Highlight
The phospholipase pPLAIIIα plays an important role in rice vegetative and reproductive growth, and constitutive, high expression of OspPLAIIIα suppresses cellulose production and cell elongation.
Key words: Cellulose, dwarf, gibberellin, longitudinal growth, phospholipase A, phospholipids, rice seeds.
Abstract
Patatin-related phospholipase A (pPLA) hydrolyses glycerolipids to produce fatty acids and lysoglycerolipids. The Oryza sativa genome has 21 putative pPLAs that are grouped into five subfamilies. Overexpression of OspPLAIIIα resulted in a dwarf phenotype with decreased length of rice stems, roots, leaves, seeds, panicles, and seeds, whereas OspPLAIIIα-knockout plants had longer panicles and seeds. OspPLAIIIα-overexpressing plants were less sensitive than wild-type and knockout plants to gibberellin-promoted seedling elongation. OspPLAIIIα overexpression and knockout had an opposite effect on the expression of the growth repressor SLENDER1 in the gibberellin signalling process. OspPLAIIIα-overexpressing plants had decreased mechanical strength and cellulose content, but exhibited increases in the expression of several cellulose synthase genes. These results indicate that OspPLAIIIα plays a role in rice vegetative and reproductive growth and that the constitutive, high activity of OspPLAIIIα suppresses cell elongation. The decreased gibberellin response in overexpressing plants is probably a result of the decreased ability to make cellulose for anisotropic cell expansion.
Introduction
Patatin-related phospholipase A (pPLA) is a major family of plant lipid acyl hydrolases that hydrolyse the acyl ester bond at either the sn-1 or sn-2 position of glycerolipids to produce free fatty acid and lysophospholipids. Recent studies in Arabidopsis have indicated that pPLAs are involved in various plant processes, including response to abiotic and biotic stresses, hormone signalling, growth, and development (Wang, 2004; La Camera et al., 2005; Yang et al., 2007, 2011; Rietz et al., 2010; Scherer et al., 2010; Li et al., 2011). The 10 Arabidopsis pPLAs are grouped into three subfamilies, pPLAI, pPLAII (α, β, γ, δ, and ε), and pPLAIII (α, β, γ, and δ). pPLAI has the highest homology to animal calcium-independent iPLA2 (Winstead et al., 2000; Holk et al., 2002) and is implicated in plant response to pathogen infection (Yang et al., 2007) and auxin (Effendi et al., 2014). AtpPLAIIα negatively regulates the production of oxylipins, such as jasmonates, and plays a role in cell death (La Camera et al., 2009) and drought tolerance (La Camera et al., 2005; Yang et al., 2012). AtpPLAIIβ has been implicated in plants coping with phosphate deficiency, and pPLAIIγ, δ, and ε in plant response to hormone treatment and nutrient starvation (Rietz et al., 2010).
Compared with pPLAI and pPLAIIs, pPLAIIIs lack the canonical S–D dyad esterase motif in their catalytic centres, the serine (S) residue in the GxSxG esterase box is replaced by glycine (G) and the aspartate (D) residue in the conserved DGG motif is also replaced by G (Li et al., 2011). Despite such changes, pPLAIIIs have been shown recently to have acyl-hydrolysing activity (Li et al., 2011, 2013). In Arabidopsis, an activation-tagged mutant of pPLAIIIδ, STURDY, exhibited stiff inflorescence stems, thicker leaves, shorter siliques, and round-shaped flowers (Huang et al., 2001). Similarly, overexpression of AtpPLAIIIβ resulted in shorter leaves, hypocotyls, petioles, and primary roots (Li et al., 2011). Further analysis revealed that altered AtpPLAIIIβ expression affected cellulose content (Li et al., 2011). Recent results indicate that pPLAIIIδ participates in plant response to auxin (Labusch et al., 2013), and overexpression of pPLAIIIδ also leads to improved oil content of seeds and the ratio of long chain fatty acids (Li et al., 2013).
In comparison, little is known about the function of pPLA genes in crop plants. Overexpression of an Oncidium pPLA OSAG78, closely related to Arabidopsis pPLAIIIδ, delayed Arabidopsis flowering by reducing gibberellin synthesis, as well as shortening the length of flowers, stamens, and pistils when compared with the wild type (WT) (Lin et al., 2011). The rice mutant DEP3 (DENSE and ERECT PANICLE 3), which was mapped in the OspPLAIIIδ gene, was isolated in a screen for alterations in panicle morphology (Qiao et al., 2011). Panicle architecture is an important agronomic and yield trait for cereal crops, and the rice genome contains six pPLAIII genes (α, β, γ, δ, ε, ζ). Thus, this study was undertaken to determine the function of pPLAIII genes in rice growth and development. The initial focus was on OspPLAIIIα because the function of OspPLAIIIα was unknown in rice and Arabidopsis. In addition, microarray and PCR analyses show that the transcript level of rice OspPLAIIIα increased under salt and drought conditions (Amarjeet et al., 2012). A gene knockout (KO) mutant was isolated and OspPLAIIIα-overexpressing (OE) rice plants were also produced, and the effect of the genetic manipulations of OspPLAIIIα on rice were analysed.
Materials and methods
Plant materials and growth condition
Oryza sativa spp. Japonica cultivar Dongjin was used as the WT for reverse transcription–PCR (RT–PCR) gene expression studies and for overexpression transformation. A mutant line of OspPLAIIIα (Os03g14950) was identified from a Dissociation (Ds) insertion transposon population derived from Dongjin and obtained from the Rice Division, Yeongnam Agricultural Research Institute, National Institute of Crop Science, Milyang, Korea. The insertion mutation was confirmed by PCR, and homozygous mutant plants were isolated following the selection of antibiotic resistance and PCR verification. Plants were grown in the field, greenhouse, or liquid medium. For gibberellin (GA; Sigma-Aldrich) treatment experiments, five-leaf stage WT, OspPLAIIIα-KO, and OspPLAIIIα-OE seedlings were sprayed with 500 μM GA3 for 13 d, once every 3 d, then leaves of the control and treated seedlings were sampled for observation and RNA extraction.
Vector construction and plant transformation
The gene region of OspPLAIIIα in the rice genome is 1407bp long, including two exons and one intron. The primer sequences used for overexpression vector construction are 5′-GGGGTACCCCGGGTTTTGGCAATTGGCATGG-3′ and 5′-CGCGGATCCGCGGCCCATCGCCGCCACCGCCGC-3′, including KpnI and BamHI restriction sites, respectively. The gene was amplified by PCR (95 °C for 40 s, 58 °C for 40 s, and 72 °C for 2min). The fragment was cloned into the pCAMBIA1301U vector with a Flag-tag fused to the C-terminus. The expression of OspPLAIIIα was under the control of the maize ubiquitin promoter. The construct was transformed into rice callus by Agrobacterium tumefaciens-mediated transformation (Lin and Zhang, 2005).
RNA extraction and quantitative real-time PCR
RNA was extracted from liquid nitrogen-frozen plant leaves using TransZol reagent according to the manufacturer’s instruction (TransGen; http://www.transgen.com.cn). DNase I was applied to remove DNA, and DNA-free RNA was used for the first-strand cDNA reverse transcription synthesis using a TIANscript RT Kit (Tiangen). Primer sequences for specific genes are listed Supplementary Table S1 available at JXB online. Quantitative real-time PCR was performed with the TransStart Tip Green Supermix (TransGen). Amplification of rice β-actin was used as a reference. The PCR conditions were: 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s for 55 cycles. Two rice genes, β-actin and ubiquitin 5 (UBQ5), were used as internal controls, and both of them were expressed at a similar level among WT, KO, and OE plants (Supplementary Fig. S1 at JXB online). The level of transcripts was normalized to β-actin.
Breaking force measurements
Breaking force is defined as the weight (g) needed to break the stem. For measurement of the force, a hoop tied to a container was placed in the middle of a stem. Water was added to the container until the stem broke. Multiple stems from each genotype were tested, and the results were analysed using Student’s t-test.
Protein extraction and immunoblotting
The proteins of rice leaves were extracted at 4 °C in pre-cooled buffer B [50mM Tris–HCl pH 7.5, 150mM NaCl, 50mM sucrose, 1mM phenylmethylsulphonyl fluoride (PMSF), 0.1% Triton X-100], containing one tablet/50ml of protease inhibitor mixture (Roche, Roche Applied Science, Mannheim, Germany). The homogenates were centrifuged at 10 000 g for 20min at 4 °C, and the supernatant was used for immunoblotting. Total proteins were separated by 8% SDS–PAGE and then transferred to a polyvinyldifluoridene (PVDF) membrane. The membrane was pre-blotted with 5% non-fat milk, incubated with Flag/GFP (green fluorescent protein) primary antibodies for 2h, and then incubated with the secondary IgG–alkaline phosphatase antibodies for 2h at room temperature. After washing with phosphate-buffered saline–Tween-20 (PBST) three times, the membrane was incubated with an alkaline phosphatase substrate containing NBT [30mg nitroblue tetrazolium+70% dimethylformamide (DMF) ml–1], 5-bromo-4-chloro-3-indolyl phosphate (15mg BCIP+1ml of 15mg ml–1 DMF), and Tris buffer (2.1g l–1 Tris+0.12g l–1 MgCl·H2O), (1:1:100, v/v/v) at room temperature for colour development.
Histological analysis
For microscopic observation, young leaves, roots, and leaf sheaths were fixed in ethanol:glacial acetic acid (9:1, v/v) for 12h at room temperature and then transferred to a solution containing chloral hydrate, glycerol, and H2O (8:1:2, w/v/v) with gentle shaking for 24h. Samples or sample sections were then observed under a microscope.
Plant cell wall fractionation and cellulose content measurements
Cellulose and hemicelluloses were extracted using the plant cell wall fractionation method (Peng et al., 2000). After pectin were removed from the crude cell wall fraction using ammonium oxalate [0.5% (w/v)] in a boiling water bath for 1h, the hemicelluloses was extracted from the remaining pellet with a solution composed of 4M KOH and 1.0mg ml–1 sodium borohydride at 25 °C for 1h. The residue was further extracted with acetic acid, nitric acid, and water (8:1:2, v/v) to separate the remaining hemicelluloses, heating for 1h at 100 °C, and the remaining pellet was cellulose. Anthrone–sulphuric acid was used to measure the cellulose content (Peng et al., 2000).
Lipid extraction and analysis by mass spectrometry
Lipids were extracted from fully expanded leaves and analysed by electrospray ionization–tandem mass spectrometry (ESI-MS/MS), and the levels of the molecular species phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), monogalactosyldiacylglycerol (MGDG), and digalactosyldiacylglycerol (DGDG) were determined as previously described (Welti et al., 2002) with modifications described by Xiao et al. (2010). Briefly, three replicates of fully expanded leaves at the tillering stage were excised and immediately immersed in isopropanol containing 0.01% butylated hydroxytoluene at 75 °C followed by four rounds of chloroform and methanol extraction. Extracts for each replicate were combined and washed with 1M KCl. After extraction, delipidated tissue was oven dried at 100 °C overnight and weighed. The phospholipids and galactolipids were quantified in comparison with phospholipid and galactolipid internal standards.
Results
The OspPLA gene family in rice
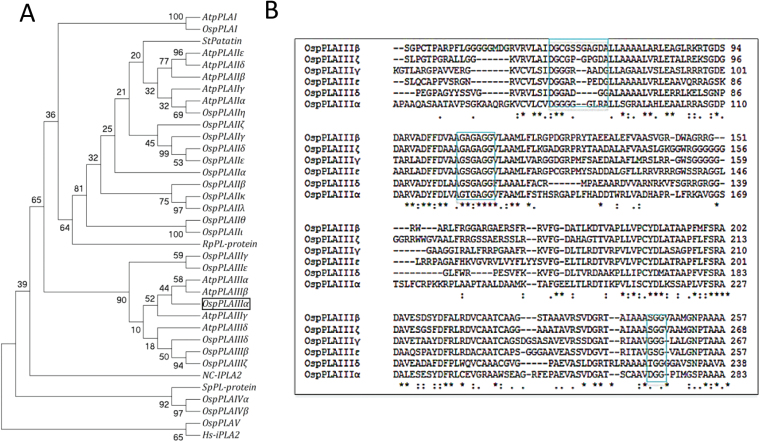
Based on the amino acid sequence similarity to the 10 Arabidopsis pPLAs and the blast result of PF01734, the ID number of potato tuber patatin protein, 21 putative pPLAs were identified and named in Oryza sativa (Fig. 1A; Supplementary Table S2 at JXB online). These OspPLAs were classified into five subfamilies according to the amino acid sequence similarity to the three AtpPLAs subfamilies, and were named as OspPLAI, OspPLAII (α, β, γ, δ, ε, ζ, η, θ, ι, κ, λ), OspPLAIII (α, β, γ, δ, ε, ζ), OspPLAIV (α, β), and OspPLAV (Fig. 1A).
Fig. 1.
Phylogenic relationship of pPLAs in Oryza sativa and other species, and sequence alignments. (A) The cladogram of putative pPLAs from Oryza sativa and their relationship with pPLAs from other species based on protein sequences was produced with MEGA4. AtPLAI, IIα, IIIα, Arabidopsis patatin-like acyl hydrolase I, IIα, and IIIα (At1g61850, At2g26560, At2g39220, Arabidopsis thaliana); StPatatin, a potato class I patatin precursor (P11768, Solanum tuberosum); Hs-iPLA2, human intracellular membrane-associated calcium-independent phospholipase A2 (EAL24384, Homo sapiens); RpPL-protein, a patatinB1 precursor from the bacterium Rickettsia prowazekii (CAA15046); SpPL-protein, a putative patatin-like protein from fission yeast (CAB16355, Schizosaccharomyces pombe); Nc-iPLA2, an iPLA2-related protein from Neurospora crassa (CAE76294); the nomenclature of OsPLA is shown in Supplementary Table S1 at JXB online. (B) Alignment of amino acid sequences of rice pPLAIII subfamilies with Clustal W2. The conserved motifs are marked with boxes; the phosphate- or anion-binding element is DGGGx(xx)G, the esterase box is GxGxG but not GxSxG, and the catalytic dyad-containing motif is GGG, SGG, TGG, TGG, or DGG.
Similar to the Arabidopsis pPLAIII subfamily, the serine (S) residue in the esterase box GxSxG in OspPLAIIIs were replaced with glycine (G). The aspartate (D) residue in the conserved DGG motif was replaced with G, Thr (T), or S (Fig. 1B). pPLAIIIα is the only member of the OspPLAIII subfamily possessing a D residue in the conserved DGG motif (Fig. 1B). OspPLAIIIα encodes a protein of 469 amino acids with the predicted pI of 6.21 and mol. wt of 49.7kDa (Fig. 1B). The function of OspPLAIIIα was analysed further because the function of OspPLAIIIα was unknown in rice or Arabidopsis.
Knockout and overexpression of OspPLAIIIα in rice
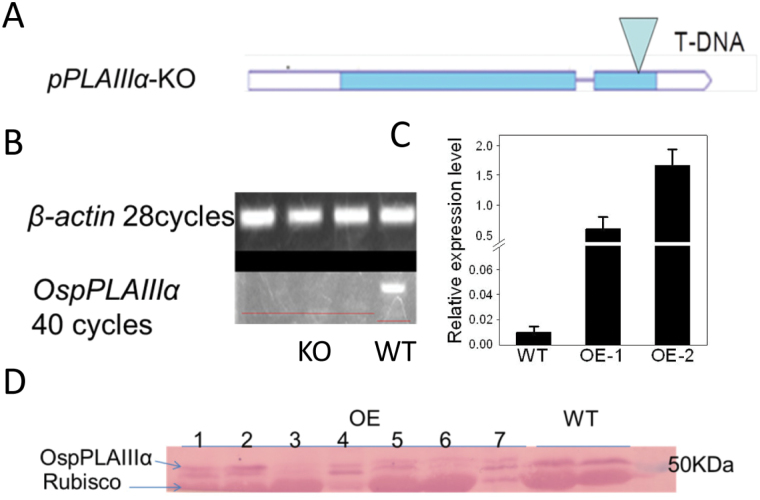
To facilitate functional studies of OspPLA genes, a homozygous mutant for this gene, OspplaIIIα-1, was identified and isolated (Fig. 2A). No transcript of full-length OspPLAIIIα was detected, indicating that the mutant is a knockout (KO) (Fig. 2B). In addition, OspPLAIIIα was overexpressed under the control of the maize ubiquitin promoter in rice. The increased expression of OspPLAIIIα in Pro ubiquitin :OspPLAIIIα (OspPLAIIIα-OE) was assessed using real-time PCR, and the transcript level of OspPLAIIIα in OE-1 and OE-2 leaves was found to be substantially higher than that of the WT (Fig. 2C). The overexpressed pPLAIIIα was fused at the C-terminus to the Flag tag. Immunoblotting using antibodies against Flag detected some non-specific bands, but the 53kDa band that was visible only in OspPLAIIIα-OE plants but not in the WT is the expected size of the product of the introduced OspPLAIIIα (Fig. 2D). The transcript and immunoblotting data together indicate that OspPLAIIIα protein is produced in the OE plants.
Fig. 2.
OspPLAIIIα T-DNA insertion mutant and overexpression in rice. (A) Schematic map showing the T-DNA insertion location (PEG_3A-11040.R) in OspPLAIIIα. (B) PCR verification of the OspPLAIIIα transcript level in the mutant. RNA was extracted from the same stage rice leaves from mutant and wild-type seedlings grown in a greenhouse. The RNA level was normalized to that of β-actin. (C) Transcript level of OspPLAIIIα in wild-type and overexpressing leaves at the tillering stage. The transcript level was measured by real-time PCR and normalized to the level of β-actin. Values are means ±SD (n=3). (D) Immunoblotting of pPLAIIIα in overexpressing and wild-type rice leaves. pPLAIIIα was fused to a Flag tag and, after SDS–PAGE separation, protein was immunoblotted with anti-Flag antibodies and visualized by alkaline phosphatase activity. The upper band was unique to OE transgenic plants.
Decreased height and cell elongation in OspPLAIIIα-OE plants
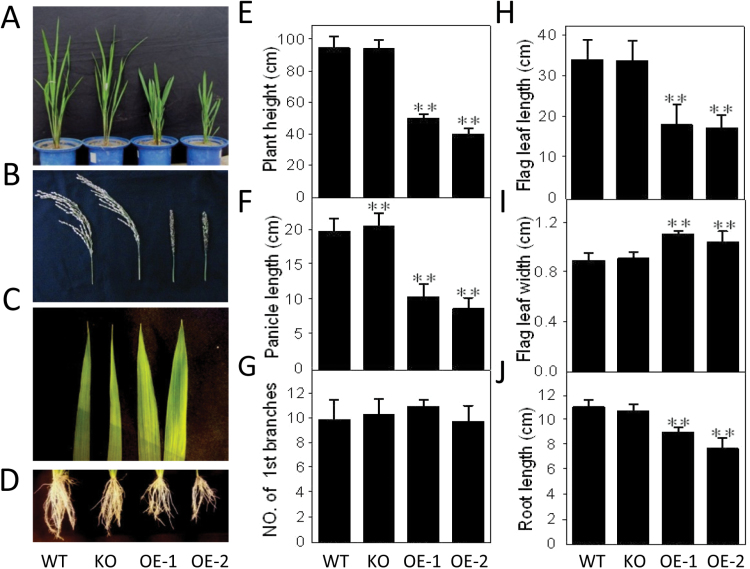
The growth of OspPLAIIIα-OE plants was stunted (Fig. 3A). Compared with the WT, the roots, stems, leaves, roots, and panicles of OE plants were shorter (Fig. 3). At the pre-flowering stage examined, the overall plant height of OE plants was ~50% and 60% shorter, respectively, for OE-1 and OE-2 (Fig. 3E), and a similar reduction in length occurred for panicles and flag leaves (Fig. 3F, H). The width of the flag leaf increased (Fig. 3I). The leaf, root, and stem length of the OspPLAIIIα-KO plant was comparable with that of the WT, but its panicles were longer (Fig. 3F). Both knockout and overexpression did not affect the number of primary tillers (Fig. 3G).
Fig. 3.
Reduced longitudinal growth in OspPLAIIIα-overexpressing plants in rice. (A–D) Morphology of seedlings, panicles, flag leaves, and roots from two independent OspPLAIIIα-overexpression lines, OE-1 and OE-2, knockout (KO), and wild type (WT) plants. Except for panicles that were collected at the maturation stage, all other tissues were sampled at the tillering stage grown in the field. (E) Plant height of wild-type, KO, and OE plants at the maturation stage. Values are means ±SD (n WT=90, n KO=90, n OE-1=4, n OE-2=4). (F and G) Length and number of first branches of the panicle. Values are means ±SD (n WT=60, n HM=60, n OE-1=28, n OE-2=28). (H and I) Flag leaf length and width at the maturation stage. Values are means ±SD (n=12) (J) Root length of primary roots of WT, KO, and OE plants. Values are means ±SD (n=12). ** indicates a significant difference compared with the wild type according to Student’s t-test, P<0.01.
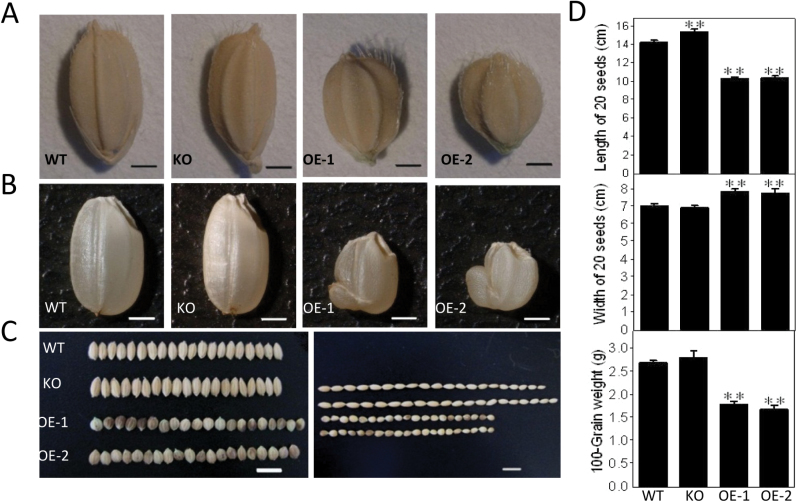
Seeds of OE lines were almost round, and were ~40% shorter and 15% wider than those of the WT (Fig. 4A, D). OspPLAIIIα-KO seeds were longer and the seed width remained the same as that of the WT. The 100-grain weight of OspPLAIIIα-OE seeds was 40% less than that of the WT (Fig. 4C, D). Endosperms of OE seeds were abnormal, with shrinkage (Fig. 4B), but OE seeds were viable.
Fig. 4.
Effect of OspPLAIIIα alterations on seeds. (A) Seed shape of WT, KO, and OE plants. (B) Seed length and width of WT, KO, and OE plants grown in the field. (C) Quantitation of length and width of 20-grain and 100-grain weight of WT, KO, and OE seeds. Values are means ±SD (n=6). ** indicates a significant difference compared with the wild type according to Student’s t-test, P<0.01.
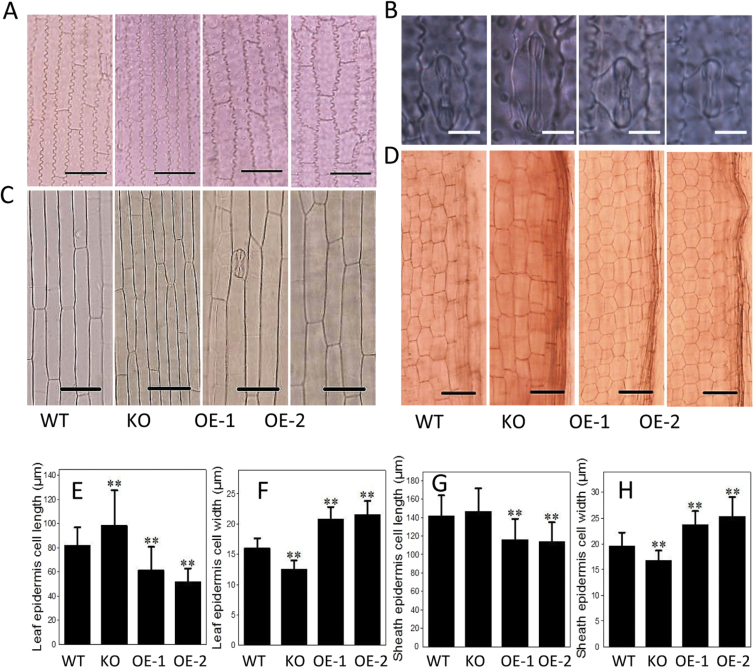
When epidermis cells and guard cells of leaves at the tillering stage were compared, OE cells were shorter and wider, whereas KO cells tended to be longer and narrower than WT cells (Fig. 5A, B, E, F). Similarly, sheath cells of OE plants at the four-leaf stage were nearly 20% shorter and 20% wider, whereas KO cells were nearly 10% longer than those of the WT (Fig. 5C, G, H). When maturation zone cells of root tips were observed and measured, the cell length of OspPLAIIIα-OE plants was shorter than that of the WT, whereas cell width was comparable between OE and WT root cells (Fig. 5D).
Fig. 5.
Cell size in roots of OspPLAIIIα-overexpressing lines and the wild type. (A) Leaf epidermis cells of WT, KO, and OE plants at the tillering stage grown in the field. Scale bar=30 μm. (B) Leaf stomata guard cells at the tillering stage. Scale bar=10 μm. (C) Sheath cells at the four-leaf stage. Scale bar=50 μm. (D) Primary root tip cells at the maturation stage. Sale bar=30 μm. (E and F) Quantitation of leaf epidermis cell length and width at the four-leaf stage. Values are means ±SD (n=40). (G and H) Sheath cell length and width. Values are means ±SD (n=50). ** indicates a significant difference compared with the wild type according to Student’s t-test, P<0.01.
Reduced mechanical strength and cellulose content in OspPLAIIIα-OE plants
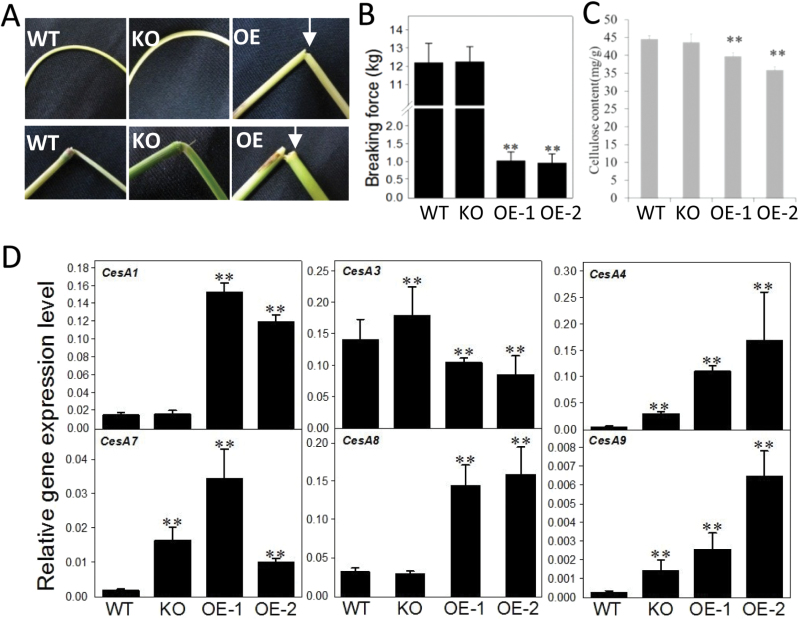
During the handling of transgenic plants, it was noticed that the leaves and stems of OE plants were brittle. When stems were bent at a 180 ° angle, OE stems were often broken but those of of the WT were flexible (Fig. 6A). The fragility was also observed for nodes of OE plants (Fig. 6B). The mechanical strength is related to the cell wall properties, such as fibre length and strength. Cellulose is an essential component of the cell wall and plays an important role in enhanced mechanical strength (Somerville, 2006). The cellulose content was 10% lower in OE-1 and 20% lower in OE-2 than in the WT, but showed no difference between WT and KO plants (Fig. 6C).
Fig. 6.
Decreased mechanical strength and cellulose content in OspPLAIIIα-overexpressing tissues. (A and B) Brittle stems and nodes of OspPLAIIIα-OE plants. (C) Breaking force of WT, KO, and OE stems. Values are means ±SD (n=20). (D) Cellulose content in mature leaves. Values are means ±SD (n=3). ** indicates a significant difference compared with the wild type according to Student’s t-test, P<0.01. (E) Transcript levels of cellulose synthase (CesA) genes of leaves at the tillering stage of plants grown in the field. The level was measured by real-time PCR and normalized to β-actin. Values are means ±SD (n=3). (This figure is available in colour at JXB online.)
To probe the effect on decreased cellulose content, the expression of cellulose synthase (CesA) genes involved in cellulose synthesis were monitored by real-time PCR using gene-specific primers (Supplementary Table S1 at JXB online). Compared with the WT, OE-1 and OE-2 seedlings had a higher mRNA level of CesA1, CesA4, CesA7, CesA8, and CesA9 but a lower level of that of CesA3 (Fig. 6E). In OspPLAIIIα-KO plants, the transcript level of CesA1 and CesA8 was comparable with that of the WT, but that of CesA3, CesA4, CesA7, and CesA9 was higher than that of the WT (Fig. 6E).
OspPLAIIIα involvement in gibberellin response
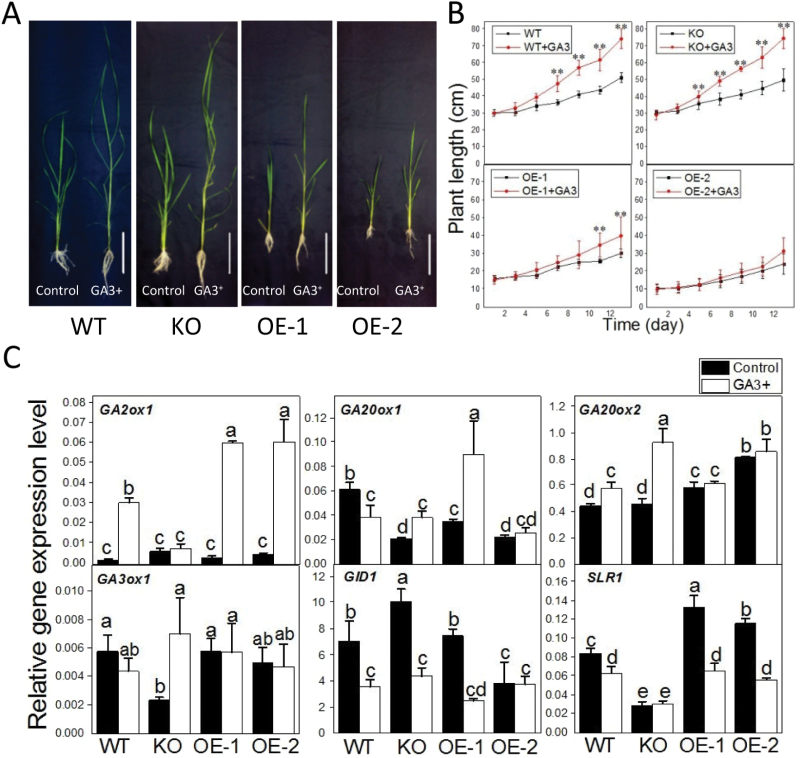
GAs are plant hormones known to promote cell elongation. WT, OspPLAIIIα-KO, and OE plants were treated with 500 μM GA3 for 13 d. The WT, KO, and OE plants all responded to GA3, and treated leaves and stems were longer and more slender than untreated controls (Fig. 7A). In the first7 d, the height of GA3-treated OE seedlings was not significantly different from that of untreated OE seedlings, but that of GA3-treated WT and KO seedlings increased nearly 30% compared with untreated seedlings. At 13 d after GA3 treatment, the height of WT and KO plants increased nearly 45% and 50%, respectively. In contrast, the height of OE-1 plants increased ~30% while that of OE-2 plants displayed little increase (Fig. 7B). These results could indicate that WT and KO seedlings are more responsive to the GA3 treatment than OE lines.
Fig. 7.
The involvement of OspPLAIIIα in gibberellin (GA) response. (A) The growth response of WT, KO, and OE seedlings grown in liquid medium without and with 500 μM GA3 for 14 d. (B) The plant height of OspPLAIIIα-alterated seedlings treated with and without GA3 for 14 d. Values are means ±SD (n=6). (C) Transcript level of genes involved in GA synthesis, degradation, and response. The level was measured by real-time PCR and normalized to β-actin. a, b, c, and d indicates different expression levels between treated and untreated plants within each genotypes according to ANOVA, P<0.05. Values are means ±SD (n=3). (This figure is available in colour at JXB online.)
To test which GA processes are affected in the OspPLAIIIα-altered plants, the expression of genes involved in GA synthesis (GA20ox1, GA20ox2, and GA3ox1), inactivation (GA2ox1), and responses [GIBBERELLIN INSENSITIVE DWARF 1 (GID1) and SLENDER 1 (SLR1)] in the WT were compared with those of OspPLAIIIα-KO and OE seedlings (Fig. 7C). Without GA treatment, the mRNA level of GA20ox1 and GA3ox1 decreased but that of GA2ox1 increased in KO plants (Fig. 7C). In contrast, overexpression of OspPLAIIIα did not result in much change in GA3ox1 whereas the level of GA20ox2 increased (Fig. 7C). OspPLAIIIα overexpression and knockout had an opposite effect on the expression of the growth repressor SLR1 in the GA signalling process. Compared with that of the WT, the mRNA level of SLR1 decreased in KO but increased in OE seedlings.
After GA3 treatment for 14 d, the mRNA level of GA2ox1 in GA synthesis increased in WT and OE plants but not in KO plants (Fig. 7C). In contrast, KO plants exhibited an increase in GA20ox2 and GA3ox1 but no such increase occurred for the WT or OE-2. Of the two genes in GA response, the RNA level of the GA receptor GID1 decreased in WT, KO, and OE-1 but not in OE-2 seedlings (Fig. 7C). After GA treatment, the expression of SLR1 was down-regulated more in OE lines than in the WT, but the level of SLR1 in the KO line was similar with or without the GA treatment (Fig. 7C).
Changes in phospholipid content and composition in OspPLAIIIα-altered plants
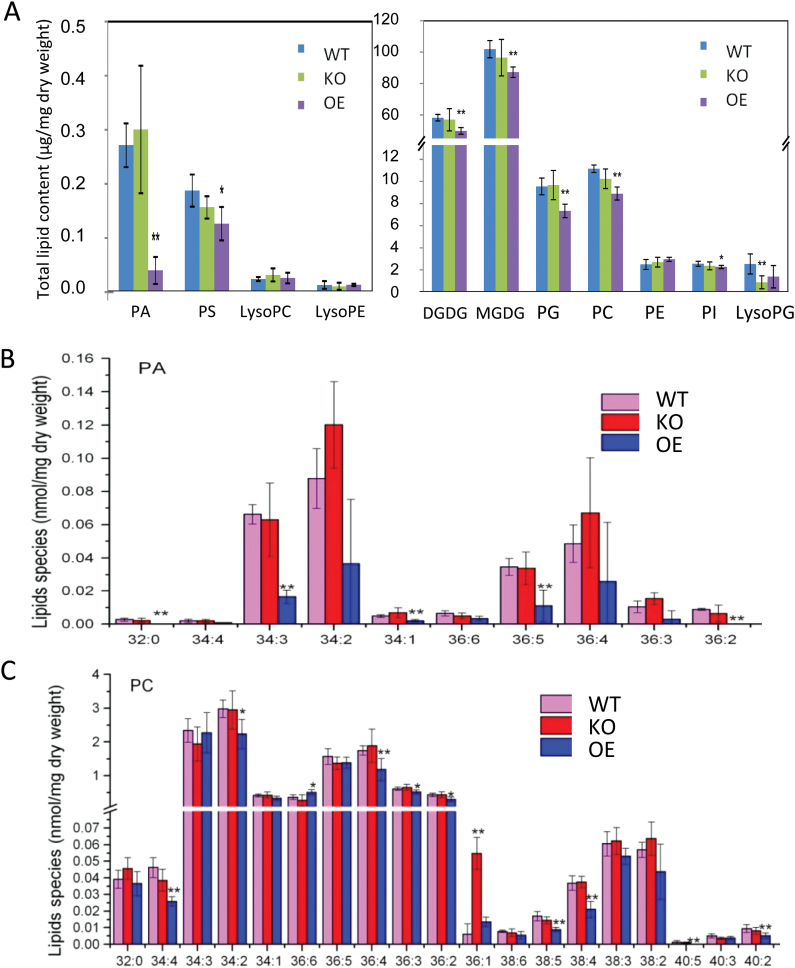
To examine the effect of OspPLAIIIα alterations on membrane lipids in rice, lipids from OspPLAIIIα-KO, OE, and WT leaves were quantitatively profiled. The content of many lipid classes in OE leaves was lower than that of the WT (Fig. 8A). In particular, the content of the minor lipid class PA in OE-1 leaves was 5-fold lower than that of WT and KO leaves (Fig. 8A, upper panel). The decrease of the major lipid classes, such as MGDG, DGDG, PC, and PG, was ~5–10% (Fig. 8A, lower panel).
Fig. 8.
The effect of OspPLAIIIα on glycerolipid content and composition.. (A) Contents of different lipid classes from fully expanded leaves at the tillering stage of plants grown in the field. Values are means ±SD (n=3). (B) PA molecular species of rice leaves. Values are means ±SD (n=3). (C) PC molecular species of rice leaves. The numbers on the x-axis in (B) and (C) denote total acyl carbons:total number of acyl carbon double bonds of PA species. * and ** indicate a significant difference between OE plants and wild-type plants with P<0.05 and P<0.01 in the Student’s t-test. Values are means ±SD (n=3).
Major PA species in rice are 34:2-PA, 34:3-PA, and 36:4-PA, and these major and many minor PA species were decreased in OE leaves (Fig. 8B). The decrease in PC was not as wide ranging as that of PA; six out of 19 PC species displayed some decline. The level of PC species in WT and KO leaves was comparable, except for 36:1-PC that was higher in KO than WT leaves (Fig. 8C). In comparison, there was no significant difference between KO and WT leaves in the membrane lipid classes examined except that lysoPG was lower in KO than in WT leaves (Fig. 8A, lower panel).
Discussion
The present analysis indicates that the pPLA family in rice is much larger than that in Arabidopsis; the rice genome has more members of OspPLAII and OspPLAIII as well as two new subfamilies (OspPLAIV and OspPLAV). Distinguishable functions have been implicated for the three subfamilies of Arabidopsis pPLAI, IIs, and IIIs, and even among the individual members of pPLAIIs and pPLAIIIs (Li et al., 2013). pPLAIII in Arabidopsis has been shown to have acyl-hydrolysing activity (Li et al., 2011). Lipid analysis indicates that knockout of OspPLAIIIα has no apparent effect on lipid composition, but overexpression led to a large decrease in PA. This raises an intriguing question of whether PA is a substrate of OspPLAIIIα in planta. PA is a central intermediate in glycerolipid metabolism and also a potent mediator affecting plant growth and stress responses (Liu et al., 2013). It will be of interest in future studies to determine whether the effect of OspPLAIIIα on PA content contributes to the cellular and phenotypic alteration of OspPLAIIIα-altered plants.
Stunted growth in OspPLAIIIα-OE plants is the most noticeable phenotype resulting from the alterations of OspPLAIIIα expression. OE plants were decreased in longitudinal growth in all tissues. In comparison, the effect of OspPLAIIIα knockout on the length of vegetative tissues was not apparent. Genetic redundancy of other PLAIII genes may explain the lack of overt growth alterations in vegetative tissues in KO plants. Indeed, the decreased longitudinal growth has been reported on AtpPLAIIIδ-OE and AtpPLAIIIβ-OE Arabidopsis and also on OspPLAIIIδ-OE Arabidopsis (Huang et al., 2001; Li et al., 2011, 2013; Lin et al., 2011). These results also indicate that the function of pPLAIII genes in decreasing elongation is evolutionarily conserved in different species.
On the other hand, the present result showed that OspPLAIIIα-KO plants had longer panicles and longer seeds. A previous study reported that mutation in OspPLAIIIδ altered panicle morphology, and that the pPLAIIIδ mutant DEP3 displayed dense and erect panicles (Qiao et al., 2011). These results indicate that OspPLAIIIα and pPLAIIIδ affect rice panicle growth and development, and yet their functions differ from one another. Panicle and seed morphologies are important yield and quality traits for cereal crops. The rice genome has six pPLAIII genes (α, β, γ, δ, ε, ζ). The availability of the mutants will help future studies of the function of the other members of the OspPLAIII subfamily in combination with OspPLAIIIα and pPLAIIIδ in rice panicle architecture and seed production.
To probe the mechanism of the decreased cell and plant elongation, the plant response to the cell elongation-promoting hormone GA was tested. Leaves and stems of GA-treated WT and KO plants were longer and more slender than those of untreated controls. OspPLAIIIα-OE seedlings responded to the GA treatment but they were much less responsive to GA than the WT and KO plants. The expression of genes involved in GA synthesis (GA20ox1, GA20ox2, and GA3ox1), inactivation (GA2ox1), and responses (GID1 and SLR1) was then compared. KO plants had decreased mRNA levels of the GA synthesis genes GA20ox1 and GA3ox1 but increased levels of the GA-inactivating gene GA2ox1 without GA treatment. These changes could be a feed-back inhibition of GA responses as epidermal cells of KO plants tended to be longer than those of the WT. In GA response, the receptor GID1 binds to GA, and GID1 interacts with DELLA proteins such as SLR1 that represses GA signalling (Hirano et al., 2012). SLR1 inhibits plant growth and this inhibition is suppressed by the GA-dependent GID1–SLR1 interaction. The expression of SLR1 was down-regulated in WT and OE lines, but not in the KO after GA treatment. Without GA treatment, OspPLAIIIα-KO plants exhibited a decrease in the mRNA level of SLR1 whereas OE plants had a higher level of SLR1 than the WT. Since SLR1 is a suppressor of cell elongation, the data are in agreement with the observation that KO plants tended to have longer epidermal cells whereas OE plants had shorter cells than the WT. After application of GA, the expression of SLR1 was down-regulated more in OE plants than in the WT, but not in KO plants. These changes in SLR1 expression in KO and OE plants could mean that SLR1 is a target of OspPLAIIIα action. The increased OspPLAIIIα expression and its protein level suppress the SLR1 transcript level in response to GA. Despite the lack of change in SLR1 level, the KO plants responded to GA-promoted elongation similarly to the WT. The lack of suppression of elongation in KO plants could mean that besides the transcript level, another mechanism, such as post-translational modification, and interaction with lipid mediators from pPLA, such as free fatty acids and lysophospholipids, are involved in the SLR1 action to suppress the GA-promoted cell elongation.
The dwarf OspPLAIIIα-OE plants also showed decreased mechanical strength. The reduced mechanical strength and plant height were linked to the decrease in cellulose content of OspPLAIIIα plants as cellulose content plays a key role in plant mechanical strength. In addition, cellulose deposition is regarded as a driving force for cell expansion through oriented deposition of cellulose microfibrils around the cell. Therefore, the change of cellulose content may influence anisotropic cell expansion and eventually exhibit a corresponding phenotype in the whole plant. Cells in OE lines were shorter than those of the WT, a probable result of a weaker driving force due to decreased cellulose, and these phenotypes were similar to those of some cellulose-deficient mutants, such as csi1 (CELLULOSE SYNTHASE INTERACTIVE PROTEIN 1) and ctl1/pom1 (Zhong et al., 2002; Gu et al., 2010). Based on the above observation, it was reasoned that pPLAIII genes regulated longitudinal growth of plants by modulating cellulose metabolism.
OspPLAIIIα-KO and WT plants displayed similar plant height and cellulose content. However, leaf epidermal cells were longer and narrower in KO than in WT plants. This discrepancy between the overall plant morphology/cellulose content and the epidermal cell length could mean that the effect of OspPLAIIIα is specific to cell types: OspPLAIIIα functions primarily in the epidermal cells, but cellulose content and plant height resulted from the measurements of whole tissue comprising various cell types. On the other hand, the overexpression of OspPLAIIIα was under the control of a constitutive ubiquitin promoter, thus resulting in a decrease in overall plant height and cellulose content.
To explore the effect of OspPLAIIIα overexpression and knockout on cellulose content, the effect of OspPLAIIIα-OE on the level of expression of several CesA genes was monitored. CesAs play important roles in cellulose synthesis, using UDP-glucose to produce β-1,4-glucan (cellulose) chain polymerization (Somerville, 2006; Cantarel et al., 2009). CesA proteins are arranged into multimeric protein complexes, and at least three different CesA proteins are required to form a functional complex (Taylor et al., 2000; Desprez et al., 2007). In Arabidopsis, CesA1 and CesA3 are required for cellulose biosynthesis during primary cell wall formation, whereas CesA4, CesA7, and CesA8 are needed for secondary cell wall formation (Taylor et al., 2000; Persson et al., 2005; Desprez et al., 2007). In OspPLAIIIα-KO plants, the mRNA level for CesA3, CesA4, CesA7, and CesA9 displayed an increase, suggesting that OspPLAIIIα-KO is a negative regulator of cell wall formation. On the other hand, in OE-1 and OE-2 plants, the mRNA level of CesA3 was decreased but those of CesA3, CesA4, CesA7, and CesA8 all showed an increase. The results appeared to be counterintuitive to the decreased cellulose content in OE plants. This could mean that the increase in CesA expression resulted from a feed-back regulation in response to decreased cellulose content.
Based on the results, it is proposed that the phenotype of decreased growth and response to GA in the overexpressors is probably a result of the decreased ability to make a cell wall, which is required for anisotropic cell expansion. The effect of OspPLAIIIα overexpression on the expression of genes in GA and cellulose synthesis results from decreased cellulose content and cell elongation. It is likely that the high activity of OspPLAIIIα and its lipid products perturbs cellulose production. To understand the role of OspPLAIII genes in plant morphology and growth, further studies are needed to determine how pPLAIIIs and associated membrane lipid changes modulate assembly and cellulose synthase activity.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Relative transcript level of UBQ5 normalized to the level of β-actin.
Table S1. Primers and sequences used for PCR.
Table S2. OspPLA family members
Acknowledgements
We thank Shaoping Lu for assistance with lipid data analysis. This work was supported by grants from the National Science Foundation of China (31470762, 31271514), the US Department of Agriculture (2007-35318-18393), and the Chinese National Key Basic Research Project (2012CB114200).
Glossary
Abbreviations:
- CesA
cellulose synthase
- DGDG
digalactosyldiacylglycerol
- GA
gibberellin
- GA2ox
GA 2-oxidase
- GA3ox
GA 3-oxidase
- GA20ox
GA 20-oxidase
- MGDG
monogalactosyldiacyglycerol
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- pPLA
patatin-related phosholipase A
- PS
phosphatidylserine.
References
- Amarjeet S, Vinay BAS, Poonam K, Rajeev R, Sandeep Y, Amita P, Sanjay K, Girdhar KP. 2012. Rice phospholipase a superfamily: organization, phylogenetic and expression analysis during abiotic stresses and development. PLoS One 7, e30947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat. 2009. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Research 37, D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. 2007. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104, 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi Y, Radatz K, Labusch C, Rietz S, Wimalasekera R, Helizon H, Zeidler M, Scherer GFE. 2014. Mutants of phospholipase A (pPLA-I) have a red light and auxin phenotype. Plant, Cell and Environment 37, 1626–1640. [DOI] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR. 2010. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proceedings of the National Academy of Sciences, USA 107, 12866–12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kouketu E, Katoh H, Aya K, Ueguchi-Tanaka M, Matsuoka M. 2012. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. The Plant Journal 71, 443–453. [DOI] [PubMed] [Google Scholar]
- Holk A, Rietz S, Zahn M, Paul RU, Quader H, Scherer GFE. 2002. Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiology 130, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Cerny RE, Bhat DS, Brown SM. 2001. Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiology 125, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labusch C, Shishova M, Effendi Y, Li M, Wang X, Scherer GFE. 2013. Patterns and timing in expression of early auxin-induced genes imply involvement of phospholipases a (pPLAs) in the regulation of auxin responses. Molecular Plant 6, 1473–1486. [DOI] [PubMed] [Google Scholar]
- La Camera S, Balagué C, Göbel C, Geoffroy P, Legrand M, Feussner I, Roby D, Heitz T. 2009. The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Molecular Plant-Microbe Interactions 22, 469–481. [DOI] [PubMed] [Google Scholar]
- La Camera S, Geoffroy P, Samaha H, Ndiaye A, Rahim G, Legrand M, Heitz T. 2005. A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. The Plant Journal 44, 810–825. [DOI] [PubMed] [Google Scholar]
- Li M, Bahn SC, Fan C, Li J, Phan T, Ortiz M, Roth MR, Welti R, Jaworski J, Wang XM. 2013. Patatin-related phospholipase pPLAIIIIδ increases seed oil content with long-chain fatty acids in Arabidopsis. Plant Physiology 162, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Bahn SC, Guo L, Musgrave W, Berg H, Welti R, Wang X. 2011. Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. The Plant Cell 23, 1107–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Chu CF, Liu PH, et al. 2011. Expression of an Oncidium gene encoding a patati-like protein delays flowering in Arabidopsis by reducing gibberellin synthesis. Plant and Cell Physiology 52, 421–435. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Liu Y, Su Y, Wang X. 2013. Phosphatidic acid-mediated signaling. Advances in Experimental Medicine and Biology 991, 159–176. [DOI] [PubMed] [Google Scholar]
- Peng L, Hocart CH, Redmond JW, Williamson RE. 2000. Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. [DOI] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. 2005. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National Academy of Sciences, USA 102, 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao YL, Piao RH, Shi JX, Lee SI, Jiang WZ, Kim BK, Lee JY, Han LZ, Ma WB, Koh HJ. 2011. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theoretical and Applied Genetics 122, 1439–1449. [DOI] [PubMed] [Google Scholar]
- Rietz S, Dermendjiev G, Oppermann E, Tafesse FG, Effendi Y, Holk A, Parker JE, Teige M, Scherer GFE. 2010. Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Molecular Plant 3, 524–538. [DOI] [PubMed] [Google Scholar]
- Scherer GFE, Ryu SB, Wang X, Matos AR, Heitz T. 2010. Patatin-related phospholipase A: nomenclature, subfamilies, and functions in plants. Trends in Plant Sciences 15, 693–700. [DOI] [PubMed] [Google Scholar]
- Somerville C. 2006. Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology 122, 53–78. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. 2000. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. The Plant Cell 12, 2529–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. 2004. Lipid signaling. Current Opinion in Plant Biology 7, 329–336. [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. 2002. Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. Journal of Biological Chemistry 277, 31994–32000 [DOI] [PubMed] [Google Scholar]
- Winstead MV, Balsinde J, Dennis EA. 2000. Calcium-independent phospholipase A2: structure and function. Biochimica et Biophysica Acta 1488, 28–39. [DOI] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML. 2010. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. The Plant Cell 22, 1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CJ, Zhang C, Lu YN, Jin JQ, Wang X. 2011. The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Molecular Plant 4, 588–600. [DOI] [PubMed] [Google Scholar]
- Yang WY, Devaiah SP, Pan XQ, Isaac G, Welti R, Wang X. 2007. AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea . Journal of Biological Chemistry 282, 18116–18128. [DOI] [PubMed] [Google Scholar]
- Yang WY, Zheng Y, Bahn SC, et al. 2012. The patatin-containing phospholipase A pPLAIIα modulates oxylipin formation and water loss in Arabidopsis thaliana . Molecular Plant 5, 452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Kays SJ, Schroeder BP, Ye ZH. 2002. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. The Plant Cell 14, 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.