activity within leaf tissues was examined in transverse sections of petioles with attached leaf blades (Fig. 2). StPTB1 prom activity was consistently observed throughout both adaxial and abaxial phloem of petioles (Fig. 2A, B, arrows). Specific StPTB1 prom activity can be observed in companion cells (CCs) but not sieve elements (SEs) of petiole phloem (Fig. 2A, arrows). In leaves, StPTB6 prom activity was not localized to vascular cells of the midvein (Fig. 2C, X and Ph), but was widespread in phloem (Fig. 2C, Ph), the epidermis (EP), and parenchyma (PP, SP) of adjacent leaf blades (Fig. 2C, arrows). In transverse sections of roots, StPTB1 prom activity was observed predominantly in the phloem (Fig. 2D, arrow), whereas StPTB6 prom activity was observed in both phloem and cortical cells (Fig. 2E, F, arrow). In general, in stems, leaves, and roots, the StPTB1 promoter was most active in phloem cells. The StPTB6 promoter exhibited activity in the phloem of roots but overall more activity than StPTB1 in ground tissues of these organs.
Transverse sections of stained internodes from stems of StPTB1 prom and StPTB6 prom lines were also visualized (Supplementary Fig. S8A at JXB online). Discrete staining was observed for the StPTB1 prom line throughout the external phloem (EP, Supplementary Fig. S8B) layers, with some staining within the internal phloem (IP, Supplementary Fig. S8B). The activity of the StPTB1 prom was specific to CCs but absent from SEs in the vascular bundles of these stem sections (Supplementary Fig. S8C, D). In contrast, StPTB6 prom activity was more widespread and irregular in the stem. StPTB6 prom activity was not as abundant in vascular bundles of stems as the StPTB1 prom construct (Supplementary Fig. S8E, F), but was observed in some cells of the internal phloem (IP, Supplementary Fig. S8F), in interfascicular regions, and in clusters of cells along the epidermis (Supplementary Fig. S8F, arrows).
Promoter activity in stolons and tubers
Because of the functional association of StBEL5 RNA and the process of tuberization, StPTB1 prom and StPTB6 prom expression was investigated during tuber development. Stolons and new tubers were harvested from mature soil-grown plants and separated into four categories based on their developmental stage (Fernie and Willmitzer, 2001). GUS staining suggested that StPTB6 prom expression was stronger in stolons than in new tubers (right column, Supplementary Fig. S9A at JXB online), whereas GUS staining of StPTB1 prom stolons and tubers was essentially the same (left column, Supplementary Fig. S9A). Manual dissection of t2 tubers revealed that both the StPTB1 prom and the StPTB6 prom were active in vascular strands of new tubers (Supplementary Fig. S9A, arrows). Quantification of the activities in these organs revealed a peak of activity for the StPTB6 prom in early, induced stolons that steadily decreased as tubers formed (Supplementary Fig. S9B, black bars), whereas StPTB1 prom activity exhibited no such developmental regulation (Supplementary Fig. S9B, grey bars).
To explain StPTB6 activation during early tuberization, the StPTB6 promoter sequence was screened for tuber and sucrose inductive motifs. Three putative tuber-specific sucrose-responsive (TSSR) elements present in the promoters of class I patatin and proteinase inhibitor II genes (Grierson et al., 1994) were identified in the promoter of StPTB6 but not StPTB1 (Supplementary Fig. S9C at JXB online). These are designated TSSR1, TSSR2, and TSSR3 (Supplementary Fig. S9C) and in the patatin promoter direct tuber-abundant expression. As an example of conservation, alignment of the putative TSSR2 motif from StPTB6 with the proximal patatin TSSR motif revealed nucleotide sequence matches in the B-box, the repression region, and the A-box of 59, 69, and 74%, respectively (Butler and Hannapel, 2012).
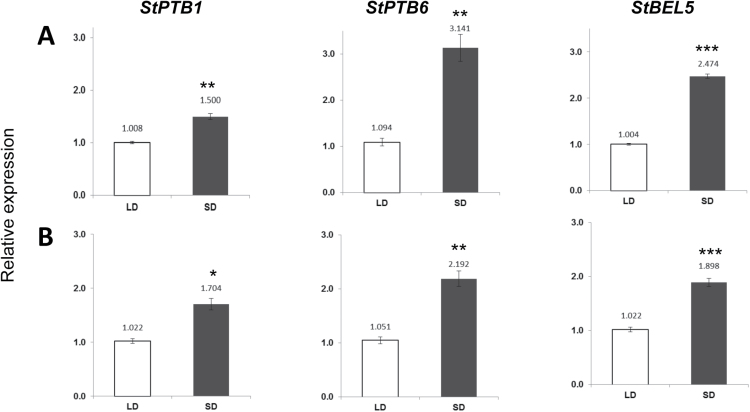
StBEL5 transcripts consistently exhibited enhanced mobility and accumulation in response to an SD photoperiod (Chen et al., 2003; Banerjee et al., 2006b ). To assess expression patterns for the StPTB genes in response to photoperiod, RNA accumulation patterns for StPTB1 and StPTB6 were monitored in the photoperiod-sensitive S. tuberosum ssp. andigena. Under long-day (LD) conditions, there were no significant differences among the five organs tested (Supplementary Fig. S10 at JXB online). Under SD conditions, however, enhanced accumulation of both StPTB1 and StPTB6 was observed concomitantly with StBEL5 accumulation in leaf mesophyll and leaf veins (Fig. 3). SD-induced accumulation was also observed in petioles for StPTB1 and in lateral roots for StPTB6 (Supplementary Fig. S11). No SD-enhanced accumulation occurred for either PTB type in stolons or stems (Supplementary Fig. S11, S12). Despite the observation that StPTB transcript accumulation was not induced by SDs in stems, levels of the StPTB proteins increased in both stems and stem exudate under SD conditions (Supplementary Fig. S12), suggesting some degree of photoperiod-mediated post-transcriptional regulation is in effect. StPTB protein levels were also enhanced by SDs in petioles, roots, and stolons (Supplementary Fig. S13).
Fig. 3.
Effect of photoperiod on transcript accumulation of StPTB1 and StPTB6 in leaf mesophyll (A) and leaf veins (B) from the photoperiod-responsive potato species, S. tuberosum ssp. andigena. Short-day (SD) plants were harvested after 14 d of SD conditions (8h light, 16h dark). Quantitative real-time RT–PCR with gene-specific primers was used to calculate the relative amounts of RNA for each StPTB gene. StBEL5 was included as a positive control. Each sample was measured in triplicate and normalized against StActin8 RNA. The fold change in expression was calculated as the 2−ΔΔCt value relative to the mean values obtained in the long-day (LD) samples. Standard errors of the means of three biological replicates are shown, with one, two, and three asterisks indicating significant differences (P<0.05, P<0.01, P<0.001, respectively) using a Student’s t-test.
Transgenic StPTB plants exhibit a tuberization phenotype
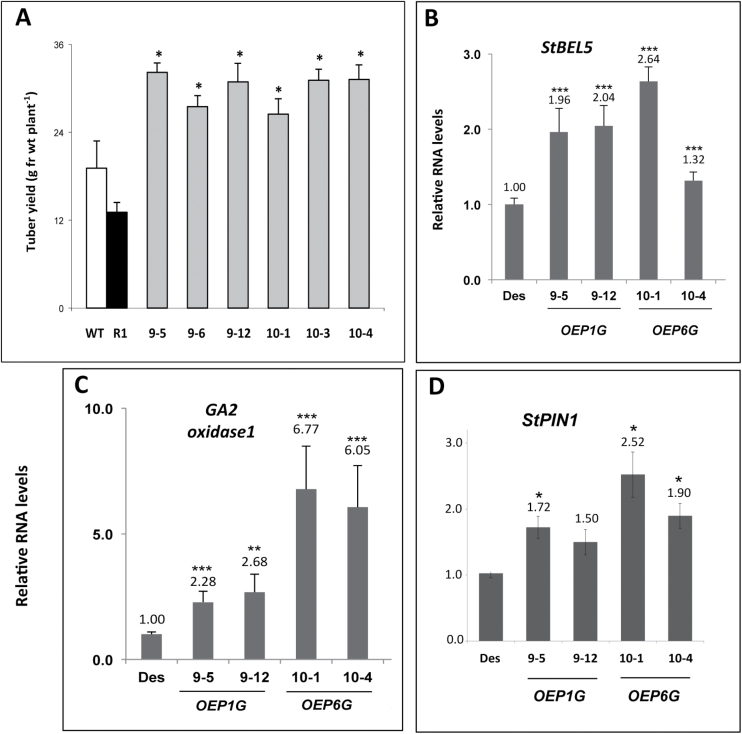
To better understand the function of StPTB1 and StPTB6, ~30 transgenic overexpression (OE) lines of potato cv. Désirée were generated for both protein types, screened, and evaluated. Several high expressing lines were identified for each construct, and three to four were used in evaluating effects on RNA metabolism and phenotypes of soil-grown plants. Overall shoot fresh weight was not affected in both OE lines, but both PTB-OE types exhibited enhanced root growth (Supplementary Fig. S14 at JXB online). With three representative lines each, the average increase for root growth was 71% for the StPTB1-OE lines and 101% for the StPTB6-OE lines. Tuber yields from both StPTB soil-grown OE lines were more than twice that of the GFP control line, R1 (Fig. 4A). Earliness, tuber numbers, and tuber yields were also enhanced in both OE lines grown under in vitro conditions (Table 1).
Fig. 4.
Tuber production (A) for independent Désirée transgenic overexpression lines for PTB1 (#9 lines) and PTB6 (#10 lines). Four to five replicates from select overexpression lines were grown in 13cm pots. Tuber yields (g FW per plant) were assessed at 45 d. Plants were grown under long-day conditions (16h light, 8h dark) with a fluence rate of 300 mmol m–2 s–1 at 24 °C during a 12h day cycle and 18 °C for the remaining time. Relative RNA accumulation of StBEL5 (B), GA2 oxidase1 (C), and StPIN1 (D) in stolons of PTB overexpression lines after 30 d in soil. RNA was calculated using qRT–PCR and is shown in relation to wild-type (WT) levels. Des, wild-type Désirée control; R1, transgenic Désirée GFP control; OEP1G, overexpression line of StPTB1; OEP6G, overexpression line of StPTB6. Standard deviations of the means of three biological replicates are shown, with one, two, and three asterisks indicating significant differences (P<0.05, P<0.01, P<0.001, respectively) using a Student’s t-test.
Table 1.
In vitro tuberization assays for StPTB1 and StPTB6 overexpression lines
Explant material was from 2-week old in vitro plantlets grown under long-day conditions on MS medium with 2% sucrose. Eighteen axillary bud explants per line were cultured in the dark on MS medium plus 8.0% sucrose for 21 d at 25 °C. Each explant was composed of one bud and an ~0.5cm stem section. Bud explants were scored for earliness (days to first tuber), shoot or tuber formation, and total tuber yield (mg).
| Transgenic line | Days to first tuber | No. of shoots | No. of tubers | Tuber yield (total mg) |
|---|---|---|---|---|
| Des | 10 | 12 | 6 | 110 |
| 9-5 | NA | 5 | 13 | 405 |
| 9-6 | 6 | 0 | 18 | 918 |
| 9–12 | 7 | 1 | 17 | 464 |
| 10–1 | 6 | 10 | 8 | 368 |
| 10–3 | 6 | 4 | 14 | 430 |
| 10–4 | 7 | 2 | 16 | 472 |
Des, Désirée control. The #9 lines are StPTB1 and the #10 lines are StPTB6.
NA, data not available.
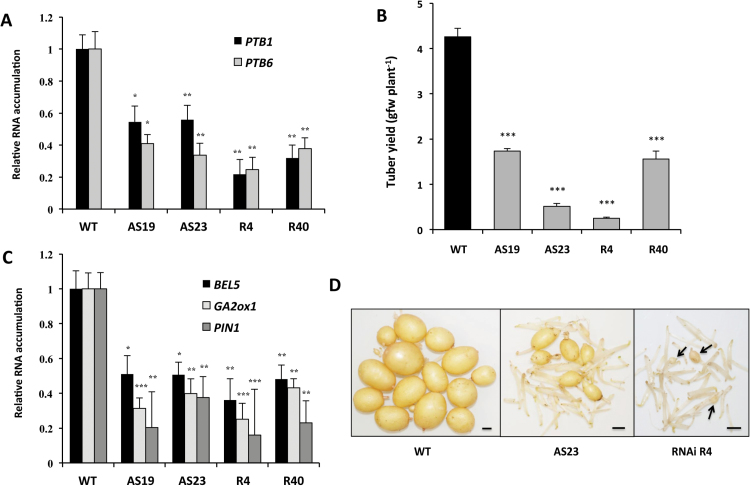
In both StPTB1- and StPTB6-OE lines, StBEL5, GA2ox1, and StPIN1 transcript levels were enhanced in stolon tips (Fig. 4B–D). Levels of GA2ox1 in StPTB6-OE lines increased by >6-fold. GA2ox1 is an important marker gene involved in reducing gibberellin levels during the onset of tuber formation (Kloosterman et al., 2007) and is induced by StBEL5 through an interaction with tandem TTGAC elements present in the promoter of GA2ox1 (Lin et al., 2013). StPIN1 is also a tuberization marker and was previously confirmed to be induced by StBEL5 (Navarro et al., 2011; Hannapel et al., 2013). To complement the OE results, ~30 transgenic suppression lines were generated and screened for reduced StPTB RNA levels. The design of the antisense/RNAi sequence provided suppression of RNA accumulation for both StPTB types (Fig. 5A). Based on StPTB levels, four lines were selected and evaluated for tuber yield and StBEL5 RNA levels. All four suppression lines exhibited a reduction in tuber yields and StBEL5 RNA levels (Fig. 5B–D). Consistent with a reduction in StBEL5 activity, two known targets of StBEL5, StGA2ox1 and StPIN1, also exhibited a decrease in transcript levels (Fig. 5C). Highlighting the specificity of StPTB activity in tuber development, yields were reduced by 8- and 16-fold for AS23 and R4, respectively (Fig. 5B, D), whereas shoot fresh weight among these lines exhibited no difference relative to the WT (Supplementary Fig. S15 at JXB online).
Fig. 5.
RNA suppression lines for StPTB1 and StPTB6 (A). Lines AS19 and AS23 expressed antisense sequence, whereas lines R4 and R40 expressed an RNAi sequence. Both types suppressed expression of both PTB types from stolons of plants after 15 d in soil (A). Four to five replicates from select RNAi lines were grown in 13cm pots. Tuber yields (g FW per plant) were assessed at 30 d (B and D). The tubers depicted in (D) are the pooled harvest of five plants. Arrows in (D) indicate the small tubers harvested from line R4 at 30 d. Plants were grown under long-day conditions (16h light, 8h dark) with a fluence rate of 300 mmol m–2 s–1 at 24 °C during a 12h day cycle and 18 °C for the remaining time. RNA accumulation for StBEL5, StGA2ox1, and StPIN1 was assessed in the same stolons from suppression lines after 15 d in soil (C). RNA was calculated using qRT–PCR and is shown in relation to wild-type (WT) levels. WT, non-trangenic Désirée control. Standard errors of the means of three biological replicates are shown, with one, two, and three asterisks indicating significant differences (P<0.05, P<0.01, P<0.001, respectively) using a Student’s t-test. The size bars in (D) are equivalent to 0.5cm.
The StPTB proteins regulate stability and movement of StBEL5 RNA
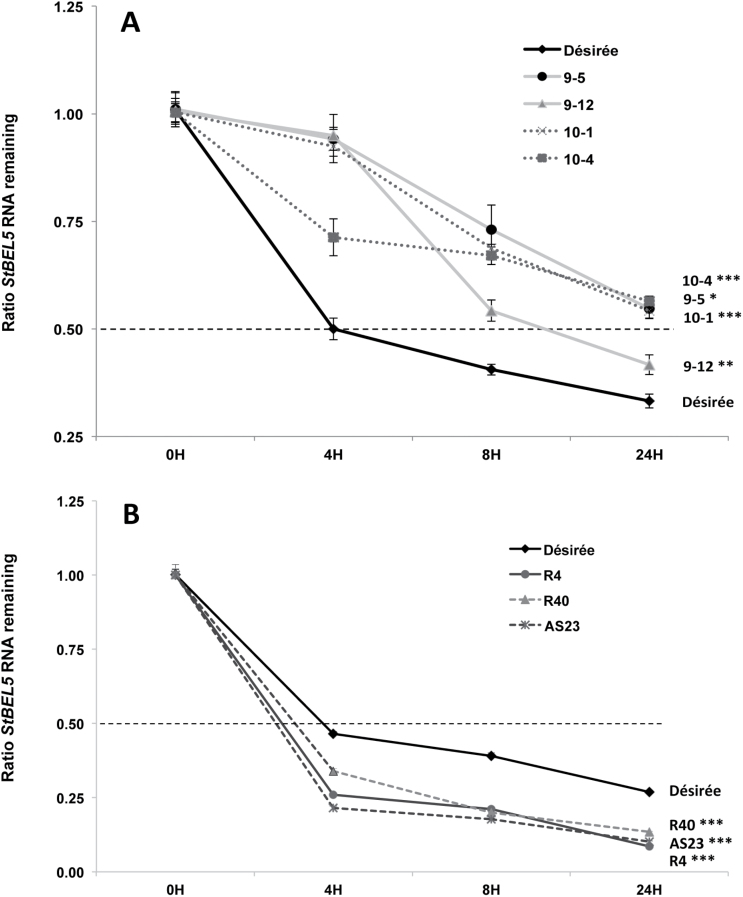
To gain insight into the mechanism of the StPTB protein interactions with StBEL5 RNA, an RNA stability assay was performed using in vitro grown shoot tips of the overexpression lines and wild-type Désirée. RNA stability of StBEL5 was enhanced in both StPTB-OE types. Significant differences were observed for the rate of StBEL5 degradation in all four StPTB-OE lines (Fig. 6A). In wild-type plants, StBEL5 reached a half-life as early as 4h, whereas, even after 8h, with a degradation ratio of 0.40 in wild-type plants, StBEL5 transcripts had not reached half-life in any of the StPTB-OE lines, with an overall average ratio of 0.66. After 24h, only one of the four StPTB-OE lines had dropped below the half-life for StBEL5 RNA (Fig. 6A). These enhanced stability rates for StBEL5 transcripts were observed in two separate experiments. In contrast, the stability of StBEL5 decreased significantly in the StPTB suppression lines relative to the wild-type control (Fig. 6B). After 24h, relative amounts of StBEL5 were at 0.27 for the wild type, whereas the suppression lines exhibited levels of 0.13 or lower. These results, coupled with the binding assays from Fig. 1, strongly suggest that StPTB1 and StPTB6 contribute to the overall stability of StBEL5 RNA in planta through a direct protein–RNA interaction and very probably contribute to the high abundance levels of StBEL5 transcripts observed throughout the potato plant (Chen et al., 2003; Sharma et al., 2014).
Fig. 6.
Stability of StBEL5 transcripts in wild-type Désirée (black solid line, A and B) and overexpression lines (A) of StPTB1 (lines 9-5 and 9–12, grey, solid lines) and StPTB6 (lines 10–1 and 10–4, dotted lines) or the three StPTB1/6 suppression lines, R40, AS23, and R4 (B). Shoot tips (1cm) from in vitro cultured plants grown under long days were harvested and incubated in a 250ml flask with incubation buffer (Seeley et al., 1992) on a shaker at 75rpm for 30min. Cordycepin was added to a final concentration of 0.6mM and the flask containing the samples was vacuum infiltrated for 45 s. Five to six shoot tips were harvested and pooled at each time interval, blotted dry, and frozen immediately in liquid nitrogen. RNA was extracted and quantitative real-time RT–PCR with gene-specific primers was used to calculate the relative amounts of StBEL5 RNA. Each sample was measured and normalized against StActin8 RNA. The ratio of RNA remaining for each sample was calculated as the 2−ΔΔCt value relative to the mean values obtained from samples at 0h. The straight dashed line indicates the half-life point. StActin8 RNA degraded at the same rate for all lines tested here. Standard errors of the means of two biological and two technical replicates are shown. One, two, and three asterisks indicate significant differences in the rate of degradation relative to the wild-type control (P<0.05, P<0.01, P<0.001, respectively) using regression analysis.
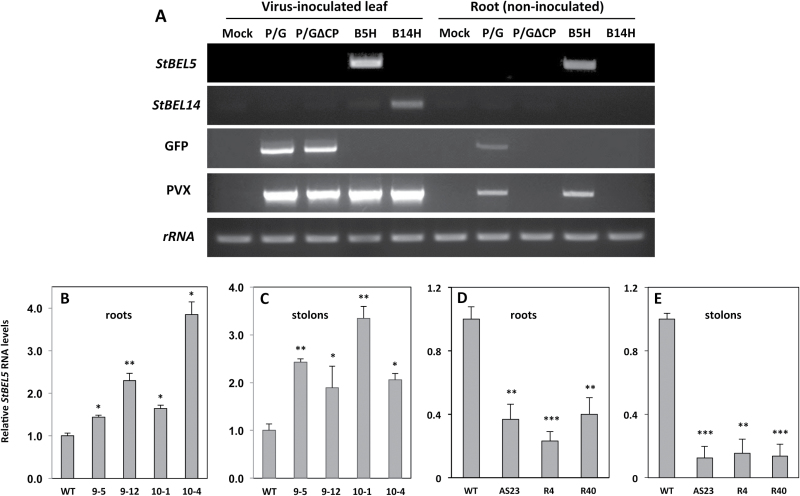
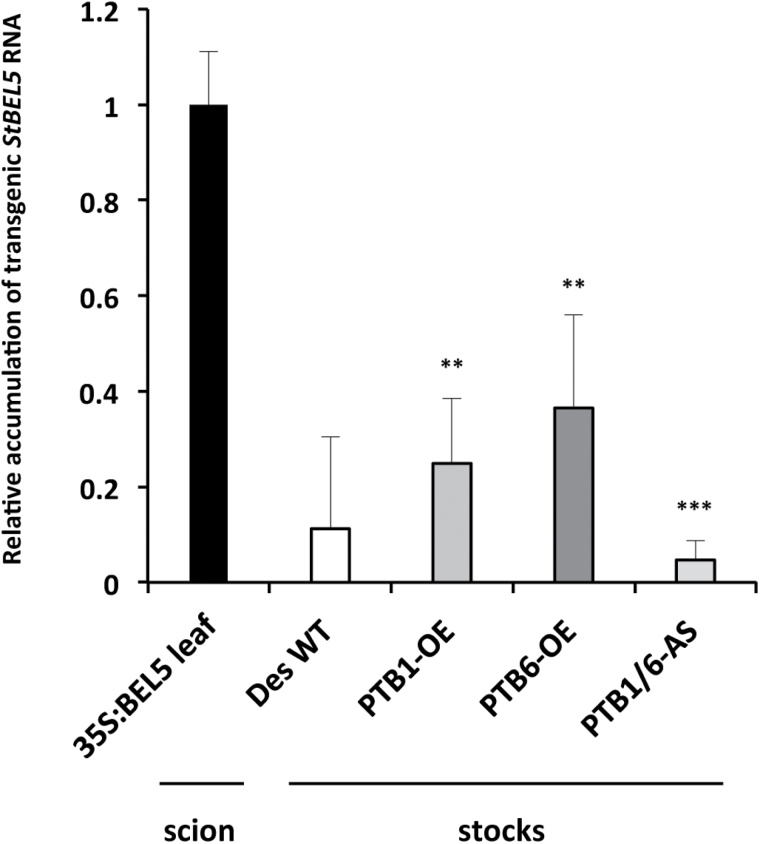
To assess the effect of these StPTBs on StBEL5 movement, RNA movement assays were performed utilizing a PVX vector system (Li et al., 2009). This whole-plant system has proven to be accurate and specific in the assessment of RNA movement over a short period of time (Li et al., 2009, 2011). As presented here (Fig. 7), by utilizing RNA-specific primers and real-time qRT–PCR, this system is especially useful for the reproducible quantification of RNA transport. After inoculation in leaves, the PVX viral RNA cannot traffic long distance without its coat protein. When full-length StBEL5 RNA replaced the coat protein sequence, transport of StBEL5 RNA from inoculated leaves into non-inoculated roots of wild-type Désirée plants was confirmed (Fig. 7A). Both the non-mobile StBEL14 RNA and GFP without a coat protein fusion were unable to transport long distance (Fig. 7A). In the StPTB-OE lines, long-distance transport of full-length StBEL5 RNA (minus the PVX coat protein) to both roots and stolons was enhanced relative to movement in wild-type plants (Fig. 7B, C). In comparison, again using the PVX system, suppression lines of StPTB1/6 repressed StBEL5 transport to both roots and stolons (Fig. 7D, E). StBEL5 movement into stolons was decreased by as much as 8-fold in suppression line AS23. To verify movement in a second system, heterografts were made in vitro with CaMV 35S:BEL5 transgenic scions grafted onto wild-type, StPTB-OE, or StPTB suppression stocks (Fig. 8). After 21 d in soil, newly formed tubers were harvested and the amount of transgenic StBEL5 RNA was measured. Consistent with the results from the PVX system, StBEL5 RNA accumulation was positively correlated with the level of the StPTB proteins (Fig. 8). In these experiments, the source (scion) of the mobile transgenic StBEL5 RNA is the same for all stocks analysed. Overall, these results suggest that steady-state levels of StBEL5 in both stolon tips and roots are due to the stability and movement of its mRNA both mediated by an interaction with StPTB proteins.
Fig. 7.
Analysis of RNA movement using a PVX vector system in wild-type (WT) Désirée (A). PVX-based StBEL5 RNA movement into roots (B and D) and stolons (C and E) of StPTB overexpression (B and C) and suppression (D and E) lines, 8 d post-inoculation. This whole-plant system provides an accurate assessment of long-distance RNA movement (Li et al., 2009). After inoculation in leaves, the PVX viral RNA cannot traffic long distance without its coat protein (A; root, non-inoculated P/G∆CP lane). When full-length StBEL5 RNA replaced the coat protein sequence, transport of StBEL5 RNA from inoculated leaves into non-inoculated roots of WT Désirée plants was confirmed (A; root, non-inoculated B5H lane). Both the non-mobile StBEL14 RNA and GFP without a coat protein fusion were unable to transport long distance (A; root, non-inoculated P/G∆CP lane). For quantification, RNA was extracted and quantitative real-time RT–PCR with gene-specific primers (Supplementary Table S5 at JXB online) was used to calculate the relative amounts of StBEL5 RNA that trafficked into roots (B and D) or stolons (C and E). Each sample was measured and normalized against StActin8 RNA. RNA values were calculated as the 2−ΔΔCt value relative to the mean values obtained from WT samples. StPTB1- and StPTB6-OE lines are designated #9 and #10, respectively. The three suppression lines are designated AS23, R4, and R40. Standard deviations of the means of two biological replicates with two technical replicates are shown, with one, two, and three asterisks indicating significant differences (P<0.05, P<0.01, P<0.001, respectively) using a Student’s t-test. For (A), Mock, inoculation with water; P/G, PVX vector with coat protein and GFP (mobile); P/G∆CP, PVX vector with GFP but no coat protein (non-mobile); B5H, PVX vector with full-length StBEL5 plus histidine tag but no GFP or coat protein; B14H, PVX vector with full-length StBEL14 plus histidine tag but no GFP or coat protein.
Fig. 8.
Movement of transgenic StBEL5 RNA from the 35S:BEL5 transgenic scion into stocks of wild-type (WT) Désirée and transgenic lines overexpressing StPTB1 (PTB1-OE line 9–12), StPTB6 (PTB6-OE line 10–1), and a line that suppresses both StPTB1 and StPTB6 (PTB1/6-AS line 23). All transgenic lines are from the potato cv. Désirée. Heterografts were performed in vitro and maintained in tissue culture for 3 weeks, and then moved to soil for 3 weeks before harvesting. For quantification, RNA was extracted from either leaves of the scion or stolon tips (new tubers) of the stocks, and qRT–PCR was used to calculate the relative amounts of transgenic StBEL5 RNA that trafficked across the graft union and down into stolon tips. Each sample was measured and normalized against StActin8 RNA. RNA values were calculated as the 2−ΔΔCt value relative to the mean values obtained from leaf samples of the 35S:BEL5 scion. Standard deviations of the means of two biological replicates with two technical replicates are shown, with two and three asterisks indicating significant differences from the WT stock stolons (P<0.01 and P<0.001, respectively) using a Student’s t-test.
Discussion
RNA binding function of StPTB1 and StPTB6
Among the six PTB proteins of potato, StPTB1 and StPTB2 are most similar in sequence to the phloem-mobile PTB protein of pumpkin, CmRBP50 (Ham et al., 2009), and to AtPTB3 of Arabidopsis (Rühl et al., 2012). CmRBP50 binds specifically to polypyrimidine tract motifs of CUCU or UCUU present in the sequence of the phloem-mobile RNA, CmGAI (Ham et al., 2009). In animals, the PTB protein contains four RRMs that bind to RNA sequence containing four separate CU motifs determined by the spatial arrangement of the β-sheet structures and RRM linkers of the protein (Oberstrass et al., 2005; Auweter and Allain, 2008; Sawicka et al., 2008; Clerte and Hall, 2009). Although the four-motif model is widely accepted, a definitive, discrete PTB-binding sequence cassette has not been established (Schmid et al., 2007). Ham et al. (2009) demonstrated binding with sequences containing two, three, or four CU motifs, but showed no relationship for any of these motifs with the capacity for movement or any effect on RNA stability. Both StPTB1 and StPTB6 bound to CU-rich sequence present in the 3′-UTR of StGAI and StBEL5 (both containing five CU motifs within 140 nucleotides of sequence, Supplementary Fig. S5 at JXB online) but not to sequence without these motifs (Fig. 1). Previous work with StBEL5 RNA movement and stability assays demonstrated that deletion of its 3′-UTR resulted in decreased mobility and stability of the BEL5 transcript (Banerjee et al., 2006b, 2009). Conversely, addition of the 3′-UTR resulted in enhanced, localized movement of another non-mobile StBEL RNA (Banerjee et al., 2009). The functional role of StPTB1 and StPTB6 in enhancing movement and stability of StBEL5 (Figs 6, 7) and their capacity for binding to 3′-UTR sequence of StBEL5 (Fig. 1; Supplementary Fig. S5) strongly suggest that this interaction is pivotal for regulating StBEL5 RNA movement. Several reports have shown that PTB proteins increase the stability of target mRNAs (Tillmar and Welsh, 2004; Pautz et al., 2006; Xu and Hecht, 2007). In mouse, a PTB protein regulates the circadian oscillation of a core clock gene by controlling its mRNA degradation through an interaction with the 3′-UTR (Woo et al., 2009). StBEL5 is a very abundant phloem-mobile RNA that functions in regulating tuber formation in potato (Banerjee et al., 2006b ). It is transcribed in the phloem of petioles and leaf veins and is transported to stolon tips to activate tuberization (Banerjee et al., 2006b ). Overexpression of the StPTBs enhanced steady-state levels of StBEL5 in stolon tips and leaf veins (data not shown) and increased tuber production, whereas suppression lines affected the opposite trend. In petioles, the promoter of StPTB1 was active in CCs of phloem but not the SEs (Fig. 2A; Supplementary Fig. S8C, D). Similar to this pattern of expression, RBP50 transcripts were detected only in CCs, whereas RBP50 protein was detected in both CCs and SEs (Ham et al., 2009). Transcriptional activity of StBEL5 was observed in leaf veins and petioles, and specifically in phloem parenchyma and CCs of petioles (Banerjee et al., 2006b ).
The role of StPTB1 and StPTB6 in regulating development
The pronounced effect on RNA accumulation levels of StPTB1 and StPTB6 mediated by SDs (Fig. 3) may help to explain some of the post-transcriptional dynamics that control patterns of accumulation for StBEL5. SDs activate tuber formation, but in leaves, StBEL5 promoter activity is activated by light with no observable effect mediated by photoperiod (Chatterjee et al., 2007). StBEL5 transcript levels in leaves and its capacity to move, however, increase under SDs (Chen et al., 2003; Banerjee et al., 2006b ). Based on binding and stability assays and the temporal and spatial control of StPTB1 and StPTB6 expression in relation to StBEL5 transcription, it would appear that both StPTB proteins can potentially interact with StBEL5 to facilitate stability and transport of its mRNA. Under an inductive photoperiod, both StPTB1 and StPTB6 may function at the leaf and petiole source to escort StBEL5 transcripts through the phloem to stolon tips and roots. Such a model is consistent with the enhanced levels of StPTB protein observed in stem exudate under SD conditions (Supplementary Fig. S12 at JXB online). Concordant with their role in animal systems (Besse et al., 2009; Karakasiliotis et al., 2010) and the repressive effect of StBEL5 UTRs on translation (Banerjee et al., 2009), these same RNA-binding proteins very probably also regulate translational control of StBEL5.
In this system, it is proposed that SD-enhanced expression of the potato PTB proteins leads to induced transport of StBEL5 to underground organs and an increase in root and tuber growth. Overexpression lines of StPTB1 and StPTB6 support this model as enhanced tuberization was observed in both lines concomitant with changes in the patterns of RNA metabolism (Fig. 4). This overexpression phenotype can be explained by an increase in StBEL5 transcripts leading to the induction of tuber identity genes. StBEL5 levels go up in stolons where StBEL5 is translated, leading to enhanced binding to its KNOX partner. In tandem, this complex activates transcription of GA2ox1 (and other genes in the tuber pathway) leading to an increase in steady-state levels of its mRNA (Lin et al., 2013) and a reduction in levels of bioactive gibberellins. The overexpression, accumulation, and movement of StBEL5 RNA have been consistently correlated with increased tuber production (Chen et al., 2003; Banerjee et al., 2006b, 2009). Here it is shown that both StPTB types bind to CU-rich RNA sequence, enhance stability and movement of StBEL5, and significantly increase tuber and root production. Through StPTB protein–StBEL5 interactions, StBEL5 RNA may be protected and localized to its functional sites in stolons and roots. Despite this observed effect on tuber development for the transgenic StPTB lines, shoot growth was essentially unchanged (Supplementary Figs S14C, D, S15A at JXB online).
PTB proteins have been observed to function in a diverse array of processes associated with RNA metabolism (Kuwahata et al., 2007; Xu and Hecht, 2007; Ham et al., 2009; Karakasiliotis et al., 2010; Rühl et al., 2012). The results presented here are consistent with the premise that PTB proteins of plants play an important role in escorting full-length mRNAs through the SE system. Despite significant research on the biochemical properties of CmRBP50, however, very little is known about how it functions in regulating development. This report presents novel results linking expression of two PTB proteins of potato to a specific phenotype, tuber formation. The mechanism of this effect on tuberization is strongly correlated to localization and stability of the mobile signal, StBEL5, and to the activity of its transcriptional targets. The use of transgenic plants in this study illuminates the function of StPTB proteins and provides an enlightening tool for understanding how they regulate a specific phase of plant growth. The phloem-associated promoter activity of StPTB1 and StPTB6, the regulated activity during early stages of tuberization, the binding affinity of both StPTB proteins for the 3′-UTR of StBEL5, and the dramatic tuberization phenotypes of the StPTB overexpression and suppression lines unequivocally support the premise that the PTB1 and PTB6 proteins of potato function prominently in post-transcriptional events associated with signalling the onset of storage organ formation.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Sequence alignment of polypyrimidine tract-binding proteins of potato.
Figure S2. Polypyrimidine tract-binding proteins of potato.
Figure S3. Phylogenetic analysis of polypyrimidine tract-binding proteins of plants.
Figure S4. Yeast three-hybrid analysis
Figure S5. RNA bait sequences used for the yeast three-hybrid and gel-shift assays.
Figure S6. Detection of purified recombinant PTB proteins using anti-PTB6 antibody.
Figure S7. Identification of reactive protein bands recognized by the anti-PTB6 antibody.
Figure S8. StPTB promoter activity in stems.
Figure S9. StPTB promoter activity in new tubers.
Figure S10. Expression profile of StPTB family members.
Figure S11. Effect of photoperiod on transcript accumulation of StPTB1 and StPTB6.
Figure S12. Immunodetection of StPTB proteins in stem exudate.
Figure S13. Immunodetection of PTB proteins in several organs.
Figure S14. Morphological analyses of StPTB1 and StPTB6 overexpression lines.
Figure S15. Shoot and root fresh weight of RNA suppression lines for StPTB1 and StPTB6.
Table S1. Primers used for RACE and full-length cDNA cloning.
Table S2. Accession numbers of PTB proteins used in Supplementary Fig. S3.
Table S3. Gene-specific primers used for RT–PCR.
Table S4. Primers used for various constructs.
Table S5. Primers for quantitative RT–PCR and mobility assay (RMA).
Acknowledgements
We thank Lukas Fischer for providing the 35S:CaMV:GFP transgenic line, Tian Lin for her help with figure design, and Dr Young-Jin Lee for assistance with the LC-MS/MS analysis. We also thank Professor Yiguo Hong for providing the PVX vector system, and Salomé Prat for providing the RNAi vector. This research was supported by the NSF Plant Genome Research Program award no. DBI-0820659 and National Research Initiative grant no. 2008–02806 from the USDA National Institute of Food and Agriculture.
References
- Auweter SD, Allain FHT. 2008. Structure–function relationships of the polypyrimidine tract binding protein. Cellular and Molecular Life Sciences 65, 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ. 2006. b . Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. The Plant Cell 18, 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Lin T, Hannapel DJ. 2009. Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiology 151, 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Prat S, Hannapel DJ. 2006. a . Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Science 170, 732–738. [Google Scholar]
- Besse F, López de Quinto S, Marchand V, Trucco A, Ephrussi A. 2009. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes and Development 23, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NM, Hannapel DJ. 2012. Promoter activity of polypyrimidine tract-binding protein genes of potato responds to environmental cues. Planta 236, 1747–1755. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Banerjee AK, Hannapel DJ. 2007. A BELL1-like gene of potato is light activated and wound inducible. Plant Physiology 145, 1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Rosin FM, Prat S, Hannapel DJ. 2003. Interacting transcription factors from the TALE superclass regulate tuber formation. Plant Physiology 132, 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerte C, Hall KB. 2009. The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry 48, 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléry A, Blatter M, Allain FH. 2008. RNA recognition motifs: boring? Not quite. Current Opinions in Structural Biology 18, 290–298. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Willmitzer L. 2001. Molecular and biochemical triggers of potato tuber development. Plant Physiology 127, 1459–1465. [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Du JS, Zabala MT, Beggs K, Smith C, Holdsworth M, Bevan M. 1994. Separate cis elements and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. The Plant Journal 5, 815–826. [DOI] [PubMed] [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cázares B, Ringgold V, Lough TJ, Lucas WJ. 2009. A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. The Plant Cell 21, 197–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannapel DJ, Sharma P, Lin T. 2013. Phloem-mobile messenger RNAs and root development. Frontiers in Plant Science 4, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasiliotis I, Vashist S, Bailey D, Abente EJ, Green KY, Roberts LO, Sosnovtsev SV, Goodfellow IG. 2010. Polypyrimidine tract binding protein functions as a negative regulator of feline calicivirus translation. PLoS One 5, e9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman B, Navarro C, Bijsterbosch G, Lange T, Prat S, Visser RG, Bachem CW. 2007. StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. The Plant Journal 52, 362–373. [DOI] [PubMed] [Google Scholar]
- Köster T, Staiger S. 2014. RNA-binding protein immunoprecipitation from whole-cell extracts. Methods in Molecular Biology 1062, 679–695. [DOI] [PubMed] [Google Scholar]
- Kuwahata M, Tomoe Y, Harada N, Amano S, Segawa H, Tatsumi S, Ito M, Oka T, Miyamoto K. 2007. Characterization of the molecular mechanisms involved in the increased insulin secretion in rats with acute liver failure. Biochimica et Biophysica Acta 1772, 60–65. [DOI] [PubMed] [Google Scholar]
- Li C, Gu M, Shi N, et al. 2011. Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Science Reports 1, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang K, Zeng X, Jackson S, Zhou Y, Hong Y. 2009. A cis element within Flowering Locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. Journal of Virology 83, 3540–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Sharma P, Gonzalez DH, Viola IL, Hannapel DJ. 2013. The impact of the long-distance transport of a BEL1-like mRNA on development. Plant Physiology 161, 760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. 2011. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. [DOI] [PubMed] [Google Scholar]
- Oberstrass FC., Auweter SD, Erat M, et al. 2005. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. 2006. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. Journal of Biological Chemistry 281, 32294–32302. [DOI] [PubMed] [Google Scholar]
- Rühl C, Stauffer E, Kahles A, Wagner G, Drechsel G, Rätsch G, Wachter A. 2012. Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. The Plant Cell 24, 4360–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. 2008. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochemical Society Transactions 36, 641–647. [DOI] [PubMed] [Google Scholar]
- Schmid N, Zagrovic B, van Gunsteren WF. 2007. Mechanism and thermodynamics of binding of the polypyrimidine tract binding protein to RNA. Biochemistry 46, 6500–6512. [DOI] [PubMed] [Google Scholar]
- Seeley KA, Byrne DH, Colbert JT. 1992. Red light-independent instability of oat phytochrome mRNA in vivo. The Plant Cell 4, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Lin T, Grandellis C, Yu M, Hannapel DJ. 2014. The BEL1-like family of transcription factors in potato. Journal of Experimental Botany 65, 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CG, Lewandowska D, Liney M, Davidson D, Chapman S, Fuller J, McNicol J, Shaw P, Brown JW. 2014. Arabidopsis PTB1 and PTB2 proteins negatively regulate splicing of a mini-exon splicing reporter and affect alternative splicing of endogenous genes differentially. New Phytologist 203, 424–436. [DOI] [PubMed] [Google Scholar]
- Stauffer E, Westermann A, Wagner G, Wachter A. 2010. Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. The Plant Journal 64, 243–255. [DOI] [PubMed] [Google Scholar]
- Terzi LC, Simpson GG. 2009. Arabidopsis RNA immunoprecipitation. The Plant Journal 59, 163–168. [DOI] [PubMed] [Google Scholar]
- Tillmar L, Welsh N. 2004. Glucose-induced binding of the polypyrimidine tract-binding protein (PTB) to the 3′-untranslated region of the insulin mRNA (ins-PRS) is inhibited by rapamycin. Molecular and Cellular Biochemistry 260, 85–90. [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Gebauer F. 1997. Post-transcriptional regulation: the dawn of PTB. Current Biology 7, R705–R708. [DOI] [PubMed] [Google Scholar]
- Wachter A, Rühl C, Stauffer E. 2012. The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Frontiers in Plant Science 3, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Okamoto T. 2009. Involvement of polypyrimidine tract-binding protein (PTB)-related proteins in pollen germination in Arabidopsis. Plant and Cell Physiology 50, 179–190. [DOI] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CWJ. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Molecular Cell 13, 91–100. [DOI] [PubMed] [Google Scholar]
- Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. 2009. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Research 37, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. 1999. A mini binary vector series for plant transformation. Plant Molecular Biology 40, 711–718. [DOI] [PubMed] [Google Scholar]
- Xu M, Hecht NB. 2007. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3′ UTR. Biology of Reproduction 76, 1025–1033. [DOI] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng A, et al. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, et al. 2009. Genome-wide analysis of PTB–-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Molecular Cell 36, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.