Abstract
In this minireview, we discuss basic aspects of germinal center biology in the context of immunity to influenza infection, and speculate on how the simultaneous evolutionary races of virus and antibody may impact our efforts to design a universal influenza vaccine.
Introduction
Influenza epidemics cause millions of infections and hundreds of thousands of deaths worldwide each year, and cost nearly $100 billion per year in the United States alone. The influenza vaccine is generally protective against the strains from which it is composed. However, effectiveness wanes as herd immunity pushes the viral envelope proteins to mutate and evolve (antigenic drift). Periodically, more antigenically distinct or virulent influenza strains arise due to recombination among zoonotic strains (antigenic shift). These strains can cause pandemics such as the 1918 Spanish flu, which had a death toll of tens of millions of people.
The primary target of anti-influenza antibodies is the hemagglutinin (HA) protein, a trimer consisting of a membrane (envelope)-embedded stalk region and an expanded globular head on which the receptor-binding site (RBS) is located. Most protective antibodies against HA bind to regions surrounding the RBS that are highly mutable, which allows antigenic drift and immune escape. However, rare antibodies have been isolated that bind functionally critical regions of HA that are much less susceptible to antigenic drift (Krammer and Palese, 2015; Schmidt et al., 2015). These antibodies bind either within the RBS, mimicking the sialic acid ligands of HA, or to regions of the HA stalk that are critical for viral fusion to host cell membranes (Fig. 1A). A major goal of vaccinologists is to develop a universal vaccine capable of eliciting protective antibodies to epitopes that are common amongst influenza strains and that are stable over time, thus circumventing antigenic variation (Krammer and Palese, 2015).
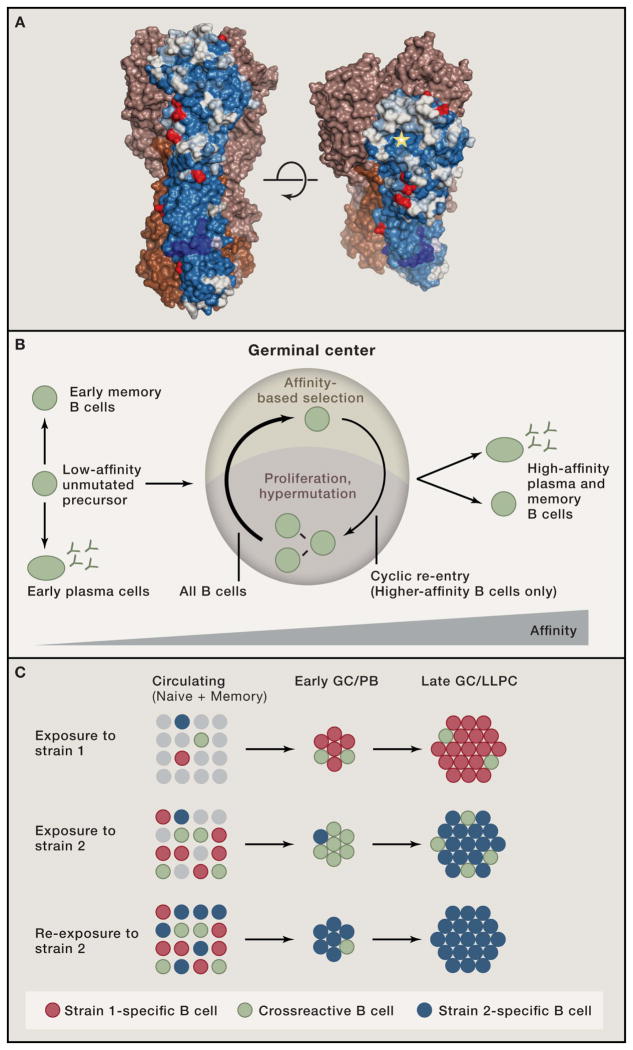
Figure 1.
(A) Residue conservation among seasonal H1 HA isolates (1975–2005). Conservation is shown for one monomer, on a scale from blue (most conserved) to white (most variable). Red residues indicate common glycosylation sites. The RBS is marked with a star. Image courtesy of Stephen C. Harrison. (B) Overview of affinity maturation in the GC. Cyclic migration of B cells between light and dark zones drives affinity maturation. Prior to GC entry and upon positive selection, B cells can differentiate into the PC or memory fates. (C) Proposed model for reestablishment of immunodominance to strain-specific epitopes. Top, exposure to Strain 1 of influenza generates a response mostly focused on immunodominant, Strain 1-specific epitopes (red), but also induces a subdominant crossreactive response (green). Middle, exposure to a divergent Strain 2 will initially reactivate cross-reactive memory cells (green) from the response to Strain 1, generating a “broad” response, but will also prime Strain 2-specific clones de novo, which will eventually outcompete the crossreactive clones. Bottom, re-exposure to Strain 2 will preferentially recall Strain 2-specific clones, reinstating immunodominance.
Antibodies attain high affinity through somatic hypermutation (SHM) of immunoglobulin (Ig) genes in B cells following exposure to antigen, in a process known as affinity maturation (Eisen, 2014). Most antibodies to influenza cloned from humans are heavily mutated, and these mutations are likely critical for broadly protective binding to the virus (Lingwood et al., 2012; Pappas et al., 2014; Schmidt et al., 2015). Affinity maturation takes place in germinal centers (GCs) (Victora and Nussenzweig, 2012), where B cells undergo SHM and are subsequently selected based on the ability of their mutant Igs to bind antigen. A fundamental constraint to this process is that GCs select for antibodies with higher affinity for antigen (or some close correlate of it), but are “agnostic” when it comes to their protective efficacy—including their ability to neutralize virus or kill infected cells and their potential to crossreact with other strains of the offending pathogen (breadth). For many infectious diseases, this “evolution by proxy” is sufficient to provide robust immunity. However, in cases like influenza, antigenic drift renders high affinity protective antibodies from one season ineffective against newly emerging strains.
GC kinetics and structure
Antigenic stimulation triggers specific B and T cells to move towards the T zone/follicle (T:B) border area of secondary lymphoid organs. There, B cells that present antigen-derived peptides to helper T cells become “authorized” to engage in a productive immune response. Successful B cells enter one of three developmental paths: they can differentiate into PC that secrete early, low-affinity antibody; they can re-establish a nonproliferative state and join the memory B cell pool; or they can enter the GC reaction (Fig. 1B) (Victora and Nussenzweig, 2012).
GCs appear several days after antigen exposure as clusters of rapidly proliferating cells in the center of B cell follicles. GCs comprise two anatomically defined areas: the dark zone (DZ), where cells proliferate and hypermutate their Ig genes, and the light zone (LZ), where antigen-driven selection takes place (Victora and Nussenzweig, 2012). Following DZ hypermutation, B cells migrate to the LZ, where antigen is deposited as immune complexes on the surface of follicular dendritic cells (FDCs). LZ B cells compete to bind and retrieve antigen from FDCs and present it to GC-resident T follicular helper (Tfh) cells. B cells that have acquired higher affinity by virtue of SHM are more likely to receive positive selection signals, triggering their return to the DZ for further proliferation and hypermutation (cyclic re-entry, Fig. 1B). GC selection is thus reminiscent of Darwinian evolution: iterative cycles of descent with modification (SHM) followed by fitness (affinity)-based selection lead to increased fitness of the population as a whole. Sporadic differentiation of positively selected LZ B cells into plasma cells (PC) and memory B cells results in the progressive increase in the affinity of serum antibodies over time and upon re-immunization (Fig. 1B).
The cues that trigger B cells to choose between cyclic re-entry and differentiation into PCs or memory B cells are unknown. High affinity for antigen appears to be a pre-requisite for PC differentiation and/or survival (Goodnow et al., 2010). However, because PC differentiation occurs after clonal expansion, diversion of part of a clone into the PC fate does not preclude further diversification of the same expanded clone in GCs. Also under scrutiny is the relative propensity of memory B cells to re-enter GCs for further diversification, rather than exclusively differentiating into secondary PC (McHeyzer-Williams et al., 2015). The ability to sequentially diversify the same clone over multiple responses is likely to be crucial to eliciting a broad response to influenza.
Selection of high-affinity mutants in the GC
Two models for how affinity-based selection operates in GCs are traditionally proposed. The first, and simplest, centers on antigen-driven signaling through the B cell receptor (BCR, comprising surface Ig, Igα, and Igβ) as the direct driver of selection. In this model, Ig with highest affinity for antigen will bind more strongly to immune complexes deposited on LZ FDCs, which triggers their return to the DZ and further proliferation (Victora and Nussenzweig, 2012). A recent development is the debate over whether BCR signaling is even active in GC B cells undergoing selection in the LZ, and the role that inhibition of BCR signaling by Fc receptors might play in this process (Espeli et al., 2012; Khalil et al., 2012).
The second model of selection proposes that, rather than competing for direct signals from antigen, GC B cells compete for limiting amounts of Tfh cell help. Here, the primary role of the BCR in selection is to trigger endocytosis: B cells acquire and present antigen in proportion to the affinity of their Ig. This maps Ig affinity—a B cell intrinsic property—onto surface peptide-MHC (pMHC) density—a feature that can be distinguished by Tfh cells. Several aspects of this model have been validated experimentally, and forcing interaction of GC B cells with Tfh is the only experimental approach so far that has been successful in triggering positive selection of GC B cells in vivo (Victora and Nussenzweig, 2012).
These two models are closely interrelated, and thus not mutually exclusive. For example, strong inhibition by Fc receptors could serve to blunt BCR signaling so that B cells rely more heavily on T cells for selection. On the other hand, signals from T cells could potentially relieve Fc-mediated repression, allowing for productive BCR signals only in selected cells.
Establishing the mechanism of GC positive selection can have important consequences to our understanding of how broadly-protective antibodies develop. A recent report by Ravetch and colleagues has shown that levels of antibodies with sialylated Fcs in trivalent influenza vaccine (TIV) recipients 7 days after vaccination predicted the affinity of the anti-HA response two weeks later (Wang et al., 2015). Vaccination of mice with sialylated (vs. non-sialylated) HA immune complexes generated antibodies capable of heterosubtypic protection in an in vivo challenge model. The authors traced this effect back to the upregulation of inhibitory Fc receptor FcγRIIB in GC B cells by sialylated Fcs, which would increase the threshold for BCR-driven selection, altering the affinity and/or specificity of the ensuing response. On the other hand, T cell priming with plasmid DNA encoding H1 prior to immunization with seasonal vaccine also increased the frequency of crossreactive antibodies directed to the HA stem (Wei et al., 2010). While the mechanistic basis for this subversion of immunodominance is not clear, a likely scenario is that increased CD4 T cell priming may have led to relaxed interclonal competition between B cells before and within GCs because T cell help became less limiting.
Clonal diversity and immunodominance in the antibody response
The naïve B cell repertoire comprises a large number of distinct V(D)J rearrangements, each expressed by only one or a few cells that proliferate to form clones upon antigenic exposure. B cells with low or even undetectable affinity for HA are capable of being recruited into GCs (Lingwood et al., 2012).
Competition between B cell clones (interclonal competition) somewhat limits the access of lower-affinity B cells to the early GC. This clonality is further restricted in mature GCs by combined competition between clones and among SHM variants of the same clone (intraclonal competition) (Eisen, 2014; Victora and Nussenzweig, 2012). Competition is thought to lead to progressive loss of clonal diversity in the responding population. Thus, only a fraction of B cells remains in the immune response for long enough to acquire the somatic mutations required to confer high affinity, leading to immunodominance. The immune system must therefore tune competition to the right level to balance affinity and diversity: if too stringent, average population affinity will increase fast, but at the expense of diversity; if too lax, diversity will remain high, but affinity will increase only slowly.
Immunodominance appears to be a key factor in preventing the emergence of broadly neutralizing influenza antibodies. Antibodies against epitopes that are conserved between different HA variants, such as the HA stem or the receptor binding site (RBS), are underrepresented when compared to antibodies to more variable regions on the HA globular head. Potential reasons for this are that antibodies that bind these conserved epitopes require particular amino acid sequence elements (Lee and Wilson, 2015; Schmidt et al., 2015), and that conserved regions represent a relatively small or inaccessible portion of the HA surface (Fig. 1A). Conversely, epitopes in the more variable regions of HA that are permissive to antigenic drift are more abundant, more accessible on the intact virion, and can be targeted in a multitude of ways. Evolutionary pressure on the virus may have led to the development of these variable but immunodominant epitopes as decoys, thus protecting conserved sites.
When exposed to a novel influenza strain for the first time, conserved epitopes are the only ones to which memory B cells exist. Thus, novel influenza strains can activate memory B cells that are crossreactive to conserved epitopes, even predominantly generating a broad response (Wrammert et al., 2011) (Ellebedy et al., 2014). However, re-exposure to a novel strain will shift the response predominantly towards antibodies on the globular head, reinstating its immunodominance (Ellebedy et al., 2014). We propose that such immunodominance is due in large part to GC (and potentially pre-GC) selection steering the antibody response away from conserved but subdominant epitopes towards more immunodominant ones (Fig. 1C). Two factors that could contribute to reestablishing immunodominance are a greater potential of epitopes on the variable portion of HA to drive affinity maturation and incomplete conservation of crossreactive epitopes between variant influenza strains. Thus, crossreactive memory B cells may have sufficient affinity to become PCs and re-enter GCs upon exposure to a novel strain, but could nonetheless be outcompeted in the course of the response by primary B cell clones undergoing de novo affinity maturation towards drifted but more immunodominant epitopes (Fig. 1C).
The importance of GC selection for immunodominance is illustrated by a recent experiment in mice. Repeated administration of a low dose of the immune-suppressant rapamycin during influenza immunization abolished the GC response, which surprisingly was followed by increased resistance to heterosubtypic challenge and a change in the HA epitopes targeted by the resulting antibodies (Keating et al., 2013). The mechanistic reasons for this shift are unclear, but are likely related to relaxed competition in the absence of a GC response. This observation suggests that immunodominance of certain regions of HA over others is not set in stone, and can potentially be overcome by optimizing vaccination strategies to skew interclonal competition.
Approaches for vaccination
Universal vaccination to influenza would require an antibody response that not only neutralizes all existent strains, but also from which no variant can escape by mutation. Epidemiological evidence suggests that responses of such type can be elicited. For example, the broadly protective responses of humans to the 2009 pandemic H1N1 strain may have caused the eradication of the previous H1N1 lineage that had infected humans for 91 years since the 1918 pandemic, but no longer circulates (Krammer and Palese, 2015). Epidemiological studies aimed at identifying individuals who are completely immune to one or more influenza subtypes may help determine what are the required features of a universally protective immune response, in a manner similar to what has been achieved in recent years by studying HIV-infected individuals (Klein et al., 2013).
A recent study by Harrison and colleagues provides a glimpse of what a universally protective response might look like (Schmidt et al., 2015). In one individual, a series of clonally unrelated antibodies were found that bind to the conserved RBS pocket from different angles. In this case, mutations in the rim of the pocket, which normally render RBS antibodies ineffective against antigenic drift, are effective in preventing neutralization by only one or a few of these antibodies, but never all of them. Thus, perhaps a “team” of neutralizing antibodies may be able perform a function that would be impossible for any single bNAb.
Several studies in the literature have suggested strategies to elicit broadly neutralizing responses (Krammer and Palese, 2015). These follow along two broad lines: the first aims at developing immunization with a variety of natural or engineered antigens designed to force the immune system to focus on crossreactive epitopes. These approaches include simultaneous or sequential immunization with different natural HA proteins, truncated (e.g., stem-only) or chimeric (e.g., conserved stem, “exotic” head) HA variants, and/or viruses of varied subtype. The rationale is to attempt to overcome immunodominance by either eliminating strain-specific immunodominant epitopes or providing a competitive advantage to B cell clones that recognize epitopes common to multiple divergent HAs. Two recent reports highlight the promise of such strategies for inducing crossreactive antibodies in multiple animal models (Impagliazzo et al., 2015; Yassine et al., 2015).
The second set of approaches relies on using standard antigens while manipulating the rules of selection in the antibody response. Some of these were discussed above (immunization with immune complexes, rapamycin treatment, and DNA priming). Strategies based on increasing Tfh help have been particularly of interest, given the great emphasis on Tfh cells as the “judges” of GC selection (Victora and Nussenzweig, 2012). A question that remains unsolved is what effect does changing Tfh numbers have on selection: while fewer Tfh cells may promote stronger competition and therefore maximize the rate of affinity maturation, more Tfh may be desirable to maximize the size, quantity, and duration of GCs, and perhaps allow for the appearance and maintenance of subdominant B cell clones. Evidence that adjuvants such as MF59 can expand the breadth of epitopes targeted by humans to include more of the conserved epitopes provides proof-of-principle evidence that manipulating the immune response can lead to increased clonal diversity (Del Giudice and Rappuoli, 2015).
Once a broadly protective response can be achieved by vaccination, a final issue that will need to be addressed is the longevity of the broadly protective response. That is, can an established broadly protective response resist challenge by immunodominant responses to drifting or non-protective epitopes eventually? Further understanding of the basic biology of recall responses and maintenance of long-lived PC will be required to address these issues.
Acknowledgments
We thank S. Harrison, M. Nussenzweig and F. Krammer for critical reading of our manuscript. G.D.V. is supported by NIH grant 5DP5OD012146. P.C.W. is supported by NIH grants 1P01AI097092-03, 2U19AI057266-11, U19AI109946, 1P01AI097092 and NIAID Center of Excellence for Influenza Research and Surveillance #HHSN272201400005C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Del Giudice G, Rappuoli R. Curr Top Microbiol Immunol. 2015;386:151–180. doi: 10.1007/82_2014_406. [DOI] [PubMed] [Google Scholar]
- Eisen HN. Cancer Immunol Res. 2014;2:381–392. doi: 10.1158/2326-6066.CIR-14-0029. [DOI] [PubMed] [Google Scholar]
- Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, et al. PNAS. 2014;111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli M, Clatworthy MR, Bokers S, Lawlor KE, Cutler AJ, Kontgen F, Lyons PA, Smith KG. The Journal of experimental medicine. 2012;209:2307–2319. doi: 10.1084/jem.20121752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- Impagliazzo A, Milder F, Kuipers H, Wagner M, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. Science. 2015 doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- Keating R, Hertz T, Wehenkel M, Harris TL, Edwards BA, McClaren JL, Brown SA, Surman S, Wilson ZS, Bradley P, et al. Nature immunology. 2013;14:1266–1276. doi: 10.1038/ni.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Cambier JC, Shlomchik MJ. Science. 2012;336:1178–1181. doi: 10.1126/science.1213368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Palese P. Nat Rev Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- Lee PS, Wilson IA. Curr Top Microbiol Immunol. 2015;386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, Nabel GJ. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams LJ, Milpied PJ, Okitsu SL, McHeyzer-Williams MG. Nature immunology. 2015;16:296–305. doi: 10.1038/ni.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. Cell. 2015;161:1026–1034. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Annual review of immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Wang TT, Maamary J, Tan GS, Bournazos S, Davis CW, Krammer F, Schlesinger SJ, Palese P, Ahmed R, Ravetch JV. Cell. 2015;162:160–169. doi: 10.1016/j.cell.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, et al. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. Nature medicine. 2015 doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]