Highlight
DIG6 encodes a large 60S subunit GTPase 1 and affects ribosome biogenesis, and auxin response and homeostasis.
Keywords: AtLSG1, Arabidopsis, auxin homeostasis, expression pattern, map-based cloning, proteomics, ribosome biogenesis.
Abstract
The circularly permuted GTPase large subunit GTPase 1 (LSG1) is involved in the maturation step of the 60S ribosome and is essential for cell viability in yeast. Here, an Arabidopsis mutant dig6 (drought inhibited growth of lateral roots) was isolated. The mutant exhibited multiple auxin-related phenotypes, which included reduced lateral root number, altered leaf veins, and shorter roots. Genetic mapping combined with next-generation DNA sequencing identified that the mutation occurred in AtLSG1-2. This gene was highly expressed in regions of auxin accumulation. Ribosome profiling revealed that a loss of function of AtLSG1-2 led to decreased levels of monosomes, further demonstrating its role in ribosome biogenesis. Quantitative proteomics showed that the expression of certain proteins involved in ribosome biogenesis was differentially regulated, indicating that ribosome biogenesis processes were impaired in the mutant. Further investigations showed that an AtLSG1-2 deficiency caused the alteration of auxin distribution, response, and transport in plants. It is concluded that AtLSG1-2 is integral to ribosome biogenesis, consequently affecting auxin homeostasis and plant development.
Introduction
Ribosomes are complexes of RNA and protein molecules that are present in the cytoplasm or attached to the surface of the rough endoplasmic reticulum. They are the primary site for protein synthesis and are thus called protein factories. Ribosomes usually consist of one small subunit and one large subunit. Production of these ribosomal subunits is a strictly regulated process. In eukaryotes, assembly begins in the nucleolus, and pre-subunits are exported to the cytoplasm where their assembly is completed. Ribosome biogenesis requires not only the factors involved in processing ribosomal RNA and proteins but also a number of trans-acting factors that organize the assembly process. These trans-acting factors include GTP- and ATP-binding proteins that are necessary for several energy-consuming steps.
Eukaryotic LSG1 genes encode a large subunit GTPase (LSG1) that is involved in the maturation of the 60S subunit. This GTPase belongs to a large circularly permuted GTPase family whose members contain a central GTP domain. The structure of this domain is different from the canonical organization of GTPases in that the order of the five motifs is rearranged from G1, G2, G3, G4, and G5, to G4, G5, G1, G2, and G3 (Reynaud et al., 2005). Many members of this GTPase family are associated with ribosome biogenesis. LSG1 is involved in the late maturation steps of pre-60S subunit assembly and participates in the nuclear export of the pre-60S subunit in yeast. However, given that in yeast, LSG1 is a cytoplasmic protein, its regulatory role on the nuclear export of pre-60S subunits is indirect. The effects of LSG1 on the nuclear export of pre-60S subunits are controlled by the nuclear export signal (NES)-bearing protein nonsense-mediated decay 3 (NMD3) (Ho et al., 2000; Gadal et al., 2001; Hedges et al., 2005). NMD3 is a nucleo-cytoplasmic shuttling protein and an adaptor for the export of 60S ribosomal subunits from the nucleus. NMD3 is recruited by LSG1 and RIBOSOMAL PROTEIN L10 (RPL10); the loss of RPL10 or LSG1 causes the accumulation of NMD3 in the cytoplasm and ultimately fewer 60S ribosomal subunits (Hedges et al., 2005). In yeast and human cells, loss of function of LSG1 is lethal (Hedges et al. 2005; Reynaud et al., 2005), while in Drosophila, the LSG1 homologue (NS3) has been reported to regulate the insulin signalling pathway and control body size (Kaplan et al., 2008). In Arabidopsis, there are two orthologues of yeast LSG1—AtLSG1-1 and AtLSG1-2 (Weis et al., 2014)—whose functions in plant development are less studied than are their yeast counterparts. A recent study showed that AtLSG1-2 is involved in the maturation process of 40S proteins such that its loss of function caused multiple phenotypes including small size and incurvature leaves. On the other hand, loss of function of AtLSG1-1 has little effect on plant development. However, simultaneous loss of both genes is lethal, suggesting that they are required for plant growth (Weis et al., 2014).
Here, the dig6 (drought inhibited growth of lateral roots) mutant was isolated and characterized. This mutant had reduced lateral root number and strong incurvature in rosette leaves. Map-based cloning identified that the dig6 mutation occurs in AtLSG1-2. The importance of AtLSG1-2 in ribosome biogenesis was further evidenced in vivo by ribosome profiling and proteomics analyses of the mutant. Because the pleotropic phenotypes of dig6 are reminiscent of auxin-related mutants, auxin distribution, response, and transport were investigated. It was found that auxin response and homeostasis were altered in the mutant. This study highlights the roles of AtLSG1-2 in ribosome biogenesis, and auxin homeostasis and response in plants.
Materials and methods
Plant materials and growth conditions
The Arabidopsis dig6 mutant was isolated for its reduced lateral root growth (Xiong et al., 2006) from an M2 population generated by ethyl methane sulphonate (EMS) mutagenized Arabidopsis in the Columbia glabrous 1 (Col-gl1) background. Seeds of T-DNA insertion lines and auxin transporter::GFP reporter lines were obtained from the Arabidopsis Biological Resource Center. Seeds were surface sterilized with a 50% bleach solution for 5min, washed with water five times and then planted on half-strength Murashige and Skoog (MS) agar-medium plates. The plates were kept at 4 °C in darkness for 2–4 d and then moved to a growth chamber (CU36-L5, Percival Scientific) at 21 °C under a photoperiod of 16h light and 8h darkness (long-day conditions) or a photoperiod of 8h light and 16h darkness (short-day conditions). For morphological and histological analyses, seedlings were transferred to soil in a growth room under the same growth conditions as in the growth chamber.
Plasmid construction
To generate the pAtLSG1-2::GUS fusion construct, a 2006bp promoter fragment from genomic DNA was amplified by PCR and first cloned into the pENTRTM/D-TOPO vector and subsequently cloned into the upstream of GUS in pMDC162 using GATEWAY technology. To construct 35S::AtLSG1-2–YFP and 35S::YFP–AtLSG1-2, the full-length AtLSG1-2 coding region was cloned into the pENTRTM/D-TOPO vector and then into vectors pEarlygate 101 or pEarlygate 104, respectively, to produce p35S::AtLSG1-2-YFP or p35S::YFP-AtLSG1-2. Using the same method, AtLSG1-1 was cloned into the pEarlygate 104 vector to construct p35S::YFP-AtLSG1-1. Information on primers used in this study is given in Supplementary Table S2 available at JXB online.
Plant transformation
Transgenic plants were generated by transferring plasmids into Arabidopsis via the Agrobacterium-mediated flower-dipping method (Clough and Bent, 1998). Protoplasts were prepared from fully expanded healthy leaves of 3–4-week-old plants. Arabidopsis protoplast transformation was performed as previously described (Yoo et al., 2007).
Quantitative real-time PCR
Total RNA was extracted from plant materials using an RNeasy Mini kit (QIAGEN). DNA was eliminated from total RNA using DNase I (NEB) and cleaned with the RNeasy Mini kit (QIAGEN). Cleaned RNA was reverse transcribed using the SuperScript Reverse III reagent (Invitrogen) according to the manufacturer’s instructions. Two micrograms of RNA were used for quantitative real-time PCR (qRT-PCR) analysis. qRT-PCR was performed on a 7900HT Fast real-time PCR system (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems). Normalization was conducted against the average of housekeeping genes UBQ10 or ACT2. Relative gene expression was calculated by the equation 2−ΔΔCT. Three biological replicates were performed.
Histochemical staining, histological analysis, and microscopy
Plant materials were incubated in ice-cold 80% acetone for 30min, rinsed with 100mM sodium phosphate buffer twice and then incubated in a 1mg/ml of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (x-gluc) solution at 37 °C for a period ranging from 1h to overnight. Stained materials were cleared overnight in 90% ethanol. Samples were examined and photographed under a microscope. For histological analysis, fully expanded fifth leaves were fixed in 2.5% glutaraldehyde with 100mM phosphate buffer (pH 7.2) followed by osmication with 2% osmium tetroxide in 100mM phosphate buffer (pH 7.2). After dehydration with an ethanol series, samples were infiltrated and embedded in Spurr’s Resin (EMS) according to the manufacturer’s instructions. Sections approximately 1 μm thick were obtained with a Leica EM UC6 microtome (Leica) and then stained with 1% Toluidine Blue O before microscopic observation. To check the expression of fluorescent fusion proteins, plant materials mounted in water or transfected protoplasts were kept in WI solution [4mM MES (pH 5.7), 0.5M mannitol, and 20mM KCl] and were examined and photographed under a confocal microscope (Zeiss LSM710).
Functional complementation of the yeast lsg1 mutant
Full-length cDNA of AtLSG1-2, AtLSG1-1, and yeast LSG1 (ScLSG1), and truncated cDNA of AtLSG1-2 were amplified by PCR and ligated into PRS415-GAD (Mumberg et al., 1995). Plasmids containing intact or truncated AtLSG1-2, AtLSG1-1, wild-type yeast LSG1 (ScLSG1), or the empty vector (PRS415-GAD) were transformed into AJY1171 (MATalpha lsg1Δ::KanMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0), which contained the plasmid pAJ626 (LSG1 URA3 CEN) (provided by Dr Arlen Johnson). Transformants were plated on Leu− medium, and colonies were diluted and dotted on Leu− medium containing 5-FOA and incubated for 5 d at 30 °C.
Ribosome profiling analysis
Ribosome profiling analysis was performed with 10-d-old seedlings as previously described (Mustroph et al., 2009).
Isobaric tags for relative and absolute quantitation analysis
Roots of 10-d-old seedlings growing on MS agar media were used for isobaric tags for relative and absolute quantitation (iTRAQ) analysis. Protein sample preparation and analysis was performed as described in Zhao et al. (2014). The final proteomic data were derived from two biological replicates with three technical replicates each.
Auxin treatment
Surface-sterilized seeds were directly plated on agar-medium plates with or without 1-N-naphthylphthalamic acid (NPA) supplement and grown for 10 d under the above-mentioned conditions. Phenotypes of the seedlings were observed and pictures were taken using a digital camera. GUS staining was performed as described above. For the auxin treatment, 5-d-old seedlings were transferred to auxin-containing medium. Length of primary roots was measured after 5 d of incubation in the growth chamber. For quantitative PCR analysis, 6-d-old seedlings were incubated in mock and 20 µM indole-3-acetic acid (IAA) for 2h.
Accession numbers
Sequence data in this article can be found in the EMBL/Genbank data libraries under accession numbers Atlg08410 (DIG6/AtLSG1-2), At2g27200 (AtLSG1-1), At3g07050 (NSN1), and At1g52980 (NUG2).
Results
Isolation of the dig6 mutant and map-based cloning of the DIG6 gene
Because drought (water deficit) stress and phytohormone abscisic acid (ABA) can inhibit lateral root development in Arabidopsis, this trait was used to screen for mutants potentially involved in the drought stress response (Xiong et al., 2006). One mutant, dig6, was isolated from an M2 population generated by EMS-mutagenized Arabidopsis in the Columbia glabrous 1 (Col-gl1) background. This mutant is constitutively defective in lateral root growth regardless of the water potential or the ABA level in the growth media. Mutant seedlings grew slower than the wild-type and had a slightly yellowish colour (Supplementary Fig. S1A, B available at JXB online). In addition, mutants had fewer emerging lateral roots (LRs; Supplementary Fig. S1C) and displayed a strong incurvature phenotype in young leaves (Supplementary Fig. S1D).
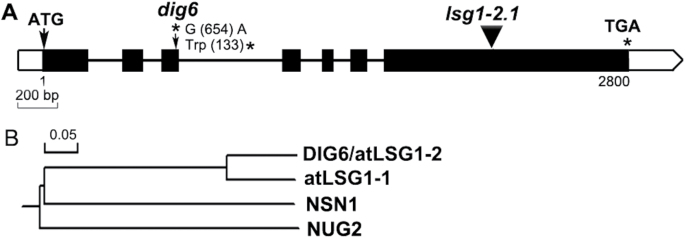
To map the mutation, the mutant was crossed with Landsberg erecta. The mutation was initially mapped to a 2.754 Mbp region on Chromosome 1 between the simple sequence length polymorphic markers Chr1-1.070M and Chr1-3.824M (sequences of the markers are provided in Supplementary Table S1). Whole-genome DNA sequencing of the mutant was subsequently conducted. In the mapping interval, a G-to-A single nucleotide change was found in the At1g08410 gene (AtLSG1-2), which was predicted to generate a premature stop codon and would produce a truncated protein with 132 amino acids instead of the intact 589 amino acid length (Fig. 1A).
Fig. 1.
Structure of the AtLSG1-2 gene and phylogenetic analysis of AtLSG1-2-related proteins in Arabidopsis. Genomic DNA structure of AtLSG1-2 and the location of AtLSG1-2 mutations. Exons, introns, and 5′or 3′UTRs are shown by black boxes, bold lines, and white boxes, respectively. Mutation sites are marked with arrows. (B) Phylogenetic tree of AtLSG1-2-related proteins generated by DNAman software.
To confirm that the phenotypes observed in the mutant can be attributed to the loss of function of the AtLSG1-2 gene, a T-DNA insertion allele (Salk_114083) in the Col-0 background was obtained. This mutant was described as lsg1-2.1 (for simplicity also referred to as lsg1-2 in this text hereafter) (Weis et al., 2014). The F1 progeny plants of dig6 and lsg1-2 crosses showed the same phenotypes as dig6 and lsg1-2 (data not shown), indicating that AtLSG1-2 is responsible for the dig6 mutant phenotypes.
Yeasts and humans have only a single copy of their respective AtLSG1 homologue, which is essential for cell viability (Hedges et al., 2005; Reynaud et al., 2005), while Arabidopsis has two homologues (Fig. 1B). AtLSG1-1 (At2g27200) encodes a protein that shares 77.3% identity with AtLSG1-2 at the protein sequence level. A Blast search using AtLSG1-2 as a query sequence also found two other circularly permuted GTPases, NUCLEOSTEMIN-LIKE 1 (NSN1) and NUCLEAR/NUCLEOLAR GTPase 2 (NUG2), with only 21.1% and 29.8 % identity with AtLSG1-2, respectively. Nevertheless, the GTPase domain region of these proteins is better conserved relative to the rest of the protein (Supplementary Fig. S2). Previous studies showed that NSN1 is required for flower and leaf development (Wang et al., 2012a, 2012b) and NUG2 is involved in pre-60S ribosomal subunit maturation. The AtNUG2 RNAi mutant has been shown to have increased sensitivity to cycloheximide treatment (Im et al., 2011).
Phenotypic characterization of the lsg1-2 mutants
Although dig6 and lsg1-2 mutants had identical morphological and developmental phenotypes (Fig. 2, Supplementary Fig. S1, see below), because of the possibility of interference from the gl1 mutation on development and potential background mutations in the EMS-mutagenized dig6 mutant, the lsg1-2 allele was used for the characterization of mutant phenotypes.
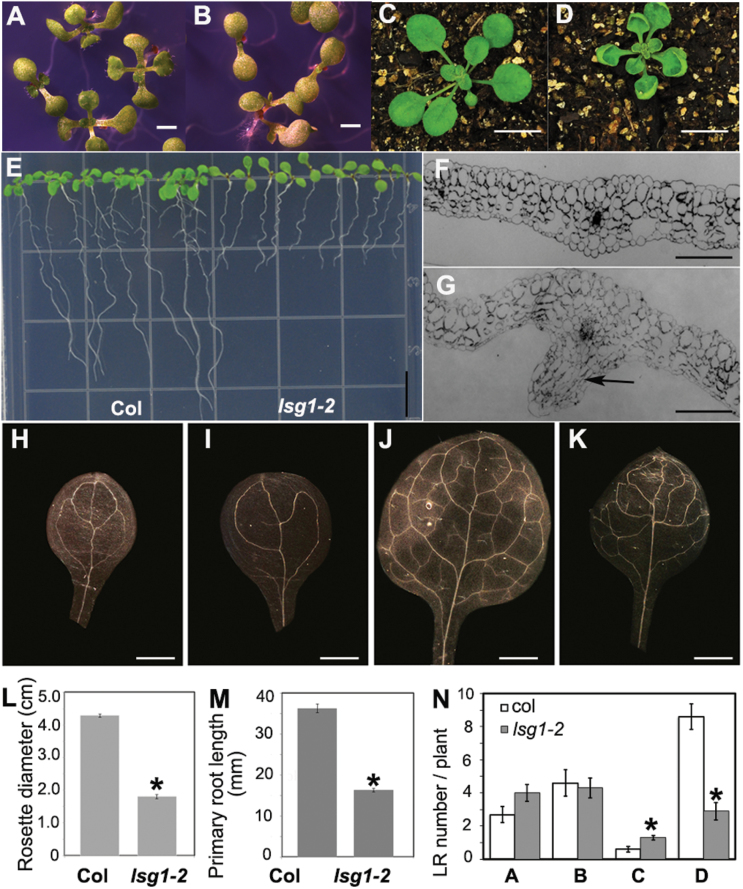
Fig. 2.
Phenotypes of the lsg1-2 mutant. (A, B) Morphology of 12-d-old seedlings of Col-0 (A) and lsg1-2 (B) growing on horizontally placed agar plates. Scale bars, 5mm. (C, D) Morphology of Col-0 (C) and lsg1-2 (D) seedlings grown in soil. Scale bars, 1cm. (E) Phenotype of 12-d-old seedlings of Col-0 (A) and lsg1-2 (B) growing on vertically placed agar plates. Scale bars, 1cm. (F, G) Transverse section of the fifth rosette leaf of Col-0 (F) and lsg1-2 (G). Arrow indicates the region where extra cell division occurred. Scale bars, 200 µm. (H–K) Venations of cotyledons and the first pair rosette leaves in Col-0 (H, J) and lsg1-2 (I, K). At least 10 plants were examined and the representative images are shown. Scale bars, 1mm. (L) The diameter of rosette leaves of 4-week-old plants growing in soil. (M) Primary root length of 12-d-old seedlings. (N) The number of LRs at different developmental stages (stages A–D) of 12-d-old seedlings. Data are means ± standard errors (n=8–16). *P<0.01, compared with wild-type plants (t-test).
Mutant leaves emerged later (Fig. 2A, B) and grew smaller (Fig. 2C, D) than wild-type leaves. Prior to bolting, lsg1-2 mutants had significantly smaller stature than the wild-type (Fig. 2C, D, L), with markedly shorter primary roots (Fig. 2E, M, and Supplementary Fig. S1) and arrested lateral root growth (Fig. 2E and Supplementary Fig. S1). In 12-d-old seedlings, LRs were visible on wild-type plants but were barely visible in lsg1-2 mutants. There are two possible explanations for decreased visible LRs in mutants: either LR growth is not being initiated or fewer LRs are elongated. By measuring the LR density, a higher density was found in the lsg1-2 mutant (Supplementary Fig. S3), indicating that fewer visible LRs in the mutant cannot be explained by the failure of initiation. Therefore, LR number was examined at different developmental stages, for which a previous study was followed: LR development is divided into four stages (A to D) (De Smet et al., 2003). In stage A, LRs are primordial with up to two cell layers; in stage B, LRs are between three cell layers and just prior to emergence; in stage C, LRs emerge but are shorter than 0.5mm; and in stage D, LRs are longer than 0.5mm (De Smet et al., 2003). Among these stages of LRs, only stage D is visible to the naked eye. After examining LR number at all stages, significantly fewer LRs were found at stage D in the lsg1-2 mutant (Fig. 2N). Thus, the reduced lateral root phenotype in the early developmental stage of the lsg1-2 mutant was a result of a delay in LR emergence (Fig. 2E, 2N and Supplementary Fig. S1).
The most notable phenotype of lsg1-2 mutants is the incurvature of rosette leaves (Fig. 2D and Supplementary Fig. S1D). Transverse thin sections were examined across the regions of fully expanded fifth leaves where the incurvature occurred and a significant increase in layers of spongy mesophyll cells was found in the abaxial side of mutant leaves (Fig. 2G), suggesting that extra cell division in the abaxial side of leaves is mainly responsible for the incurvature. Interestingly, the leaf incurvature phenotype was mainly exhibited at the early stages of plant development. When the inflorescence emerged, this incurvature phenotype could occasionally be seen in the newly emerged axillary leaves (Supplementary Fig. S1F). On the other hand, old rosette leaves and cauline leaves showed no sign of the incurvature phenotype. Noticeably, flatness of the cotyledon was not affected by AtLSG1-2 deficiency (Fig. 2B).
Because vascular structure is closely related with leaf development, the structure of the wild-type and lsg1-2 mutants was investigated. As shown in Fig. 2H–K, the vascular structure of cotyledons and the first pair of leaves in the lsg1-2 mutant were very different from that of the wild-type plant. Vein loops were often open in the cotyledons of lsg1-2 mutants, while those of the wild-type were typically closed (Fig. 2H, I). In the first pair of leaves, the distal part of the midvein was bifurcated in the lsg1-2 mutant unlike the wild-type. Furthermore, the lsg1-2 mutant had considerably fewer tertiary and quaternary veins compared with the wild-type (Fig. 2K).
Expression patterns of AtLSG1-2
The expression of AtLSG1-2 and AtLSG1-1 was examined using qRT-PCR on RNA extracted from different organs of wild-type plants. AtLSG1-2 was highly expressed in the flowers, moderately expressed in the roots, axillary leaves, rosette leaves, and siliques of young seedlings, and marginally expressed in the stalks of inflorescence and cauline leaves (Supplementary Fig. S4). AtLSG1-1 was highly expressed in siliques, moderately expressed in roots, the flowers, and rosette leaves, and marginally expressed in the stalks of inflorescence, and cauline and axillary leaves of young seedlings (Supplementary Fig. S4). Generally, expression levels of AtLSG1-2 were notably higher than those of AtLSG1-1 in all the organs examined (Supplementary Fig. S4 available at JXB online).
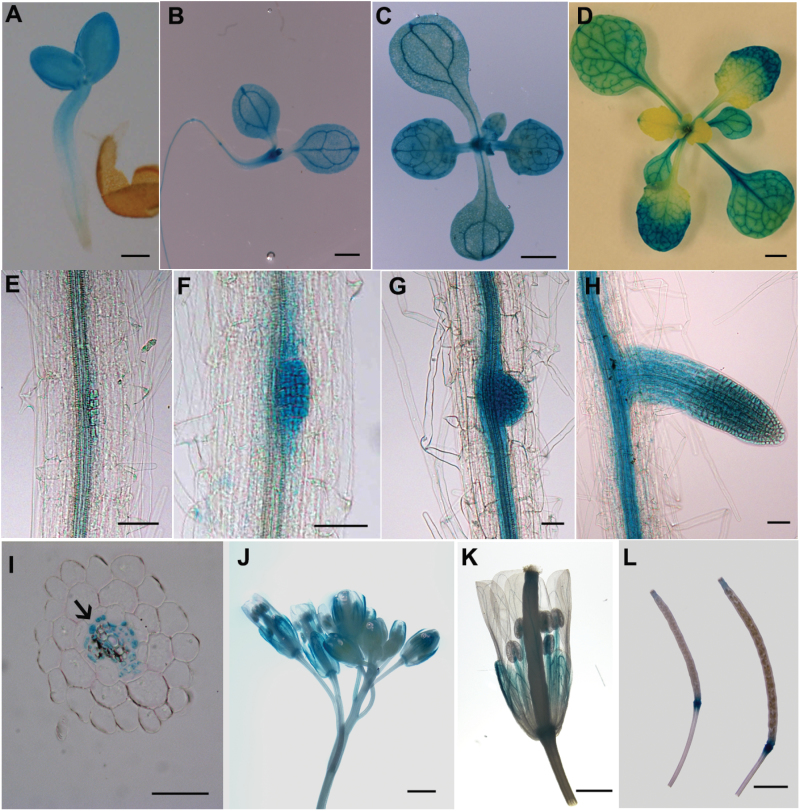
The expression pattern of AtLSG1-2 during plant development was further studied by using the promoter-driven GUS reporter gene (Fig. 3A–L). Strong GUS signals in the entire cotyledon (Fig. 3A–D) and in emerging leaves (Fig. 3B) were observed. Strong signals were detected in all first pairs of leaves (Fig. 3A–D), but only in the distal part and with a concave pattern in second leaves (Fig. 3D). The GUS signal was highly expressed in the marginal region but minimally in the midvein region. This expression pattern was further demonstrated by qRT-PCR (Supplementary Fig. S5). In the primary root, AtLSG1-2 was specifically expressed in vascular bundles along elongation and differentiation zones (Fig. 3A, E–I). Transverse section analysis showed that AtLSG1-2 was mainly expressed in the stele but was excluded from the xylem (Fig. 3I). Because prominent LR phenotypes were observed in lsg1-2 mutants, its expression was investigated in detail (Fig. 3E–H). It was found that AtLSG1-2 is strongly expressed in lateral root cells of all stages examined (Fig. 3E–H) and in the inflorescence (Fig. 3I–J). In flowers, higher expression in filaments and sepals was observed (Fig. 3K). Strong signals were detected at the junction between the silique and the pedicel, while only low signals were detected at the tip of siliques (Fig. 3L). Despite the high expression level of AtLSG1-2 in inflorescence or the silique, no visible phenotype was observed in these organs (Supplementary Fig. S6), probably because of the functional redundancy between AtLSG1-2 and AtLSG1-1. Thus, it is evident that AtLSG1-2 is highly expressed in newly emerged leaves, leaf veins, the vascular structure of the roots, and lateral root primordia where auxin also accumulates.
Fig. 3.
The expression pattern of AtLSG1-2. pAtLSG1-2::GUS expression was detected in 2- (A), 8- (B), and 12-d-old seedlings (C); 26-d-old soil-grown plant (D); LRs at stage A (E), C (F, G), and D (H); inflorescence (J, K); and siliques (L). (I) Transverse section of GUS-stained primary roots. Arrow indicates the xylem. Ten GUS-positive transgenic plants were checked and similar expression patterns were observed. A representative image is shown. Scale bars, 100 µm (A), 1mm (B–E), 50 µm (F–I), and 500 µm (J– L).
AtLSG1-2 and AtLSG1-1 are localized in the cytosol in stably transformed plants
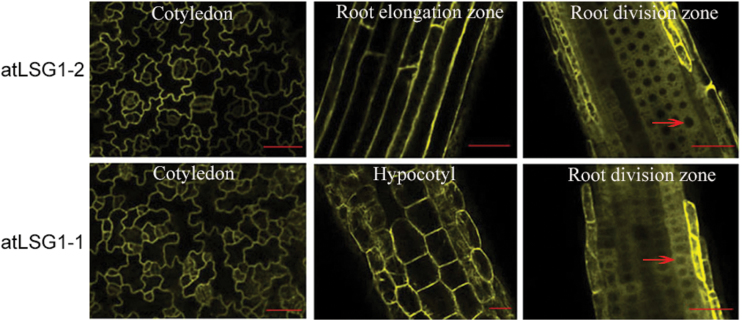
Previous studies have shown that LSG1 proteins have different subcellular localization in yeast, human, and Drosophila cells (Gadal et al., 2001; Reynaud et al., 2005; Hartl et al., 2013). To determine where they localize in plant cells, 35S::AtLSG1-2-YFP and 35S::YFP-AtLSG1-2 constructs were created and transformed into the protoplasts of wild-type plants, and dig6 and lsg1-2 mutants. These constructs fully complemented the mutant phenotypes (Supplementary Fig. S7), suggesting that they were fully functional. In transiently expressed protoplasts, these proteins were localized in the nucleus and cytosol (data not shown), which is consistent with a recent study (Weis et al., 2014). In stably transformed plants, the localization of fluorescence-tagged proteins was examined in different organs including the cotyledon, and the root elongation and division zones. Surprisingly, the localization of both AtLSG1-2 and AtLSG1-1 was limited to the cytosol in these organs (Fig. 4). This is particularly evident in the root division zone, where fluorescence signals were excluded from the hollow areas (i.e., nuclei) and were found in the cytoplasm (Fig. 4).
Fig. 4.
Subcellular localization of AtLSG1-2 and AtLSG1-1. The YFP signal was detected in the cytosol of cells in cotyledon, root division zone, root elongation zone, and hypocotyl of plants transformed with 35S::YFP-AtLSG1-1 or 35S::AtLSG1-2-YFP. Arrows indicate nuclei. Scale bars, 50 µm. At least 10 transformed plants were examined.
Loss of function of AtLSG1-2 affects ribosome biogenesis
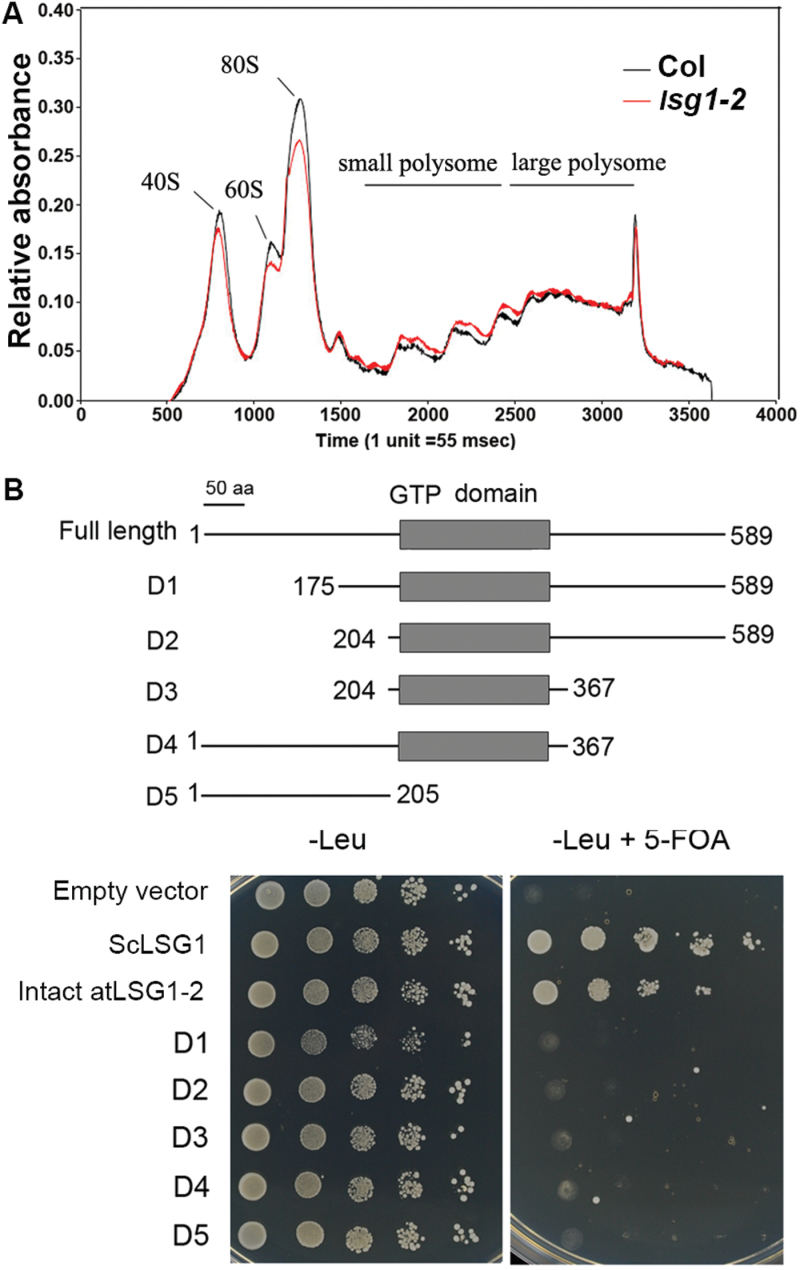
To investigate the impact of AtLSG1-2 deficiency on ribosome biogenesis in plants, ribosome levels were examined in 10-d-old mutant seedlings using ribosome profiling assays. The lsg1-2 mutants were found to have decreased levels of 60S and 40S ribosomal subunits, and 80S monosomes, although polysome production was not affected (Fig. 5A). Thus, AtLSG1-2 is important for maintaining normal 60S and 40S ribosome biogenesis and 80S monosome assembly in plants.
Fig. 5.
Ribosome profiling analysis and deletion analyses. (A) Ribosome profiling analysis using 10-d-old seedlings of Col-0 and the lsg1-2 mutant. This experiment was repeated twice with similar results. (B) Deletion analysis. Various deletion fragments (D1 to D5, upper panel) of AtLSG1-2 used in yeast complementation assays are shown (lower panel). Empty vector (PRS415-GAD) and yeast LSG1 (ScLSG1) were used as negative and positive controls, respectively. At least three clones per construct were examined.
The AtLSG1-2 protein includes an N- and a C-terminus and a GTP domain (Supplementary Fig. S2). In a previous study, certain regions were shown to be important for LSG1 function in yeast; for example, the G1 motif is essential for GTP binding (Hedge et al., 2005), and a mutation in the predicted coil-coil region affects yeast growth (Hedge et al., 2005). To investigate which regions are important for AtLSG1-2 function, intact and a series of deletion constructs were made and these proteins were expressed in the yeast lsg1 mutant. As shown in Fig. 5B, full-length cDNA from AtLSG1-2 complemented the phenotype of the yeast lsg1 mutant, similar to results from a recent study (Weis et al., 2014). However, neither truncated protein rescued the yeast lsg1 mutant, indicating that all the examined regions are essential for AtLSG1-2 function in ribosome biogenesis.
Changes in the proteomic profile in the lsg1-2 mutant
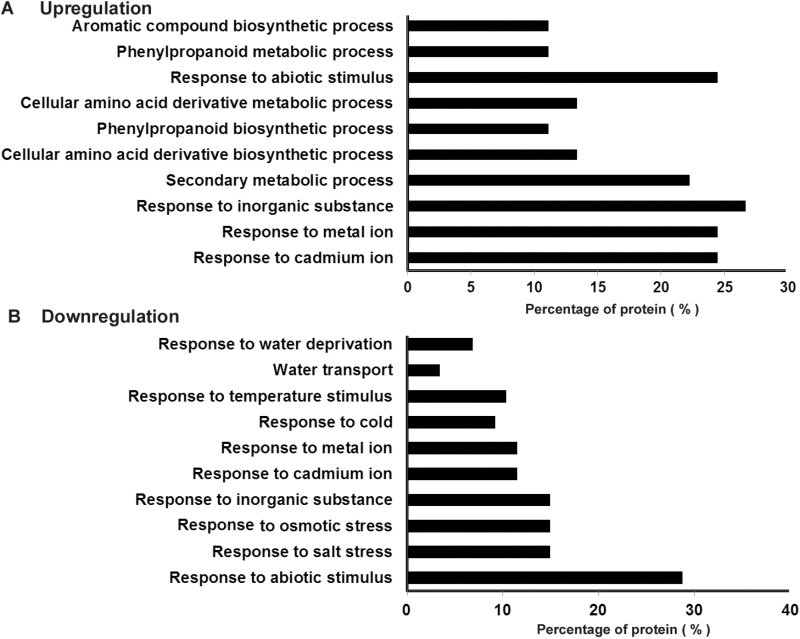
Because AtLSG1-2 is involved in ribosome biogenesis and lsg1-2 mutants exhibited multiple developmental defects, it was anticipated that there would be changes in the proteome of the mutants. To probe changes in the proteomic profile of lsg1-2 mutants, iTRAQ analysis was used to compare the global protein profile of 10-d-old-seedling roots of the wild-type and the lsg1-2 mutant. Using a fold change greater than 1.3 and P<0.05 as the criteria for significance, 132 proteins in total were found to be differentially regulated (Supplementary Table S1 available at JXB online). Among them, 45 were significantly up-regulated and 87 were down-regulated in the mutant. A functional classification of differentially regulated proteins was made using the software DAVID (Huang et al., 2009), and it was found that the up-regulated group was mainly associated with secondary metabolic process and in response to abiotic stimuli (Fig. 6A). The down-regulated group was enriched in proteins involved in response to abiotic stimuli and water transport (Fig. 6B). Interestingly, the iTRAQ analysis identified that 15 proteins involved in ribosome biogenesis were differentially regulated, which accounted for 11.3% of total differentially regulated proteins in the lsg1-2 mutant. Among these, five have higher expression levels in the lsg1-2 mutant (Table 1). These included two NOP56-LIKE pre-mRNA processing ribonucleoproteins (RNP), NUCLEOLIN LIKE 1 (ATNUC-L1), a 60S ribosomal protein, and NUCLEOPORIN 57 (NUP57). ATNUC-L1 is involved in rRNA processing, ribosome biosynthesis, and vascular pattern formation (Petricka and Nelson, 2007). All of the 10 down-regulated proteins are ribosomal proteins, including seven 60S and three 40S ribosomal proteins (Table 1). This is evidence that AtLSG1-2 loss of function affects the level of proteins involved in ribosome biogenesis. It was also noted that most of the down-regulated proteins involved in water transport are aquaporins. Among the 17 major aquaporin proteins in Arabidopsis (Péret et al., 2012), seven were simultaneously repressed in the lsg1-2 mutant. Notably, aquaporin proteins are regulated by auxin and are involved in lateral root emergence (Péret et al., 2012).
Fig. 6.
Functional classification of differentially expressed proteins in lsg1-2 compared with wild-type Col-0. Proteins in the up-regulated (A) and down-regulated (B) groups were classified by DAVID software. The top 10 functional annotations ordered by the enrichment scores are presented. Two biological repeats and three technique repeats were performed for the iTRAQ analysis.
Table 1.
Differentially regulated proteins involved in ribosome biogenesis in the lsg1-2 mutant
| Accession number | Protein | Fold change (lsg1-2 vs Col) | P value |
|---|---|---|---|
| Up-regulated protein | |||
| AT5G27120 | NOP56-like pre-mRNA processing RNP | 2.066 | 0.005 |
| AT3G57150 | Putative pseudouridine synthase (NAP57) | 1.96 | 0.016 |
| AT1G48920 | Nucleolin like 1 (AtNUC-L1) | 1.698 | 0.007 |
| AT3G60245 | 60S ribosomal protein L37a-2 | 1.64 | 0.043 |
| AT3G05060 | NOP56-like pre-mRNA processing RNP | 1.618 | 0.016 |
| Down-regulated protein | |||
| AT3G09200 | 60S acidic ribosomal protein P0-2 | 0.744 | 0.013 |
| AT5G47930 | 40S ribosomal protein S27-3 | 0.727 | 0.022 |
| AT5G64140 | 40S ribosomal protein S28-2 | 0.718 | 0.023 |
| AT4G34670 | 40S ribosomal protein S3a-2 | 0.618 | 0.018 |
| AT4G29410 | 60S ribosomal protein L28-2 | 0.616 | 0.009 |
| AT2G44120 | 60S ribosomal protein L7-3 | 0.611 | 0.011 |
| AT3G55280 | 60S ribosomal protein L23a-2 | 0.606 | 0.024 |
| AT3G28900 | 60S ribosomal protein L34-3 | 0.395 | 0.006 |
| AT1G61580 | 60S ribosomal protein L3-2 | 0.362 | 0.041 |
| AT3G44590 | 60S acidic ribosomal protein P2-4 | 0.101 | 0.001 |
Values are means of three technical replicates expressed in relative fold changes.
AtLSG1-2 deficiency affects auxin distribution and response
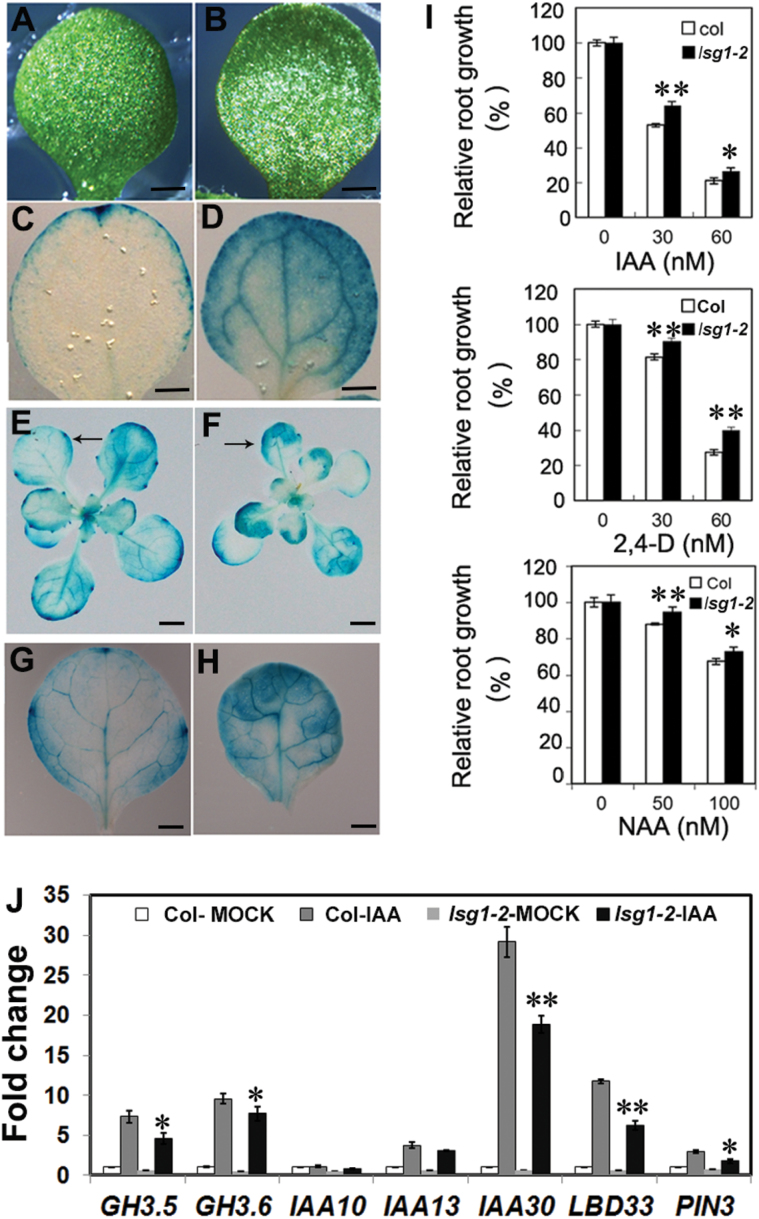
The lsg1-2 mutants exhibited similar developmental defects as auxin-related mutants such as abnormal vein structure in leaves and cotyledons, short primary roots, and delayed LR emergence. The DR5::GUS reporter was employed to monitor the distribution of auxin in lsg1-2 mutant leaves. The reporter gene was introduced into lsg1-2 by genetic crossing and its expression was examined. The GUS staining pattern in the mutant was distinctly different from that in the wild-type. High expression of the DR5::GUS reporter gene was observed in the leaf border of lsg1-2 mutant leaves (Fig. 7F, H), similar to NPA-treated seedlings (Fig. 7C, D). The accumulation of auxin or an increased auxin response may have triggered the extra cell division activity that was observed in the abaxial side of lsg1-2 mutant leaves, causing the leaf incurvature phenotype in lsg1-2 mutants.
Fig. 7.
Auxin distribution in lsg1-2 mutant leaves and auxin responses of the lsg1-2 mutant. (A, B) Cotyledon phenotype of wild-type (Col-0) seedlings grown on MS medium without (A) or with (B) NPA. (C, D) DR5::GUS expression in the cotyledons of NPA untreated (C) and treated (D) wild-type seedlings. (E, F) DR5::GUS expression in the whole plant of Col-0 and lsg1-2. Arrows indicate first leaves. (G, H) Close-up view of DR5::GUS expression in the first leaf of Col-0 (g) and lsg1-2 (h). (I) Auxin response of the lsg1-2 mutant. Five-day-old seedlings grown on half-strength MS medium were transferred to media supplemented with the indicated concentrations of auxins (IAA, 2,4-D, and NAA) and grown for an additional 5 d. Primary root length was then measured and expressed relative to that under auxin-free conditions. Data are means ± standard errors (n=18). (J) Reduced levels of auxin-induced genes in the lsg1-2 mutant as revealed by qRT-PCR analysis. *P<0.05, **P<0.01 (t-test) compared with wild-type plants. Scale bars, 1mm (A–D, G, H) and 2mm (E, F).
The sensitivity of lsg1-2 mutants to auxin treatments was investigated. Primary root growth of the wild-type and the lsg1-2 mutant was examined with seedlings growing on MS medium containing either IAA, 2,4-Dichlorophenoxyacetic acid (2,4-D), or 1-naphthaleneacetic acid (NAA). Auxin influx carriers mediate the influx of exogenous IAA and 2,4-D into the roots, whereas NAA can freely penetrate into the cells in a carrier-independent manner (Estelle, 1998). As shown in Fig. 7I, the lsg1-2 mutant consistently exhibited reduced sensitivity to these auxins.
Next, the response of AtLSG1-2 to auxin was examined in terms of auxin-inducible gene expression. Six-day-old wild-type and lsg1-2 seedlings were treated with 20 μM IAA for 2h and the expression levels of several auxin-responsive genes were analysed using qRT-PCR. Although the expression levels of most genes were induced to various extents by auxin, they were induced to a lesser extent in the lsg1-2 mutant than in the wild-type (Fig. 7J).
The lsg1-2 mutant had reduced levels of auxin influx and efflux carriers, and impaired auxin transport
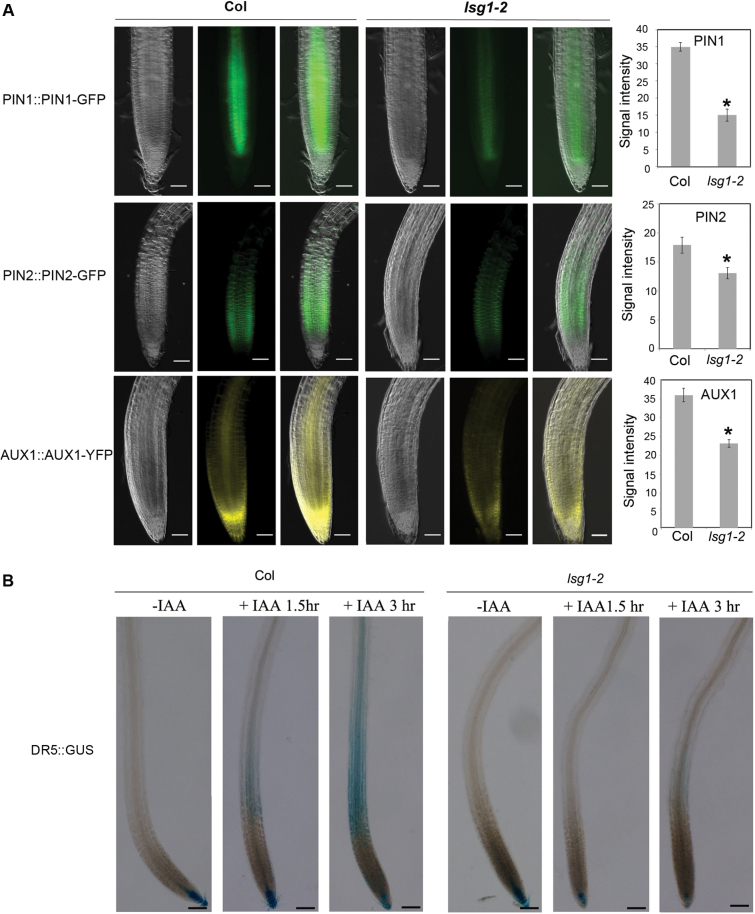
The movement of auxin within plants is mediated by a group of transporters. Thus, it was examined whether the expression of auxin transporters was affected in the lsg1-2 mutant. The GFP-tagged auxin efflux carriers pPIN1::PIN1-GFP and pPIN2::PIN2-GFP, and the auxin influx carrier pAUX1::AUX1-GFP were introduced into the lsg1-2 mutant by genetic crossing. It was found that the expression levels of PIN1, PIN2, and AUX1 were significantly lower in lsg1-2 mutants compared with those in the wild-type (Fig. 8A). Using DR5::GUS as a reporter, auxin transport between the wild-type and mutants was compared. After root tips were treated with IAA for 1.5h, DR5::GUS signals became easily visible in the upper part of the wild-type but not in mutant roots (Fig. 8B). After an additional 1.5h, more signals were detected in the upper part of the wild-type root but only faint signals were detected in mutant roots (Fig. 8B). These data indicated that auxin transport was also impaired in the mutants.
Fig. 8.
The lsg1-2 mutant has reduced levels of auxin transporters and displays auxin transport defects. (A) Expression of pPIN1::PIN1-GFP, pPIN2::PIN2-GFP, and pAUX1::AUX1-YFP in roots of Col-0 and lsg1-2, and quantification of the fluorescence intensity. For each genotype- reporter, the left panel presents a bright-field image of the root; the middle panel presents the GFP or YFP image; and the right panel shows the merged image of bright-field and GFP/YFP images. Scale bars, 50 µM. Fluorescence intensities were quantified using Image J. Data represent means ± standard error (n = 10). *P<0.01 (t-test) compared with wild-type plants. (B) The auxin transport process was monitored using DR5::GUS reporter in wild-type (Col-0) and lsg1-2 mutant roots. Root tips were treated with IAA for 1.5 or 3h. Roots were then stained for reporter gene expression. Scale bars, 100 µM. At least 10 plants were examined with similar results. Representative images are shown.
Loss of AtLSG1-2 activates compensatory expression of AtLSG1-1
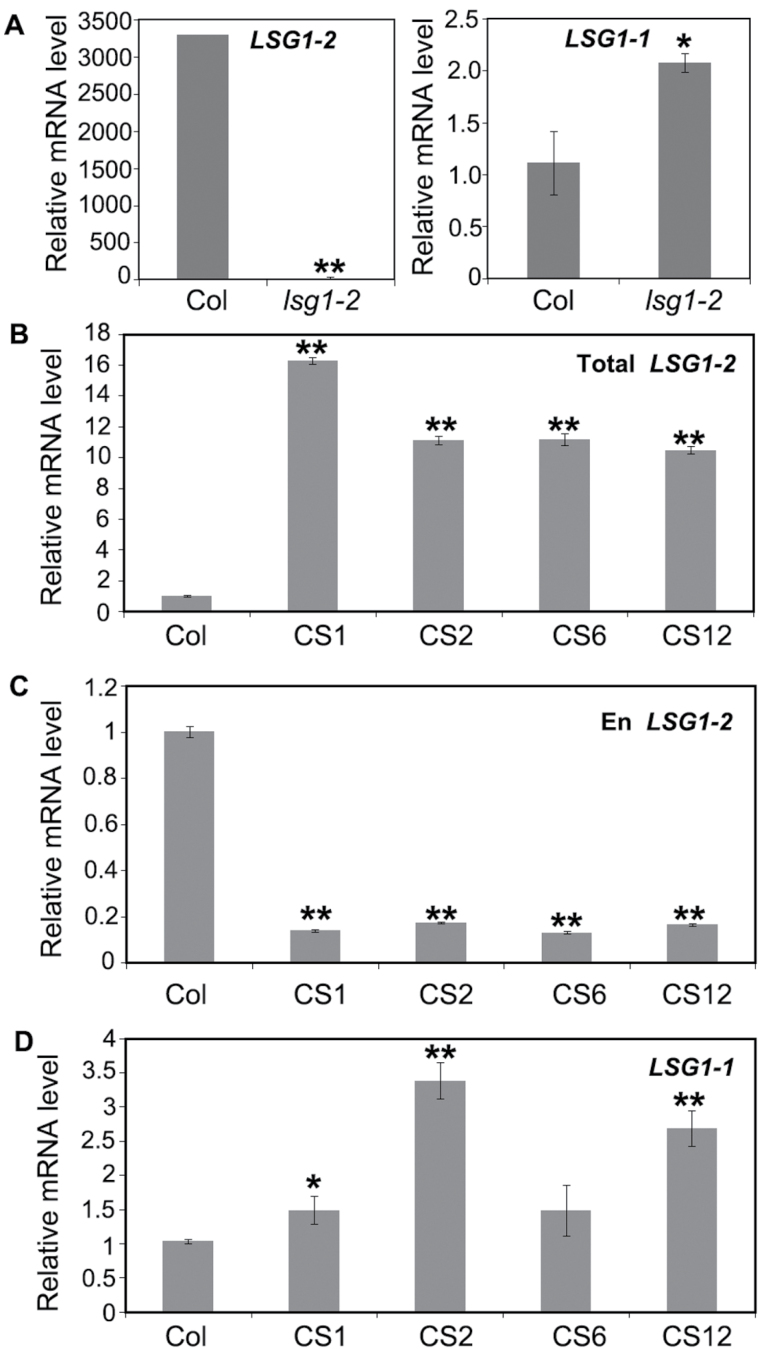
AtLSG1-2 and atLSG1-1 have a high degree of amino acid identity and also share overlapping gene expression patterns. The relatively mild phenotypes of lsg1-2 mutants suggest that AtLSG1-1 might partly compensate for the loss of AtLSG1-2. The expression level of AtLSG1-1 was examined in the leaves of AtLSG1-2 knockout line by qRT-PCR and was found markedly increased in lsg1-2 (Fig. 9A). Next, the expression levels of AtLSG1-1 in AtLSG1-2 cosuppression lines that were transformed with the AtLSG1-2 overexpression construct were investigated yet showed phenotypes similar to those of lsg1-2 mutants, likely due to silencing of the endogenous gene. In these plants, the total expression level of AtLSG1-2 from both the endogenous gene and the transgene was significantly higher (Fig. 9B), but the expression level of endogenous AtLSG1-2 was greatly decreased (Fig. 9C). In contrast, the expression level of AtLSG1-1 in these plants increased (Fig. 9D), consistent with the notion that AtLSG1-2 loss of function could induce increased expression of AtLSG1-1.
Fig. 9.
Expression levels of atLSG1-2 and atLSG1-1 in the wild-type, lsg1-2, and cosuppression plants. (A) Relative expression levels of atLSG1-2 and atLSG1-1 in wild-type and lsg1-2 plants. (B, C) Relative expression level of atLSG1-2 from both the endogenous gene and the transgene (B) and from the endogenous gene only (C). (D) Relative expression level of atLSG1-1 in (B–D), Col, wild-type Col-0; CS1, CS2, CS6, and CS12 independent transgenic plants expressing 35S::atLSG1-2-YFP that showed cosuppression phenotypes. Gene expression level was measured by qRT-PCR and expressed relative to that of the UBQ10 gene. Data are means ± standard error from three biological replicates. *P<0.05, **P<0.01 (t-test) compared with the wild-type plants.
Discussion
In this study, a genetic screen and position cloning identified AtLSG1-2 as an important protein involved in several developmental processes in Arabidopsis. Both here and in a previous study, AtLSG1-2 is shown to share similar conserved functions in ribosome biogenesis as its homologues in other organisms. However, this study showed that AtLSG1 proteins also have some distinct functions. The ribosome profiling experiments revealed that the lsg1-2 knockout mutant had lower levels of 40S and 60S ribosome subunits, and the 80S monosome, while the polysome level was not affected (Fig. 5A), which is different from yeast lsg1 dominant-negative mutants and temperature-sensitive mutants where polysomes were also affected (Kallstrom et al., 2003; Hedge et al., 2005). This might be explained by differences between species or the presence of AtLSG1-1 in Arabidopsis. In the lsg1-2 single mutant, some phenotypes are similar to the Drosophila ns3 weak allele mutant, such as growth arrest and small stature, although these two proteins localize in different compartments. This phenotype similarity is probably related to defective ribosome biogenesis. Interestingly, the growth defects of the ns3 mutant can be rescued by the overexpression of AKT1 (Kaplan et al., 2008), a kinase that is the central effector of the insulin pathway. It is still unclear if the insulin pathway exists in plants, and no AKT1 homologue has been found in Arabidopsis. Thus, AtLSG1-2 probably uses a different pathway to control body size in Arabidopsis.
This study showed that lsg1-2 mutants exhibited some auxin-related phenotypes including stunted root growth, delayed LR emergence, and altered auxin response and transport. These phenotypes imply that certain auxin-related or auxin-regulated proteins might be differentially regulated. Nonetheless, auxin-related proteins were not detected in the proteomics data, perhaps because the expression level of these proteins was too low to be detected under these experimental conditions, where likely only abundant proteins could be detected. Interestingly, many aquaporin proteins, which are negatively regulated by auxin (Péret et al., 2012), were significantly down-regulated in the lsg1-2 mutant. Because LR emergence depends on the spatial and temporal distribution of aquaporins, knocking out or overexpressing the aquaporin gene PIP2;1 causes delayed LR emergence (Péret et al., 2012). This particular gene was found to be down-regulated in the lsg1-2 mutant. Thus, altering the expression of these aquaporins may contribute to the delayed LR emergence observed in lsg1-2 mutants.
Previous studies have shown that several ribosome mutants display auxin-related phenotypes (Nishimura et al., 2005; Rosado et al., 2012). For example, loss of function of the ribosome protein RPL24 causes defects in gynoecium development, similar to the auxin response factor mutants ettin and monopteros (Nishimura et al., 2005). The rpl4d and rpl5a mutants showed altered vascular structure in leaves and stunted primary root growth (Rosado et al., 2012), phenotypes that have been associated with the translational control of auxin response factors by their upstream ORFs (uORFs) (Nishimura et al., 2005; Rosado et al., 2012). This study demonstrated the roles of AtLSG1-2 in ribosome biogenesis and found that the level of 10 ribosomal proteins was simultaneously decreased in the atlsg1-2 mutant. Collectively, these defects may compromise the ability of ribosomes to translate certain uORF-containing regulatory proteins. It is possible that the regulation of the auxin response by AtLSG1-2 is also through similar mechanisms by affecting the translation of uORF-containing regulatory genes.
A recent study showed that AtLSG1-2 is localized in the nucleus and cytosol in transiently transformed protoplasts (Weis et al., 2014). Here, its localization pattern was examined in stably transformed plants and it was found that, similar to yeast LSG1, both AtLSG1-2 and AtLSG1-1 are restricted to the cytoplasm (Fig. 4). Because the constructs used here complemented the lsg1-2 mutant phenotypes, the GFP-fused AtLSG1 protein is functional. The different localization patterns found in protoplasts transiently expressing the AtLSG1 proteins and transgenic plants stably expressing these proteins likely result from different expression levels of their fusion proteins. It is known that transient protoplast transformation requires a high concentration of plasmid DNA, thus the fluorescence fusion proteins are probably excessively expressed in these cells. Alternatively, differences in cellular state between isolated protoplast cells and intact cells from living plants could play a role. To more accurately identify the subcellular localization of AtLSG1 proteins in plants, one may need to use their native promoter-driven fluorescent proteins expressed in stably transformed plants or use subcellular fractionation approaches in the future.
In yeast, Drosophila, and humans, LSG1 deficiency is typically lethal. However, in Arabidopsis there are two LSG1 genes, and the loss of either AtLSG1 gene does not cause death, indicating that the two LSG1 genes have redundant functions. Although AtLSG1-1 showed low expression levels and atlsg1-1 displayed subtle phenotypes (Weis et al., 2014), increased levels of AtLSG1-1 were observed in the lsg1-2 knockout or knockdown mutants, suggesting there may exist a compensatory mechanism in these mutants. In fact, two compensatory mechanisms have been found in other organisms (Gu et al., 2003; Hanada et al., 2011). The first mechanism is associated with duplicate genes because sequence similarity is positively correlated with compensation frequency (Gu et al., 2003); and the second mechanism is associated with alternative pathways or regulatory networks. This investigation shows that AtLSG1 follows the first type of mechanism because of the high similarity of the two protein sequences. However, compensation does not completely fulfil the requirement for LSG1 in the lsg1-2 mutant because the mutant still showed mild phenotypes. The compensatory mechanism has also been experimentally demonstrated in other mutants; for example, in the ribosome protein mutant rpl4a, its paralogue RPL4D protein levels were found to accumulate and vice versa (Rosada et al., 2012), and the knockout of Rpl22 resulted in the upregulation of Rpl22-like1, the paralogue of Rpl22 in mice (O’Leary et al., 2013). This compensatory mechanism may contribute to the plasticity of adaptation of organisms.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The phenotypes of the dig6 mutant.
Fig. S2. Multiple sequence alignment of AtLSG1-2-related proteins in Arabidopsis.
Fig. S3. Lateral root density.
Fig. S4. Expression of AtLSG1-2 and AtLSG1-1 in different organs examined by qRT-PCR.
Fig. S5. Expression of AtLSG1-2 in leaf middle and margin regions.
Fig. S6. The lsg1-2 mutant displayed no visible phenotype at late development stages.
Fig. S7. 35S::YFP-AtLSG1-2 complements the phenotypes of the lsg1-2 mutant.
Table S1. Differentially regulated proteins identified by iTRAQ analysis.
Table S2. List of primers used in this study.
Acknowledgements
This study was supported by King Abdullah University of Science and Technology (KAUST) grant OCRF-2014-CRG3-62140394. We thank Yuqin Qiu for isolating the dig6 mutant and the initial genetic analysis of the mutation. We are grateful to Dr Natasha Raikhel (University of California, Riverside) for supporting the ribosome profiling assays in her lab and Dr Arlen Johnson (The University of Texas at Austin) for kindly providing the yeast AJY1171 strain and plasmids PAJ626, pRS415-GPD, and pRS425-GPD.
References
- Clough SJ, Bent AF.1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inze D, Foyer CH, Zhang HM. 2003. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. The Plant Journal 33, 543–555. [DOI] [PubMed] [Google Scholar]
- Estelle M. 1998. Polar auxin transport. New support for an old model. The Plant Cell 10, 1775–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Molecular and Cellular Biology 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66. [DOI] [PubMed] [Google Scholar]
- Hanada K, Sawada Y, Kuromori T, Klausnitzer R, Saito K, Toyoda T, Shinozaki K, Li WH, Hirai MY. 2011. Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Molecular Biology and Evolution 28, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Ni J, Cao J, Suyama KL, Patchett S, Bussiere C, Gui DY, Tang S, Kaplan DD, Fish M, Johnson AW, Scott MP. 2013. Regulation of ribosome biogenesis by Nucleostemin 3 promotes local and systemic growth in Drosophila . Genet ics 194, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson A W. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. The EMBO Journal 24, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. The Journal of Cell Biology 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CH, Hwang SM, Son YS, Heo JB, Bang WY, Suwastika IN, Shiina T, Bahk JD. 2011. Nuclear/nucleolar GTPase 2 proteins as a subfamily of YlqF/YawGGTPases function in pre-60S ribosomal subunit maturation of mono- and dicotyledonous plants. The Journal of Biological Chemistry 286, 8620–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallstrom G, Hedges J, Johnson A. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Molecular Cellular Biology 23, 4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DD, Zimmermann G, Suyama K, Meyer T, Scott MP. 2008. A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size. Genes & Development 22, 1877–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J. 2009. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods in Molecular Biology 553, 109–126. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K. 2005. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. The Plant Cell 17, 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MN, Schreiber KH, Zhang Y, et al. 2013. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genetics. 9, e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, et al. 2012. Auxin regulates aquaporin function to facilitate lateral root emergence. Nature Cell Biology 14, 991–998. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Nelson TM. 2007. Arabidopsis nucleolin affects plant development and patterning. Plant Physiology 144, 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud EG, Andrade MA, Bonneau F, Ly TB, Knop M, Scheffzek K, Pepperkok R. 2005. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biology 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV. 2012. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proceedings of the National Academy of Sciences 109, 19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gingrich DK, Deng Y, Hong Z. 2012a. A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Molecular Biology of Cell 23, 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xie B, Zhu M, Zhang Z, Hong Z. 2012b. Nucleostemin-like 1 is required for embryogenesis and leaf development in Arabidopsis. Plant Molecular Biology 78, 31–44. [DOI] [PubMed] [Google Scholar]
- Weis BL, Missbach S, Marzi J, Bohnsack MT, Schleiff E. 2014. The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana . The Plant Journal 80, 1043–1056. [DOI] [PubMed] [Google Scholar]
- Xiong L, Wang RG, Mao GH, Koczan JM. 2006. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiology 142, 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1567. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhang H, Cui P, Ding F, Wang G, Li R, Jenks MA, Lü S, Xiong L. 2014. The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in Arabidopsis. Plant Physiology 165, 1255–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.