Abstract
Class IA phosphotidylinositol-3-kinases (PI3Ks) are a family of p85/p110 heterodimeric lipid kinases that are important in regulating signaling events in B and T cells. However, their role in natural killer (NK) cells is not understood. Here, using mice that lack the regulatory p85α subunit and its alternatively spliced variants p55α/p50α (collectively termed as p85α−/−), we defined the role of PI3K in NK cell development and function. p85α−/− mice had impaired lineage commitment leading to reduced NK cellularity in the bone marrow and liver. p85α−/− NK cells showed a defective Ly49 subset specification and a decreased expression of CD43. Lack of p85α severely reduced the NK-mediated cytotoxicity against tumor cells representing ‘induced-self’ and ‘missing-self’. More importantly, NKG2D and NK1.1 receptor-mediated cytokine and chemokine generation was significantly compromised in p85α−/− NK cells. These results reveal a previously unrecognized role of p85α in the development, terminal maturation, cytokine/chemokine generation and tumor clearance of NK cells.
Keywords: NK cell, NKG2D, PI3K, p85α
Introduction
Natural killer (NK) cells are critical mediators of innate immunity that defend against many viral or bacterial infections as well as tumor growth.1 NK cells are granular, bone marrow (BM)-derived lymphocytes capable of executing ‘natural cytotoxicity’ of target cells without prior sensitizations.2 NK cells bridge innate and adaptive immune responses through the secretion of a variety of cytokines and chemokines. Effector functions of NK cells are regulated by the coordinated interaction of activating and inhibitory receptors.1 NK cells use multiple nonpolymorphic activating receptors such as NKG2D, NK1.1, FcRγIII, Ly49D, Ly49H and NCRs.3
NKG2D, is a type-II lectin expressed on all human and murine NK cells and it recognizes MIC-A/B4 and ULBP-1/2/35 in human; H60,6,7 Rae-1α/β/γ/δ/ε7 and Mult-18 in mouse. NKG2D employs Src family protein tyrosine kinases (PTKs) to initiate two distinct signaling pathways. In the first pathway, activated PTK phosphorylates DAP10 at Tyr-Ile-Asn-Met (YINM) motif, which in turn recruits PI3K-p85.9 In the second pathway, the Src family PTKs phosphorylates DAP12 in the immunoreceptor tyrosine-based activation motif, which subsequently triggers Syk and ZAP70.10–13 Lack of DAP10, DAP12 or Syk/ZAP7013 do not significantly affect the NKG2D-mediated cytotoxicity. This suggests that DAP10-PI3K-p85 and DAP12-Syk/ZAP70 pathways are redundant in mediating cytotoxicity. However, lack of DAP12 or Syk/ZAP70, but not DAP10, severely impairs NKG2D-mediated cytokine production.12 This indicates an exclusive role for the DAP12-Syk/ZAP70 pathway in cytokine generation. NK1.1 (Nkrp1c) is a unique cell marker expressed on NK and NKT cells.14 Although the activating ligands for Nkrp1c have yet to be determined, the inhibitory ligands for its related family members Nkrp1d and Nkrp1f have been defined as the Clr family of C-type lectins.15 NK1.1 physically associates with FcRγ to mediate its signal.16
The interplay between the activating and inhibitory receptors regulates NK cell functions. Murine NK inhibitory receptors such as Ly49A, Ly49C, Ly49G2 and Ly49I recognize classical major histocompatibility complex (MHC) class I molecules.17 Upon recognition, they recruit phosphatases to the immunoreceptor tyrosine-based inhibitory motif in their cytoplasmic domains.18 Thus, NK cells use a complex set of receptors and signaling pathways to achieve their intended effector functions.
Although it is well established that NKG2D/DAP10 complexes recruit p85, the specific isoform that is used and its precise downstream functions are not known. Class 1A phosphotidylinositol-3-kinases (PI3Ks) are a family of p85/p110 heterodimeric lipid kinases that generate critical second messengers downstream of T- and B-cell antigen receptors.19–21 PI3K-p85α gene gives rise to two other alternatively spliced isoforms, p55α and p50α, which also participate in p85α-mediated signaling. p85α is a ubiquitously expressed subunit, which binds to one of the p110 catalytic isoforms (p110α, p110β or p110δ).22 The p110α and p110β are also ubiquitously expressed, whereas the p110δ is predominantly expressed in hematopoietic cells.23 PI3K binds to lipid substrates in the membrane and generates PIP2 and PIP3 that interact with pleckstrin homology (PH) domain containing proteins. Akt, PDK1 and Btk are the major downstream signaling molecules of PI3K signalosomes.24–26 Most of the mice deficient of p85α, p55α and p50α die at birth.20
In this study, we investigated the role of p85α/p55α/p50α subunits in NK cells. We show that these isoforms are highly expressed in both freshly isolated and interleukin (IL)-2-cultured NK cells. p85α gene deficiency caused a number of defects in NK cells. Specifically, lack of p85α reduced the lineage commitment of BM-derived progenitors into NK cells. In the absence of p85α, percentages of Ly49A+, Ly49D+ and Ly49G2+ NK subsets were severely compromised. Further, p85α−/− NK cells fail to express normal levels of CD43, implying additional defects in the terminal maturation of NK cells. Functionally, p85α−/− NK cells exhibited reduced NKG2D-mediated cytotoxicity compared to that of wild type (WT). Cytotoxic potential of p85α−/− NK cells against RMA/S cells was also significantly reduced. Particularly, p85α−/− NK cells had severely impaired NKG2D and NK1.1-mediated cytokine and chemokine generation. These data, for the first time, provide important insights into the role of p85α/p55α/p50α in the development and functions of NK cells.
Results
Generation of p85α−/− NK cells
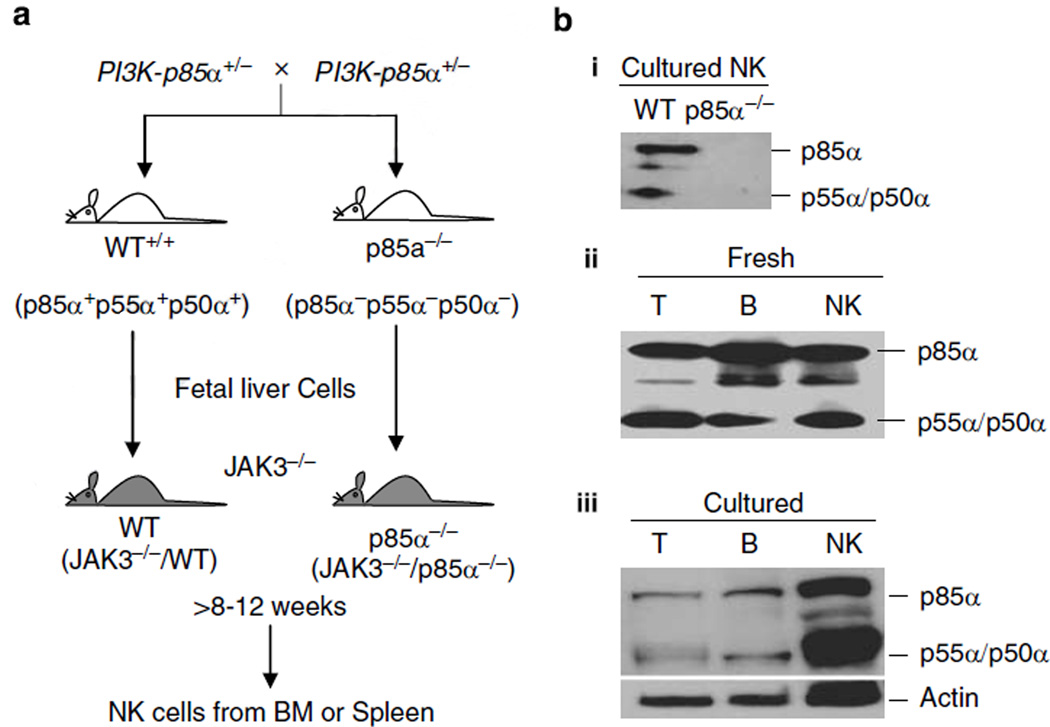
PI3K-p85α−/− mice were generated by deleting the last five exons that are common to p85α, p55α and p50α subunits, in which the last exon also includes the putative polyadenylation signal.20 Mice deficient for these three regulatory subunits exhibit a high degree of perinatal lethality.20 Therefore, we generated chimeric mice by transplanting fetal liver cells from 13- to 14-day-old p85α−/− embryos into 4- to 6-week-old, sublethally irradiated NK-null JAK3−/− mice27 (Figure 1a). JAK3−/− mice used in this study are of C57BL/6 background (H-2b). After 8–12 weeks, NK cells that were exclusively derived from donor fetal liver cells in the chimeric mice were analyzed for their development and effector functions. We examined the population of CD3−NK1.1+ NK cells derived from BM, spleen and liver of reconstituted JAK3−/− mice. For simplicity, these cell preparations from reconstituted JAK3−/− mice are denoted as WT and p85α−/−, respectively.
Figure 1.
Transfer of p85α−/− fetal liver cells into JAK3−/− recipients results in the generation of p85α−/− natural killer (NK) cells. (a) p85α−/− mice deficient for all three p85α/p55α/p50α regulatory subunits exhibit a high degree of perinatal lethality with very few animals surviving beyond 1 week of age. Therefore, we generated chimeric mice by transplanting fetal liver cells from 13- to 14-day-old p85α−/− embryos into sublethally irradiated NK-null JAK3−/− mice. Irradiated JAK3−/− recipients (300 rad) received 2 × 106 nucleated fetal liver cells from wild type (WT) or p85α−/− through retro-orbital injections. After 2 months, development and functions of NK cells generated from bone marrow, spleen and liver of the transplanted mice were examined. (b–i) p85α/p55α/p50α subunits were absent in p85α−/−, but were abundantly expressed in WT NK cells. Interleukin (IL)-2-cultured NK cells were harvested on day 6, lysed and used for western blot analyses. (b–ii) Sorted fresh T, B and NK or (b–iii) 24 h lipopolysaccharide (LPS)-stimulated T or B and IL-2-activated sixth day NK cells from WT mice express abundant p85α, p55α and p50α proteins. Blots were probed with anti-p85α antibody, stripped and reprobed with anti-actin monoclonal antibodies (mAb) as loading control.
p85α, p55α and p50α are highly expressed in NK cells
Regulatory p85α along with the catalytic p110δ subunit is important in the development and functions of T and B cells.19–21 Deletion of p85α/p55α/p50α severely impaired the functions of these immune cells. To assess its potential role in NK cells, we first examined its protein expression. NK cells were prepared and used on day 6 or 7 of IL-2 culture throughout this study. NK purity was tested with anti-CD3 and -NK1.1 monoclonal antibodies (mAbs) (Supplementary Figure 1). Cell lysates from IL-2-activated NK cells indicated the absence of all three subunits in the p85α−/− compared to the WT (Figure 1b–i). Fresh T, B and NK cells were purified through cell sorting from the spleens of WT mice. Cell lysates were analyzed by western blotting, which demonstrated that p85α and its splice variants p55α/p50α were abundantly expressed in fresh NK cells (Figure 1b–ii). Expression levels of these subunits in WT NK cells were comparable to that of T and B cells. Expression of p85α/p55α/p50α in IL-2-activated WT NK cells was readily detectable and comparable to lipopolysaccharide (LPS)-activated T and B cells (Figure 1b–iii).
NK cell number is reduced in the BM and liver of p85α−/− mice
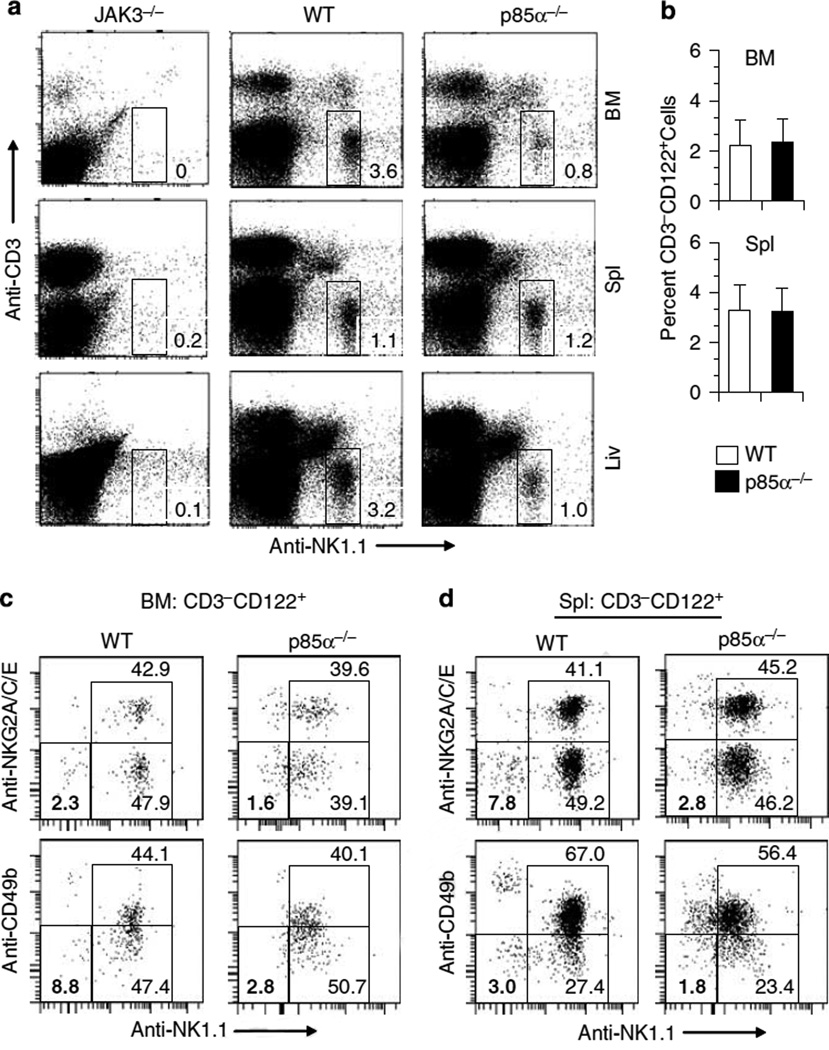
Role of p85α deficiency on NK cell development was investigated by analyzing cell-surface markers.28 The total number of lymphocytes in the BM, spleen and liver were comparable between WT and p85α−/− mice as previously reported20 (data not shown). Lymphocytes were divided into T (CD3+NK1.1−), NKT (CD3+NK1.1+) and NK (CD3−NK1.1+) cells based on their staining patterns. Nonreconstituted, JAK3−/− mice possessed both T and B cells (data not shown) that was consistent with previous studies.29,30 However, as reported earlier, the CD3−NK1.1+ NK cell number was negligible.29,30 In the reconstituted JAK3−/− mice, total number of CD3+NK1.1− T cells did not statistically differ between the p85α−/− and WT mice (data not shown). However, in the BM of p85α−/− mice, percentages of CD3−NK1.1+NK cells were significantly reduced (absolute number—WT: 0.72 × 106; p85α−/−: 0.16 × 106; P<0.05). Similarly, liver-derived NK cell numbers were reduced in the p85α−/− mice, whereas the spleen-derived NK cell number were comparable (absolute number— WT: 0.42 × 106; p85α−/−: 0.35 × 106; Figure 2a). These results demonstrate that JAK3−/− environment can support the development of NK cells from the fetal liver of p85α−/− mice. However, the commitment and maturation of NK cells in the BM are partially dependent on p85α/ p55α/p50α subunits.
Figure 2.
Early natural killer (NK) cell development in p85α−/− mice. (a) CD3−NK1.1+ NK cell number is reduced in the bone marrow (BM) and liver of p85α−/− mice. Single-cell suspensions prepared from the indicated organs of JAK3−/−, wild-type (WT) and p85α−/− mice were stained for NK1.1 and CD3ε. NK cells were gated for CD3−NK1.1 + phenotype. Numbers indicate the percent CD3−NK1.1+ NK cells in respective organs. Data shown are from one representative of 3–4 mice in each category. (b) Percentages of CD3−CD122+ cell numbers are not altered in p85α−/− compared to WT mice. BM and spleen cells were stained for CD3ε and CD122 markers and the CD3−CD122+ cell percentages were enumerated. Data presented are the averages of three independent analyses with standard deviation. (c, d) Ex vivo analyses of CD3−CD122+NK1.1−CD49b− and CD3−CD122+NK1.1−NKG2A/C/E− immature NK precursors (NKPs) in WT and p85α−/− mice. Fresh BM (c) and spleen (d) cells were exclusively gated for CD3−CD122+ cells and separated for NK1.1+ and NK1.1− populations to differentiate between immature and committed NKPs. These subpopulations were further divided based on their CD49b or NKG2A/C/E positivity Immature NKPs were defined either by CD3−CD122+NK1.1−CD49b− or CD3−CD122+NK1.1−NKG2A/C/E− phenotype and their percentages are highlighted by bold numbers.
Early commitment of HSCs into CD3−CD122+NK1.1−CD49b− immature NKPs in the BM requires p85α
Depending on the expression of different cell-surface markers, NK development in the BM can be defined into five distinct stages.31 In the first stage of NK development, commitment of hematopoietic stem cells (HSCs) into NK precursors (NKPs) occur by the expression of IL-2/IL-15 receptor β chain, CD122.32 Total percentages of CD3−CD122+, which include both NK and NKT cells were not affected in the BM and spleens of p85α−/− mice (Figure 2b). CD3−CD122+ immature NKPs are the earliest known progenitors committed to become NK cells, defined by the absence of mature markers such as NK1.1, NKG2D, NKG2A/C/E, Ly49, CD49b and CD11b.31,33 Our results show that the lack of p85α resulted in the reduction of CD3−CD122+NK1.1−CD49b− progenitors in the BM and spleen. Similar reductions were also observed in CD3−CD122+ NK1.1−NKG2A/C/E− progenitors (Figures 2c and d). On the basis of these results, we conclude that normal commitment of HSCs into immature CD3−CD122+ progenitors depends on p85α.
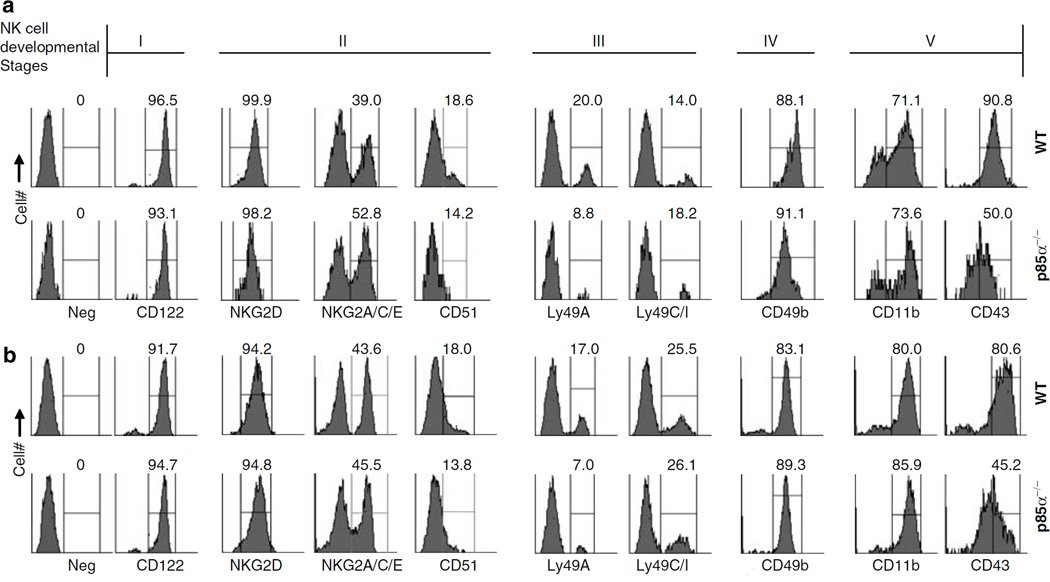
Lack of p85α reduces the total number of mature NK cells but does not affect the acquisition of NKG2D, NKG2A/C/E and CD51 receptors
Our results demonstrate that the functions of p85α are partially required for the commitment of HSCs into early CD3−CD122+NKPs. To determine the role of p85α for further NK cell development, we analyzed the expression of additional markers on fresh NK. Gated CD3−NK1.1+ cells were analyzed for the expression of CD122, NKG2D, NKG2A/C/E and CD51. Acquisition of these markers occurs at the second stage of NK development (Figures 3a and b). Our results show that the expression of these receptors between p85α−/− and WT NK cells were comparable (Figures 3a and b). On the basis of these, we conclude that there are defects in the early stage of NK development in p85α−/− mice. However, once they become CD122 positive, they can proceed through the second stage by successfully acquiring NK1.1, NKG2D, NKG2A/C/E and CD51.
Figure 3.
Acquisition of developmental markers by p85α−/− natural killer (NK) cells. (a) Fresh bone marrow (BM) and (b) spleen cells were stained ex vivo with anti-CD3ε and -NK1.1 monoclonal antibodies (mAbs). Gated CD3−NK1.1 + population were analyzed for the expression of CD122 (stage I), NKG2D, NKG2A/C/E, CD51 (stage II); Ly49A, Ly49C/I (stage III); CD49b (stage IV); CD11b, CD43 (stage V) receptors. Percent positive NK cells for each receptor are shown. Developmental stages in which the acquisition of these receptors occurs are indicated on the top. Original gates used to calculate percent positive cells for each staining are shown. Data presented were representatives of one independent experiment out of three.
Lack of p85α impairs the Ly49 subset specification and CD43 acquisition
Acquisition of Ly49 receptors by CD3−NK1.1+NKG2+ NK cells marks the third developmental stage. Expression of distinct Ly49 receptors establish a ‘repertoire’ of NK cell subsets.34,35 However, the molecular events that regulate Ly49 gene transcriptions are far from understood. Murine Ly49 receptors can be activating (Ly49D and Ly49H) or inhibiting (Ly49A, Ly49C, Ly49I and Ly49G2). Ly49 receptors recognize MHC class I molecules as their physiological ligands. Expression of one or more inhibitory Ly49 receptors is a key step by which the developing NK cells achieve full functional competency36,37 Our preliminary ex vivo analyses of fresh BM-derived NK cells revealed a significant reduction of Ly49A+ subset (44.0%) compared to WT. Similar reductions of Ly49A+ subset (41.1%) were seen in the spleen-derived fresh NK cells (Figures 3a and b).
Fourth stage of NK cell development is defined by a reduction in CD51 and an increase in CD49b expression. Results presented in Figures 3a and b indicate that the expression levels of CD51 and CD49b were comparable between the WT and p85α−/− NK cells.
In the fifth and final stage of development, NK cells increase the expression of CD11b and CD43.31 This provides the functional competency to mediate cytotoxicity and cytokine generation. Fresh BM and splenocytes were stained and CD3−NK1.1+ cells were gated and analyzed for the expression of CD11b and CD43. Our results demonstrate that the expression of CD11b was comparable between the p85α−/− and WT (Figures 3a and b). However, the expression levels of CD43 in p85α−/− NK cells were drastically reduced both in BM (MFI: 1366 ± 128 of p85α−/− compared to 5648 ± 494 in the WT) and in spleen (MFI: 9123 ± 6334 of p85α−/− compared to 18,290 ± 1051 in the WT). Reductions in CD43 expression were also found in IL-2-activated p85α−/− NK cells (data not shown).
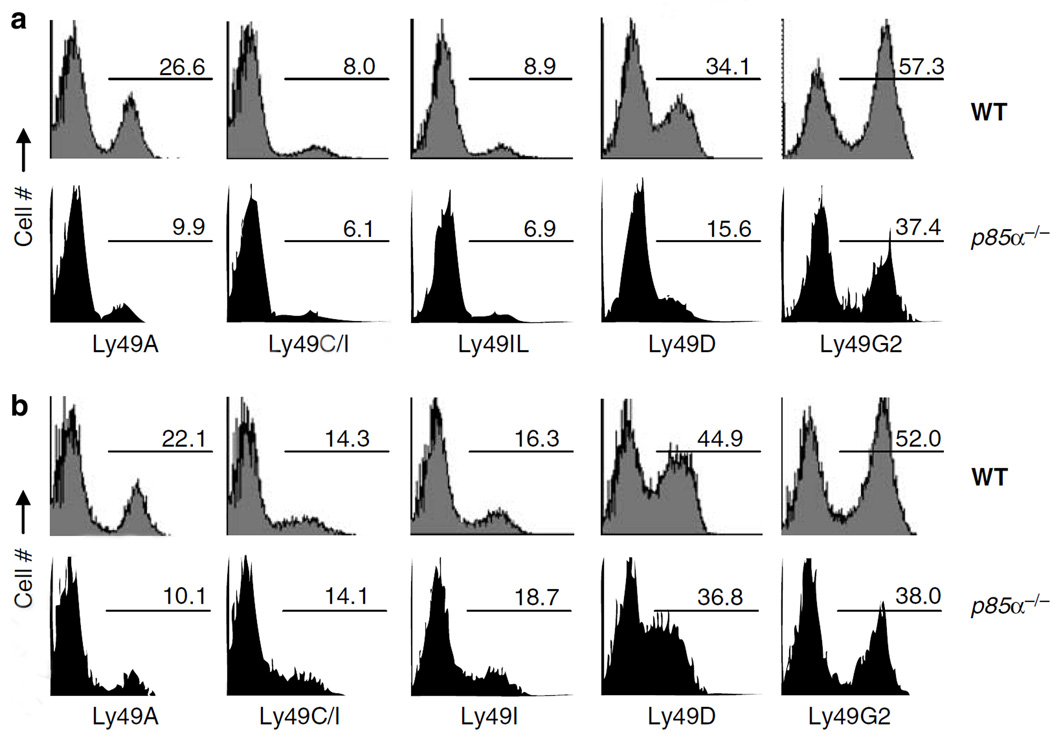
We extended our Ly49 analyses to IL-2-activated NK cells. Our results show that Ly49A-, Ly49D- and Ly49G2-expressing subsets were reduced in the BM-derived IL-2-activated NK cells from p85α−/− mice (64, 55 and 35%, respectively; Figure 4a). IL-2-activated NK cells from p85α−/− spleen also showed comparable reductions (Figure 4b). However, both BM- and spleen-derived NK cells contained normal numbers of Ly49C+ or Ly49I + NK subsets. It is interesting to note that Ly49A, Ly49D and Ly49G2 receptors recognize H-2Dd as their ligand. Collectively, we conclude that p85α is important in subset development and terminal maturation of NK cells.
Figure 4.
Lack of p85α impairs Ly49 subset specification of natural killer (NK) cells. Expression of Ly49 markers on p85α−/− NK cells were analyzed by flow cytometry. (a) Bone marrow (BM)- and (b) spleen-derived, interleukin (IL)-2-activated NK cells from wild-type (WT) and p85α−/− mice were stained for NK1.1 and CD3ε (CD3−NK1.1+). NK preparations from sixth day of IL-2-culture were used for this analysis. The CD3−NK1.1+ NK cells were analyzed for Ly49A, Ly49C/I, Ly49I, Ly49D and Ly49G2 expression. Data shown are percent Ly49+ subsets. One representative set of flow data are shown from a total of three independent experiments.
NKG2D-mediated recognition of ‘induced-self’ is impaired in p85α−/− NK cells
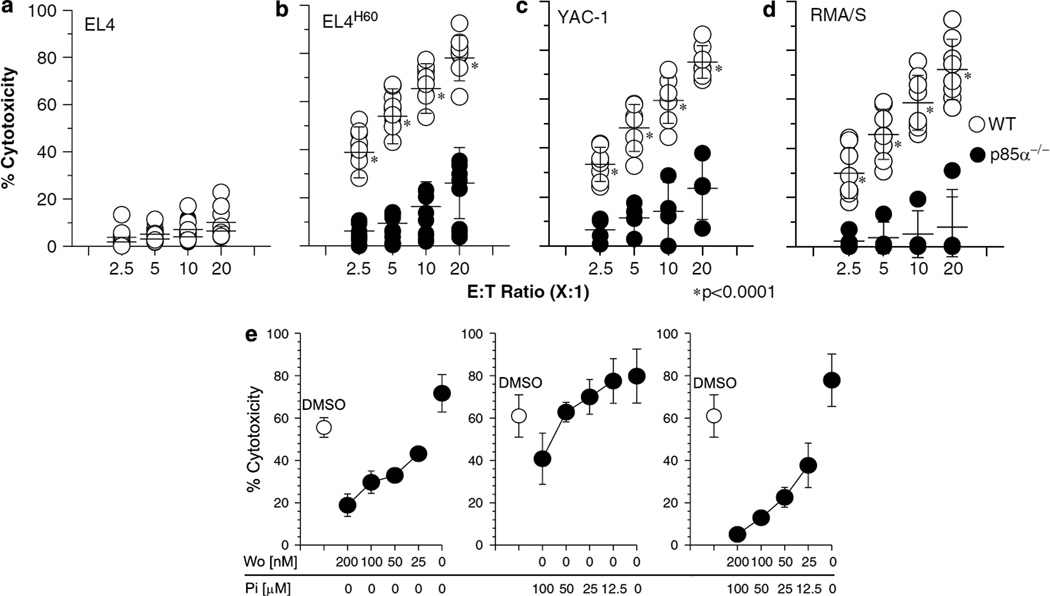
Expression of ligands for NKG2D receptor are induced by cellular stress.38 Recognition of these ligands by NK cells leads to target cell lysis. Upon ligand binding, NKG2D/DAP10 complex recruits the p85 subunit.9 To determine its specific role, we evaluated the ability of p85α−/− NK cells to mediate cytotoxicity. We used EL4 cell lines stably expressing H60 (EL4H60) as a model of induced-self. IL-2-activated p85α−/− NK cells were significantly impaired in their ability to lyze EL4H60 compared to WT NK cells (a reduction of 82% at 20:1 E:T Ratio; Figures 5a and b). Another tumor cell, YAC-1 that naturally expresses H60 and Rae-1 ligands was also lyzed significantly less by the p85α−/− NK cells (Figure 5c; a reduction of 53% at 20:1 E:T Ratio). These results demonstrate that the NKG2D-mediated recognition of induced-self is dependent on p85α-mediated signaling events. Similar observations were made using BM-derived NK cells (Supplementary Figure 2). These results further imply that a complete compensatory effect from the NKG2D/DAP12 complexes did not occur in p85α−/− NK cells and that PI3K may be playing a more significant role than predicted.
Figure 5.
Induced-self and missing-self recognitions of natural killer (NK) cells are significantly impaired in the absence of p85α. Interleukin (IL)-2-activated NK cells from sixth or seventh day of culture were tested against 51Cr-labeled (a) EL4, (b) EL4H60, (c) YAC-1 or (d) RMA/S cells at indicated E:T Ratios. In total 6–8 mice were used for each category. Open and closed circles represent the NK cells from individual wild-type (WT) and p85α−/− mice, respectively. (e) Inhibition of WT NK cells with pharmacological agents recapitulates the functional impairment of p85α−/− NK cells. WT NK cells were treated with varying concentrations of inhibitors for phosphotidylinositol-3-kinase (PI3K) (Wortmannin), Syk (Piceatannol) or both. Data shown are the average percent cytotoxicity of three independent assays.
Recognition of ‘missing-self’ is severely impaired in p85α−/− NK cells
Cells that lack or have reduced expression of self MHC class I molecules are susceptible to NK-mediated cytotoxicity.39 The lack of engagement by the inhibitory Ly49 receptors to respective MHC class I relieves the NK cells from inhibition. However, the signaling events that positively regulate this cytotoxicity are not fully understood. To test whether p85α is involved in missing-self recognition, we measured the cytotoxic potential of IL-2-activated NK cells against RMA/S tumor cell. Figure 5d demonstrates that the ability of p85α−/− NK cells were about eightfold reduced compared to the WT in mediating this cytotoxicity (a reduction of 73% at 20:1 E:T Ratio). Thus, we conclude that although missing-self recognition does not require activation by NKG2D/ DAP10 complexes, p85α is required for this effector function. The exact molecular mechanism and the receptor–ligand interactions that utilize p85α in this process need to be investigated.
To further confirm the role of p85α in NK-mediated cytotoxicity, we used pharmacological agents Wortmannin and Piceatannol, which inhibit PI3K and Syk, respectively. These inhibitors were chosen because NKG2D/DAP10/PI3K pathway is primarily thought to be responsible for cytotoxicity, whereas NKG2D/ DAP12/Syk pathway can mediate both cytokine gene transcription and cytotoxicity. Wortmannin is known to block all isoforms of PI3K.40 Earlier studies have shown that the cytotoxicity mediated by NKG2D/DAP10/PI3K pathway can be blocked by Wortmannin.41,42 IL-2- cultured WT NK cells were preincubated with varying concentrations of inhibitors, washed and used in cytotoxicity. Treatment with Wortmannin resulted in a significant reduction in the ability of NK cells to mediate cytotoxicity against EL4H60 in a dose-dependent manner (Figure 5e). However, only at the highest concentration (100 µm), Piceatannol could inhibit the lysis of EL4H60. Wortmannin and Piceatannol together inhibited the cytotoxicity in a dose-dependent manner, similar to that of Wortmannin alone (Figure 5e). Thus, our observations with p85α−/− NK cells and Wortmannin treatment of WT NK cells demonstrate that p85α is important in regulating NK-mediated cytotoxicity.
Lack of p85α significantly impairs NK-mediated cytokine and chemokine generation
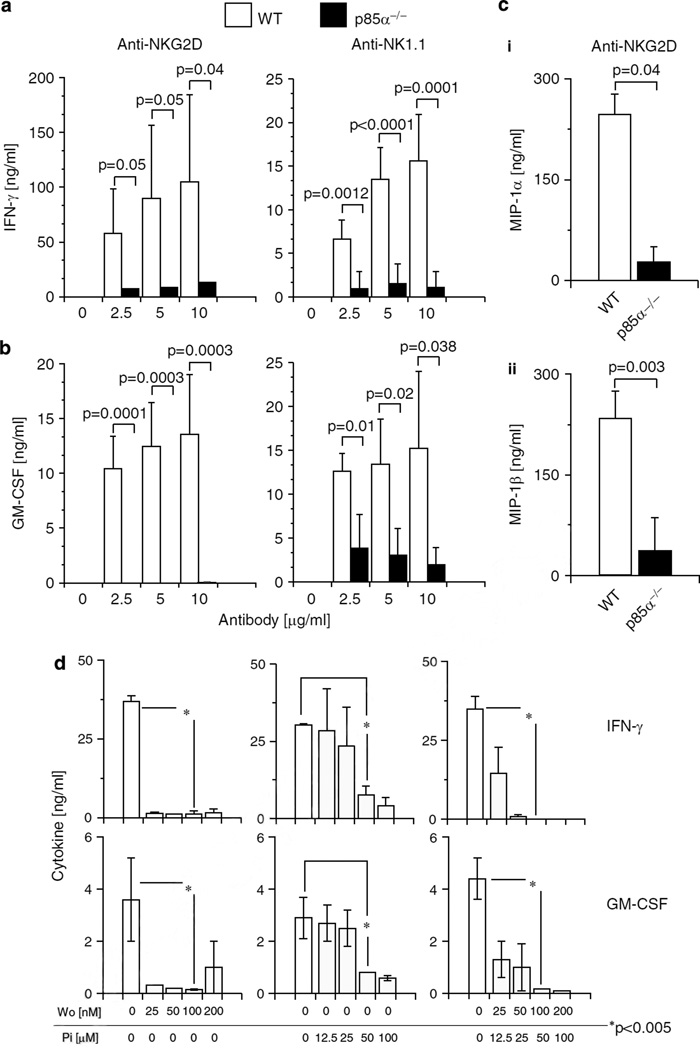
Signaling events that initiate the gene transcription of inflammatory cytokines and chemokines in NK cells are not well understood. Therefore, we analyzed the role of p85α in the generation of cytokines and chemokines. IL-2-cultured NK cells on day 6 were treated with titrated concentrations of plate-bound anti-NKG2D (A10) or -NK1.1 (PK136) mAbs. Supernatants were collected and the levels of interferon (IFN)-γ and granulocyte-macrophage colony-stimulating factor (GM-CSF) were measured by enzyme-linked immunosorbent assay (ELISA). WT NK cells produced significant amounts of IFN-γ and GM-CSF, in a dose-dependent manner. In contrast, p85α−/− NK cells were significantly impaired in their ability to produce IFN-γ and GM-CSF (Figures 6a and b). Further, generation of chemokines such as MIP-1α and MIP-1β were also significantly reduced (Figure 6c–i and –ii). On the basis of these observations, we conclude that p85α is a critical regulator for signaling events that are initiated by NKG2D/DAP10 or NK1.1 complexes for the generation of cytokines and chemokines. Stimulation of NK cells with pharmacological agents, phorphol myristate acetate (PMA) and ionomycin (Iono) can bypass the requirement of PLCγ2 or its second messengers, DAG and IP3 28 However, activation of p85α−/− NK cells with PMA and Iono, either individually or in combinations did not rescue the generation of IFN-γ (Supplementary Figure 3). This indicates that further studies are required to determine the additional defects in p85α−/− NK cells. One possibility is that rather than downstream of PI3K, PMA (or protein kinase C, PKC) could uniquely function upstream for the activation of Src family kinases that are needed to phosphorylate p85α.43
Figure 6.
Lack of p85α significantly impairs the ability of natural killer (NK) cells to generate cytokines or chemokines. Generation of (a) interferon (IFN)-γ and (b) granulocyte-macrophage colony-stimulating factor (GM-CSF) in interleukin (IL)-2-activated NK cells from p85α−/− and wild-type (WT) mice in response to anti-NKG2D or -NK1.1 monoclonal antibody (mAb)-mediated activations. Open and filled histograms represent WT and p85α−/− NK cells, respectively. (c) Generation of chemokines MIP-1α and MIP-1β in response to anti-NKG2D mAb from IL-2-activated NK cells. NK cells from sixth or seventh day of culture were used and the data presented are an average of 3–6 mice in each group. (d) Blocking phosphotidylinositol-3-kinase (PI3K) function in WT NK cells with pharmacological agents abrogate cytokine generation. WT NK cells were treated with varying concentrations of inhibitors for PI3K (Wortmannin), Syk (Piceatannol) or both. Data presented are representative of three independent experiments.
To assess whether cell death was involved, we stimulated the NK cells with anti-NKG2D mAb for 16 h and analyzed the viability with anti-Annexin-V mAb and a fluorescent DNA stain, 7-amino actinomycin-D (7-AAD). We did not observe any significant differences in the staining of either Annexin-V or 7-AAD between activated WT and p85α−/− NK cells (data not shown). Therefore, we conclude that the activation-induced cell death is not a probable cause for the lower level of cytokine generation in p85α−/− NK cells. Next, we analyzed the ability of the inhibitors in cytokine generation. Wortmannin blocked the generation of both IFN-γ and GM-CSF (Figure 6d). Even at a lower concentration, the level of inhibition on cytokine generation was significantly high (P<0.005). Conversely, only at a higher concentration (50 µm), Piceatannol inhibited the cytokine generation. Both the inhibitors together significantly reduced the generation of IFN-γ and GM-CSF (P< 0.005; Figure 6d). Analyses of NK cells after inhibitor treatment did not reveal significant cell death, indicating that any toxic effects of these drugs did not account for our observations (Supplementary Figure 4). Taken together, our results demonstrate that p85α plays a critical and nonredundant role in cytokine and chemokine generation.
Discussion
In this study, we show p85α is important in regulating NK-mediated effector functions. Alternative splicing of p85α messages generate p55α and p50α proteins that also participate in p85α-mediated signaling. The essential tasks of p85α in T- and B-cell development and function have been extensively characterized.19–21 Using p85α−/− mice, we demonstrate that p85α, p55α and p50α are crucial for NK cell lineage commitment, terminal maturation, cytokine/chemokine generation and cytotoxicity.
In recent years, multiple p85 and p110 gene knockout mice have been generated to understand the functions of PI3K complexes.20,44–46 Compared to p85α−/− mice used in our study, a different p85α-deficient mice was generated by disrupting the first exon. This resulted in the exclusive deletion of p85α but not p55α and p50α proteins, which helped the mice to survive.21 Moreover, NK cell development in these p85α-only deficient mice were unaffected.21 Thus, the presence of p55α and p50α splice variants can rescue multiple cellular functions in the absence of p85α subunit. The mice used in the present study were generated by removing the last five exons of the p85α gene, resulting in the loss of p85α, p55α and p50α isoforms.20 As these mice deficient for all three regulatory subunits die perinatally,20 we generated chimeric mice by transplanting fetal liver cells from 14-day-old p85α−/− embryos into sublethally irradiated NK-null JAK3−/− mice.27
Class 1A PI3Ks are heterodimeric proteins made up of regulatory and catalytic subunits. Five regulatory class 1A subunits have been defined and they are p85α, p55α, p50α, p85β and p55γ.23 Each of these regulatory subunits has the ability to associate with any of the three catalytic subunits, p110α, p110β and p110δ.23 Upon receptor-mediated activation, p85/p110 complexes are recruited to ‘YINM’ motif containing proteins such as DAP10, CD28, CD19, BCAP, Gab and insulin receptor substrate-IRS.47 Membrane-recruited PI3K complexes phosphorylate PIP2 at D3 position to generate PIP3. A multitude of proteins with PH domains are activated by PIP3, thereby regulating several critical signaling pathways.
Our results demonstrate WT NK cells abundantly express p85α, p55α and p50α and as expected, p85α−/− NK cells lacked all three subunits. One of the major defects due to the lack of p85α is a severe reduction in the NK cell number in the BM and liver. Multiple defects at distinct developmental stages of NK cells can account for this reduction. First, we analyzed the status of early progenitors that are generated from HSCs. In general, the development of NK cells in the BM is divided into five distinct stages.31 Early lymphoid precursors (ELPs) that give rise to T, B and NK cells have been found in the BM.48 First stage of NK cell development is defined by generation of NKPs from multipotent HSCs and ELPs. NKPs exclusively mature into NK cells and their presence in the adult BM and fetal thymus have been confirmed.32 This earliest developmental stage of NKPs is defined by the expression of IL-2/IL-15 receptor-β chain, CD122.32 Both IL-2 and IL-15 cytokines are important in the development and maturation of NK cells.49 Also, expression of CD122 by early progenitors uniquely marks the committed NKPs.33 Both the initiation of CD122 expression and its level in p85α−/− NK cells were not affected, indicating that p85α is dispensable for this developmental step. Also, percentages of CD3−CD122+ cells were not altered in p85α−/− mice. However, it is important to note that the total CD3−CD122+ cells include both NK and NKT cells, consisting of different maturation status. Therefore, to determine developmental defects in the early progenitors, we analyzed one specific subset of CD3−CD122+ cells that have yet to acquire NK1.1, NKG2A/C/E or CD49b. Our results showed the CD3−CD122+NK1.1−CD49b− progenitors were considerably reduced in the BM of p85α−/− mice. We conclude that one of the major reasons for the reduction in the mature NK cell number in p85α−/− mice is due to a decrease in the commitment of early progenitors.
Mechanistically, as the PI3K/Akt pathway is important in cell survival,50 lack of p85α could have rendered the NKPs more susceptible to proapoptotic cell death. As well, a partial blockade of HSCs committing to become NKPs cannot be ruled out. In addition, recent studies have shown that activation and proliferation of certain CD8+ T cells by IL-15 through CD122 require PI3K/Akt pathway51 Analogous to this, lack of p85α could have resulted in a reduced proliferative rate of CD3−CD122+NK1.1−CD49b− progenitors. Present observations with p85α−/− mice are distinct compared to our previous studies using PLCγ2−/− mice, where we saw a twofold increase in the numbers of CD3−CD122+NK1.1−CD49b− progenitors.28 This indicates that p85α and PLCγ2 may be involved in the commitment of early NK progenitors.
In the second stage of development, CD122+ NKPs acquire NKG2D, NK1.1 NKG2A/C/E and CD51 in a sequential developmental process.31 Our results show that NK cells from both p85α−/− and WT expressed comparable levels of these receptors. Therefore, we conclude that the p85α does not play any obligatory role at this stage of NK development.
Ly49 subset specification occurs in the third developmental stage. Here, the ‘immature’ NKPs start acquiring different Ly49 receptors, whose expression determines the specific repertoire of NK subsets.32 Analyses of Ly49 expression on p85α−/− NK cells indicated an exclusive reduction in Ly49A, Ly49G2 and Ly49D subsets. Other NK subsets such as Ly49C and Ly49I were unchanged. These reductions can be either due to an early NK cell developmental defect, owing to the absence of p85α or because of a missing regulation of Ly49 expressions by functional PI3K complexes. The p85α−/− mice used in this study was backcrossed to C57BL/6 for seven generations and are of H-2b haplotype, thereby excluding any strain background effects on these observations. Our previous studies using PLCγ2−/− mice demonstrated a significant reduction in all the Ly49+ NK subsets, irrespective of their MHC class I ligand preferences.28 This observation along with our present study indicates that both PLCγ2 and p85α are important for subset specifications. Both PLCγ2 and PI3K are known to regulate a number of transcription factors. However, their role in regulating transcription factors such as T-bet,52 GATA-3,53 IFN-regulatory factor-1,54 IRF-2,55 ID2,56 Ets-1,57 MEF58 and PU-159 that are involved in NK cell development and maturation has yet to be elucidated. Indeed, earlier studies from Di Santo laboratory have shown that lack of GATA-3 reduces the levels of Ly49A, Ly49C/I and Ly49D NK subsets, whereas Ly49G2 and NKG2A/CD94 were unaffected.53 Thus, a lack of transcription factors such as GATA-3 could partially explain the reduction in Ly49 subsets in p85α−/− mice.
Also, PI3K may regulate the expression of these Ly49 receptors after subset specification. ITIM motifs that are located in the cytoplasmic domains of inhibitory Ly49 receptors recruit protein phosphatases such as SHP1, SHP2.60 These phosphatases regulate the activation pathways by dephosphorylating substrates and are expected to be part of the activation complexes. Therefore, a reduction in Ly49 levels may indicate that p85α is a key player in the dynamic interaction between DAP10/ PI3K complexes, its second messengers and inhibitory Ly49 receptors bound to phosphatases. Evidence for such a relationship has been recently documented in NKT cells. Gene deletion of lipid phosphatase, PTEN, whose major substrate is PIP3, leads to an increased Ly49C and Ly49I expression in NKT cells.61 Interestingly, this abnormal increase can be reversed by backcrossing the PTEN−/− mice with p110δD910A/D910A mice.61 Thus, it is possible that PI3K regulates the Ly49 expression to provide a signaling balance and to prevent activation of NK against ‘self’ targets.
Fourth stage of NK cell development is defined by a concomitant reduction in CD51, an increase in CD49b expression and an augmented proliferation.31 Lack of p85α did not alter these expected phenotypic changes.
In the fifth and final developmental stage, NK cells increase the levels of CD11b and CD43.31,32 This terminal maturation is an essential step for the NK cells to achieve full functional competency. NK cells from p85α−/− mice expressed significantly lower levels of CD43 compared to that of WT, whereas the expression of CD11b was unaffected. CD43 is a highly glycosylated, transmembrane mucin expressed in all hematopoietic cells except resting B cells and erythrocytes.62 Galectin-1 is one of the natural ligands for CD43.63 Why is the expression level of CD43 affected in p85α−/− NK cells? Recent studies show CD43 contains a putative SH3 domain-binding site and through this it can interact with Fyn kinase.64 Fyn is also one of the major Src family members that phosphorylates and activates p85α. A synthetic peptide derived from the SH3 domain-binding site of p85α completely blocked the interaction between CD43 and Fyn, indicating that both CD43 and p85α compete for Fyn.64 These studies indicate that CD43 can regulate receptor-mediated activation by direct physical inhibition of p85α. On the basis of these, we predict the existence of a counterbalance that regulates the expression and function of CD43 by p85α. A similar CD43 reduction in T-bet−/− splenocytes52 provides another molecular explanation, where T-bet could be one of the target transcription factors downstream of p85α. Collectively, our observations set the stage for a detailed analysis that would determine the exact cause for the CD43 reduction.
Natural cytotoxicity mediated by NK cells can kill tumor cells without prior sensitization. Significant reduction observed in p85α−/− NK recognition of induced-self and missing-self demonstrate p85α is important in NK cytotoxicity. Murine NKG2D associates with DAP10 or DAP12. DAP10 contains a YINM motif, through which p85α is recruited.10 DAP12 recruits Syk/ ZAP70 and activates PLCγ2.12 However, the role of p85α has not been elucidated in any of these pathways. In the absence of DAP10, the NKG2D/DAP12 pathway has been suggested to compensate.65 However, we found a significant reduction in the overall ability of p85α−/− NK cells to lyze target cells, indicating that NKG2D/DAP12 does not fully compensate for the impaired NKG2D/ DAP10 pathway. Thus, we conclude that NKG2D/ DAP12 pathway may also partially depend on PI3K to mediate cytotoxicity. Support for this comes from earlier studies, where Syk has been demonstrated to activate and regulate PI3K-mediated degranulations.42 Although missing-self recognition does not utilize NKG2D/DAP10 complexes, our results demonstrate that p85α is required for the cytotoxicity against RMA/S cells. Further studies are needed to distinguish between ‘impaired signaling’ and ‘hyporesponsiveness’ of NK cells.
NK cells bridge innate and adaptive immune responses through the secretion of a variety of cytokines and chemokines.66 Previous studies demonstrated that the lack of DAP12, or Syk/ZAP70 but not DAP10, severely impaired NKG2D-mediated cytokine production.13,65,67 In contrast, a significant reduction in cytokine and chemokine generation was observed in p85α−/− NK cells, when activated through NKG2D. Similar reductions were also observed when p85α−/− NK cells were activated by NK1.1 receptor that signals through FcRγ.16 Our earlier studies have shown that combinations of PMA and Iono were required for the generation of IFN-γ from NK cells.28 However, this combination failed to rescue IFN-γ generation in p85α−/− NK cells. One possible explanation for this observation is the existence of potential requirement of Src kinases that are activated by PMA. In fact, earlier studies have shown that PMA was unable to induce colocalization or activation of cSrc in cells that lacked the p85α and -β.43 Thus, additional defects in p85α−/− NK cells may account for their failure to generate IFN-γ. Together, we conclude that p85α is important in the cytokine generation by NK cells. Our results are consistent with recent observations using gene knockout mice for catalytic subunits of PI3K, p110γ and p110δ, where the NK-mediated cytotoxicity and cytokine generations were impaired.68,69
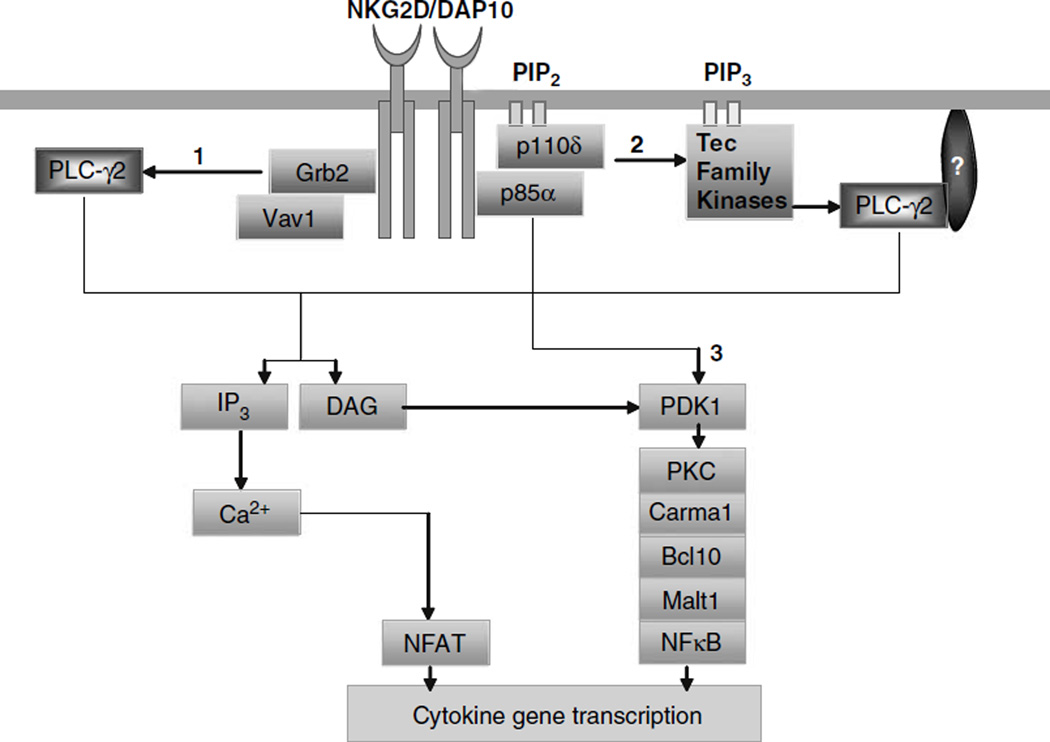
Reductions in the ability of p85α−/− NK cells in generating inflammatory cytokines need further explanations. Multiple mechanistic models can explain our findings (Figure 7). Recent studies in NK and other cell types have indicated potential cross talks between PI3K and PLCγ2 pathways.70,71 Leibson laboratory has shown that binding of PI3K-p85 along with Grb2-Vav1 complexes was necessary for full calcium release and cytotoxicity70 NKG2D/DAP10 complexes can recruit combinations of p85α and Grb2 molecules, both of which bind to DAP10 by the YINM motif. Further, in this model, coupling of Grb2 complexes to DAP10 was necessary and sufficient for the recruitment and activation of Vav1, SLP76 and PLCγ2.70 This observation explains how NKG2D, without using adaptors such as LAT can effectively recruit PLCγ2 leading to cytotoxicity. However, this study does not explain how the upstream events initiated from NKG2D/DAP10/p85 regulate cytokine generation. Our recent study has shown that the Carma1/Bcl10/Malt-1/NF-κB signaling axis is critical for NKG2D-mediated generation of cytokines.72 DAG, generated by PLCγ2 regulates Carma1 activation by PKC.73 Thus, we predict DAP10/p85α/Grb2-Vav1 complexes can connect to Carma1/Bcl10/Malt-1/NF-κB axis by DAG and PKC.
Figure 7.
Signaling pathways that are potentially regulated by p85α in the generation of cytokines. We propose that p85α can regulate cytokine gene transcriptions by: (1) a direct recruitment of PLCγ2 by Grb2/Vav-1 complexes. Grb2 is an adapter protein that binds to DAP10 through the YINM motif. Activation and recruitment of Grb2 to NKG2D/DAP10 complexes has been shown to initiate phosphorylation of PLCγ2.70 PLCγ is one of the major regulators of protein kinase C (PKC) by DAG. Thus, DAP10/p85α/Grb2-Vav1 complexes can signal either through Carma1/Bcl10/Malt-1/nuclear factor (NF)-κB or PLCγ2/IP3/Ca2+/NFAT, which lead to cytokine gene transcriptions. (2) Activation of PLCγ2 by phosphotidylinositol-3-kinase (PI3K) by Tec kinases. Btk is a Tec family protein tyrosine kinase, which can bind to PI3K product, PtdIns-3,4,5-P3 leading to the recruitment and phosphorylation of PLCγ2.26 This in turn can also result in the activation of PKC and Carma1/ Bcl10/Malt-1/NF-κB or PLCγ2/IP3/Ca2+/NFAT signaling pathways. (3) Direct link between PI3K and Carma1/Bcl10/Malt-1/NF-κB signaling axis. Recent studies have shown that PI3K-dependent kinase, PDK1, has the ability to activate PKC.71 Thus, the activation of PKC can link the Carma1/Bcl10/Malt-1/NF-κB signaling axis to PI3K.
In the second model (Figure 7), we predict P13K can activate PLCγ2 using Tec family PTKs such as Btk as described in B cells.26 Phosphorylated Btk is recruited to the plasma membrane and binds to PI3K product, PtdIns-3,4,5-P3.74 Using this product, Btk generates PtdIns-4,5-P2, which is hydrolyzed by PLCγ2 into DAG and IP3.75 DAG can activate PKCθ; thus connecting the PI3K/Btk/PtdIns-3,4,5-P3 pathway to Carma1/Bcl10/ Malt-1/NF-κB signaling axis.76 In addition, PLCγ2 may also regulate cytokine gene transcriptions through the production of IP3/Ca2+/NFAT axis. Although the expression of Btk is primarily restricted to B cells,26 its presence and function has yet to be explored in NK cells. Further work is needed to analyze Btk or similar kinases that belong to Tec family in NK cells.
In the third model, we propose the involvement of PDK1 in cytokine generation (Figure 7). PI3K activates PDK1, which phosphorylates PKCθ. Activated PKCθ can directly recruit IKK complexes into the lipid rafts.71 PDK1 has also been shown to recruit Carma1/Bcl10/ Malt-1 complex or the membrane-bound Bcl10 to activate IKK.71 Our recent studies showed NK cells from Bcl10−/−72 or Carma1−/− mice (H Chu and S Malarkannan, unpublished) also had severe impairment in generating cytokines in response to NKG2D-mediated activations. These studies provide support to our notion that both Carma1 and Bcl10 are obligatory signaling molecules, irrespective of PI3K → Btk → PLCγ2 or PI3K → PDK1 → PKCθ-mediated cytokine generations. In conclusion, our current observations establish a strong basis to further explore PI3K-mediated cytokine generation in NK cells.
Materials and methods
Mice and cell lines
PI3K-p85α−/− and WT mice were previously described.20,27 All mice used in this study were maintained in pathogen-free conditions at the Biological Resource Center (BRC), Medical College of Wisconsin (MCW), Milwaukee, WI, USA. All the animal protocols used were approved by the BRC, MCW, Milwaukee, WI, USA. EL4, EL4H60, RMA/S and YAC-1 cells and their culture conditions were described.77
Fetal liver cell transplantation
PI3K-p85α−/− mice were originally generated by transfecting mouse embryonic stem cell line TC-1 (129Sv) with targeting constructs and clones identified to contain the heterozygous gene disruption was injected into C57BL/6 blastocysts.20 Chimeric male mice were bred with C57BL/6 mice. At the time of usage, original 129Sv lines were backcrossed to C57BL/6 to a minimum of seven generations. As less than 5% of the homozygous gene-deleted newborns survive beyond 7 days of age, both WT and the p85α−/− were generated by crossing p85α+/− × p85α+/−. Irradiated JAK3−/− recipients (300 rad; 4- to 6-week old) of C57BL/6 background (H-2b) were transplanted with 2 × 106 nucleated fetal liver cells from 13- to 14-day-old embryos of WT or p85α−/− through retro-orbital injections.27 After 2–3 months, development and functions of NK cells from BM, spleen and liver of the transplanted mice were examined (Figure 1a). Reconstituted JAK3−/− mice appeared normal without any external symptoms of any autoimmune pathology that has been previously reported in JAK3−/− mice.29,30 Wherever indicated, WT and p85α−/− represent JAK3−/− mice reconstituted with liver cells from p85α+/+ or p85α−/− fetuses, respectively. All the mice used in this study were of H-2b haplotype (data not shown).
Flow cytometry and cell sorting
Cell preparations were stained with fluorescent-labeled mAbs as described.77 Abs for NK1.1 (PK136), CD3ε (145-2C11), NKG2D (A10), NKG2A/C/E (20d5), CD11b (M1/ 70), CD43 (1B11), CD49b (DX5), CD51 (RMV-7), CD122 (5H4) and Ly49I (YLI-90) were obtained from e-Bioscience (San Diego, CA, USA). Abs for Ly49A (A1), Ly49C/I (5E6), Ly49D (4E5) and Ly49G2 (4D11) were obtained from BD Biosciences (San Jose, CA, USA). Standard flow cytometry analyses were carried out in LSR-II using FACSDiva software (Becton Dickinson, CA, USA). Splenocytes from WT mice were stained with anti-CD3ε, -B220 or -NK1.1 mAbs and subjected to cell sorting using FACSAria (Becton Dickinson) to isolate T, B and NK cells, respectively.
Western blotting
Immunoblots of p85α in T, B and NK cell lysates were performed as previously described.27 Whole-cell lysates (40 µg) were resolved using 12% SDS–polyacrylamide gel electrophoresis (PAGE) gels and transferred to nitrocellulose membranes and probed with indicated Abs. Antimouse p85α (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-β-Actin (Boehringer Mannheim, Germany) antibodies were used and signals were detected using ECL kit (GE Healthcare, Piscataway, NJ, USA).
NK cell preparation
NK cells were purified as previously described.72 Briefly, single-cell suspensions from spleen and BM were passed through nylon wool columns to deplete adherent populations consisting of B cell and macrophages. Nylon wool nonadherent cells were cultured with 1000 Uml−1 of IL-2 (NCI-BRB—Preclinical Repository, Maryland, MD, USA). Nonadherent cells were removed on days 4, 5 and 6 and the remaining adherent cells were replenished with IL-2-containing medium to generate highly enriched NK cells. NK cell preparations were tested for purity and all experiments were carried out using 6- or 7-day-old NK cells. NK preparations with more than 90% of NK1.1+ cells were used (Supplementary Figure 1).
Cytotoxicity assays
NK-mediated cytotoxicity was quantified using 51Chromium (51Cr)-labeled target cells at varied Effector to Target ratio (E:T Ratio). Percent-specific lysis was calculated using amounts of absolute, spontaneous and experimental 51Cr-release from target cells. Briefly, IL-2-cultured NK cells on day 6 or 7 were incubated with target cells for 4 h at 37 °C and the percent cytotoxicity was assayed by quantifying the amount of 51Cr released into the supernatant.
Secretion and quantification of cytokines and chemokines through ELISA and Bioplex assays
IL-2-cultured, NK cells on day 6 were harvested, Fcblocked and were activated with titrated concentrations of plate-bound anti-NKG2D (A10) or -NK1.1 (PK136) mAbs. Nunc Immunolon plates were coated with varying concentration of mAbs for overnight, washed thrice with phosphate-buffered saline before the addition of NK cells. Culture supernatants were collected between 16 and 18 h and used for cytokine and chemokine quantification. Standard curves generated using recombinant cytokines were used to calculate the concentrations of IFN-γ and GM-CSF in the culture supernatants using ELISA kits (eBioscience, San Diego, CA, USA). MIP-1α and MIP-1β were quantified using Bioplex kits (Bio-Rad, Richmond, CA, USA).
Drug inhibition assays
IL-2-activated NK cells (sixth day) were Fc-blocked and treated with pharmacological inhibitors for PI3K (Wort-mannin; Sigma-Aldrich, USA) and Syk (Piceatannol; AG scientific Co., San Diego, CA, USA) separately or in combination for 1 h at room temperature. Treated cells were washed twice and used in cytotoxicity or cytokine assays as described.72 Cytotoxicity was performed against EL4H60 at 20:1 E:T ratio. For cytokine generation, treated and untreated cells were added to Nunc Immunolon plates that were precoated with 5 µg ml−1 of anti-NKG2D (A10) mAb. Culture supernatants were collected between 16 and 18 h and assayed for IFN-γ through ELISA.
Statistical analysis
Statistical analysis was performed by two-tailed, paired, Student’s t-test using Microsoft Excel 2003 software to compare the differences between WT and p85α−/− mice. P-values of <0.05 were considered significant.
Supplementary Material
Acknowledgements
We thank David A Fruman and Lewis C Cantley for p85α−/− mice. This work was supported in part by ACS Scholar grants RSG-02-172-LIB (to SM); RSG CCG-106204 (to DW); ROTRF grant no. 111662730 (to SM); and NIH grants R01 A1064826-01, U19 AI062627-01, NO1-HHSN26600500032C (to SM); R01 AI52327 (to RW) and R01 HL073284 (to DW).
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Karre K, Schneider G. Immunology. The footprint of a killer. Nature. 2000;405:527–528. doi: 10.1038/35014724. [DOI] [PubMed] [Google Scholar]
- 3.Chiesa S, Mingueneau M, Fuseri N, Malissen B, Raulet DH, Malissen M, et al. Multiplicity and plasticity of natural killer cell signaling pathways. Blood. 2006;107:2364–2372. doi: 10.1182/blood-2005-08-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 6.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 7.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 8.Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J Immunol. 2002;169:4079–4083. doi: 10.4049/jimmunol.169.8.4079. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Cherwinski H, Spies T, Phillips JH, Lanier LL. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J Exp Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol. 2002;3:1150–1155. doi: 10.1038/ni857. [DOI] [PubMed] [Google Scholar]
- 12.McVicar DW, Taylor LS, Gosselin P, Willette-Brown J, Mikhael AI, Geahlen RL, et al. DAP12-mediated signal transduction in natural killer cells. A dominant role for the Syk protein-tyrosine kinase. J Biol Chem. 1998;273:32934–32942. doi: 10.1074/jbc.273.49.32934. [DOI] [PubMed] [Google Scholar]
- 13.Zompi S, Hamerman JA, Ogasawara K, Schweighoffer E, Tybulewicz VL, Di Santo JP, et al. NKG2D triggers cytotoxicityin mouse NK cells lacking DAP12 or Syk family kinases. Nat Immunol. 2003;4:565–572. doi: 10.1038/ni930. [DOI] [PubMed] [Google Scholar]
- 14.Carlyle JR, Martin A, Mehra A, Attisano L, Tsui FW, Zuniga-Pflucker JC. Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J Immunol. 1999;162:5917–5923. [PubMed] [Google Scholar]
- 15.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 16.Arase N, Arase H, Park SY, Ohno H, Ra C, Saito T. Association with FcRgamma is essential for activation signal through NKR-P1 (CD161) in natural killer (NK) cells and NK1.1+ T cells. J Exp Med. 1997;186:1957–1963. doi: 10.1084/jem.186.12.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Held W, Roland J, Raulet DH. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- 18.Raulet DH, Held W, Correa I, Dorfman JR, Wu MF, Corral L. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol Rev. 1997;155:41–52. doi: 10.1111/j.1600-065x.1997.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 19.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, et al. Impaired B and Tcell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 20.Fruman DA, Snapper SB, Yballe CM, Davidson L, Yu JY, Alt FW, et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, et al. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 22.Vanhaesebroeck B, Leevers SJ, Panayotou G, Waterfield MD. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 23.Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Kane LP, Weiss A. The PI-3 kinase/Akt pathway and T cell activation: pleiotropic pathways downstream of PIP3. Immunol Rev. 2003;192:7–20. doi: 10.1034/j.1600-065x.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Jee K, Kim D, Koh H, Chung J. Cyclic AMP inhibits Akt activity by blocking the membrane localization of PDK1. J Biol Chem. 2001;276:12864–12870. doi: 10.1074/jbc.M001492200. [DOI] [PubMed] [Google Scholar]
- 26.Saito K, Tolias KF, Saci A, Koon HB, Humphries LA, Scharenberg A, et al. BTK regulates PtdIns-4,5-P2 synthesis: importance for calcium signaling and PI3K activity. Immunity. 2003;19:669–678. doi: 10.1016/s1074-7613(03)00297-8. [DOI] [PubMed] [Google Scholar]
- 27.Dai X, Chen Y, Schuman J, Hua Z, Adamson JW, Wen R, et al. Distinct roles of phosphoinositide-3 kinase and phospholipase Cgamma2 in B-cell receptor-mediated signal transduction. Mol Cell Biol. 2006;26:88–99. doi: 10.1128/MCB.26.1.88-99.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regunathan J, Chen Y, Kutlesa S, Dai X, Bai L, Wen R, et al. Differential and nonredundant roles of phospholipase Cgamma2 and phospholipase Cgamma1 in the terminal maturation of NK cells. J Immunol. 2006;177:5365–5376. doi: 10.4049/jimmunol.177.8.5365. [DOI] [PubMed] [Google Scholar]
- 29.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, et al. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 30.Saijo K, Park SY, Ishida Y, Arase H, Saito T. Crucial role of Jak3 in negative selection of self-reactive T cells. J Exp Med. 1997;185:351–356. doi: 10.1084/jem.185.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 32.Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. 2001;31:1900–1909. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Vosshenrich CA, Ranson T, Samson SI, Corcuff E, Colucci F, Rosmaraki EE, et al. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J Immunol. 2005;174:1213–1221. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 34.Fahlen L, Lendahl U, Sentman CL. MHC class I-Ly49 interactions shape the Ly49 repertoire on murine NK cells. J Immunol. 2001;166:6585–6592. doi: 10.4049/jimmunol.166.11.6585. [DOI] [PubMed] [Google Scholar]
- 35.Held W, Dorfman JR, Wu MF, Raulet DH. Major histocompatibility complex class I-dependent skewing of the natural killer cell Ly49 receptor repertoire. Eur J Immunol. 1996;26:2286–2292. doi: 10.1002/eji.1830261003. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005 Jul 3; doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002;55:221–228. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- 40.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, et al. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang K, Zhong B, Gilvary DL, Corliss BC, Hong-Geller E, Wei S, et al. Pivotal role of phosphoinositide-3 kinase in regulation of cytotoxicity in natural killer cells. Nat Immunol. 2000;1:419–425. doi: 10.1038/80859. [DOI] [PubMed] [Google Scholar]
- 42.Jiang K, Zhong B, Gilvary DL, Corliss BC, Vivier E, Hong-Geller E, et al. Syk regulation of phosphoinositide 3-kinase-dependent NK cell function. J Immunol. 2002;168:3155–3164. doi: 10.4049/jimmunol.168.7.3155. [DOI] [PubMed] [Google Scholar]
- 43.Walker VG, Ammer A, Cao Z, Clump AC, Jiang BH, Kelley LC, et al. PI3K activation is required for PMA-directed activation of cSrc by AFAP-110. Am J Physiol Cell Physiol. 2007;293:C119–C132. doi: 10.1152/ajpcell.00525.2006. [DOI] [PubMed] [Google Scholar]
- 44.Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, Gray A, et al. A crucial role for the p110delta subunit of phosphatidy-linositol 3-kinase in B cell development and activation. J Exp Med. 2002;196:753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Carpino N, et al. Essential, nonredundant role for the phosphoinositide 3-kinase p110delta in signaling by the B-cell receptor complex. Mol Cell Biol. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 47.Okkenhaug K, Vanhaesebroeck B. PI3K-signalling in B- and T-cells: insights from gene-targeted mice. Biochem Soc Trans. 2003;31(Part 1):270–274. doi: 10.1042/bst0310270. [DOI] [PubMed] [Google Scholar]
- 48.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 49.Ranson T, Vosshenrich CA, Corcuff E, Richard O, Laloux V, Lehuen A, et al. IL-15 availability conditions homeostasis of peripheral natural killer T cells. Proc Natl Acad Sci USA. 2003;100:2663–2668. doi: 10.1073/pnas.0535482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franke TF, Kaplan DR, Cantley LC. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 51.Kim HR, Hwang KA, Kang I. Dual roles of IL-15 in maintaining IL-7Ralphalow. J Immunol. 2007;179:6734–6740. doi: 10.4049/jimmunol.179.10.6734. [DOI] [PubMed] [Google Scholar]
- 52.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 53.Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CA, Colucci F, et al. GATA-3 promotes maturation, IFN-gammaproduction, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 54.Ogasawara K, Hida S, Azimi N, Tagaya Y, Sato T, Yokochi-Fukuda T, et al. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature. 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 55.Lohoff M, Duncan GS, Ferrick D, Mittrucker HW, Bischof S, Prechtl S, et al. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J Exp Med. 2000;192:325–336. doi: 10.1084/jem.192.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 58.Lacorazza HD, Miyazaki Y, Di CA, Deblasio A, Hedvat C, Zhang J, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 59.Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 60.Vely F, Olivero S, Olcese L, Moretta A, Damen JE, Liu L, et al. Differential association of phosphatases with hematopoietic co-receptors bearing immunoreceptor tyrosine-based inhibition motifs. Eur J Immunol. 1997;27:1994–2000. doi: 10.1002/eji.1830270825. [DOI] [PubMed] [Google Scholar]
- 61.Kishimoto H, Ohteki T, Yajima N, Kawahara K, Natsui M, Kawarasaki S, et al. The Pten/PI3K pathway governs the homeostasis of Valpha14iNKT cells. Blood. 2007;109:3316–3324. doi: 10.1182/blood-2006-07-038059. [DOI] [PubMed] [Google Scholar]
- 62.Sperling AI, Green JM, Mosley RL, Smith PL, DiPaolo RJ, Klein JR, et al. CD43 is a murine T cell costimulatory receptor that functions independently of CD28. J Exp Med. 1995;182:139–146. doi: 10.1084/jem.182.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baum LG, Pang M, Perillo NL, Wu T, Delegeane A, Uittenbogaart CH, et al. Human thymic epithelial cells express an endogenous lectin, galectin-1, which binds to core 2 O-glycans on thymocytes and T lymphoblastoid cells. J Exp Med. 1995;181:877–887. doi: 10.1084/jem.181.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedraza-Alva G, Merida LB, Burakoff SJ, Rosenstein Y. CD43-specific activation of Tcells induces association of CD43 to Fyn kinase. J Biol Chem. 1996;271:27564–27568. doi: 10.1074/jbc.271.44.27564. [DOI] [PubMed] [Google Scholar]
- 65.Colucci F, Schweighoffer E, Tomasello E, Turner M, Ortaldo JR, Vivier E, et al. Natural cytotoxicity uncoupled from the Syk and ZAP-70 intracellular kinases. Nat Immunol. 2002;3:288–294. doi: 10.1038/ni764. [DOI] [PubMed] [Google Scholar]
- 66.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 67.Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D-DAP10 triggers human NK cell-mediated killing via a Syk-independent regulatory pathway. Nat Immunol. 2003;4:557–564. doi: 10.1038/ni929. [DOI] [PubMed] [Google Scholar]
- 68.Tassi I, Cella M, Gilfillan S, Turnbull I, Diacovo TG, Penninger JM, et al. p110gamma and p110delta phosphoinositide 3-kinase signaling pathways synergize to control development and functions of murine NK cells. Immunity. 2007;27:214–227. doi: 10.1016/j.immuni.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 69.Kim N, Saudemont A, Webb L, Camps M, Ruckle T, Hirsch E, et al. The p110delta catalytic isoform of PI3K is a key player in NK cell development and cytokine secretion. Blood. 2007;110:3202–3208. doi: 10.1182/blood-2007-02-075366. [DOI] [PubMed] [Google Scholar]
- 70.Upshaw JL, Arneson LN, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. NKG2D-mediated signaling requires a DAP10-bound Grb2-Vav1 intermediate and phosphatidylinositol-3-kinase in human natural killer cells. Nat Immunol. 2006;7:524–532. doi: 10.1038/ni1325. [DOI] [PubMed] [Google Scholar]
- 71.Lee KY, D’Acquisto F, Hayden MS, Shim JH, Ghosh S. PDK1 nucleates T cell receptor-induced signaling complex for NF-kappaB activation. Science. 2005;308:114–118. doi: 10.1126/science.1107107. [DOI] [PubMed] [Google Scholar]
- 72.Malarkannan S, Regunathan J, Chu H, Kutlesa S, Chen Y, Zeng H, et al. Bcl10 plays a divergent role in NK cell-mediated cytotoxicity and cytokine generation. J Immunol. 2007;179:3752–3762. doi: 10.4049/jimmunol.179.6.3752. [DOI] [PubMed] [Google Scholar]
- 73.Shinohara H, Yasuda T, Aiba Y, Sanjo H, Hamadate M, Watarai H, et al. PKC beta regulates BCR-mediated IKK activation by facilitating the interaction between TAK1 and CARMA1. J Exp Med. 2005;202:1423–1431. doi: 10.1084/jem.20051591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scharenberg AM, El Hillal O, Fruman DA, Beitz LO, Li Z, Lin S, et al. Phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P3)/ Tec kinase-dependent calcium signaling pathway: a target for SHIP-mediated inhibitory signals. EMBO J. 1998;17:1961–1972. doi: 10.1093/emboj/17.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol. 1999;17:555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- 76.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 77.Regunathan J, Chen Y, Wang D, Malarkannan S. NKG2D receptor-mediated NK cell function is regulated by inhibitory Ly49 receptors. Blood. 2005;105:233–240. doi: 10.1182/blood-2004-03-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.