Abstract
Colorectal cancer (CRC) is a disease whose genesis may include metabolic dysregulation. Cancer stem cells are attractive targets for therapeutic interventions since their aberrant expansion may underlie tumor initiation, progression, and recurrence. To investigate the actions of metabolic regulators on cancer stem cell-like cells (CSC) in CRC, we determined the effects of soybean-derived bioactive molecules and the anti-diabetes drug metformin (MET), alone and together, on the growth, survival, and frequency of CSC in human HCT116 cells. Effects of MET (60 μM) and soybean components genistein (Gen, 2 μM), lunasin (Lun, 2 μM), β-conglycinin (β-con, 3 μM), and glycinin (Gly, 3 μM) on HCT116 cell proliferation, apoptosis, and mRNA/protein expression and on the frequency of the CSC CD133+CD44+ subpopulation by colonosphere assay and fluorescence-activated cell sorting/flow cytometry were evaluated. MET, Gen, and Lun, individually and together, inhibited HCT116 viability and colonosphere formation and, conversely, enhanced HCT116 apoptosis. Reductions in frequency of the CSC CD133+CD44+ subpopulation with MET, Gen, and Lun were found to be associated with increased PTEN and reduced FASN expression. In cells under a hyperinsulinemic state mimicking metabolic dysregulation and without and with added PTEN-specific inhibitor SF1670, colonosphere formation and frequency of the CD133+CD44+ subpopulation were decreased by MET, Lun and Gen, alone and when combined. Moreover, MET + Lun + Gen co-treatment increased the pro-apoptotic and CD133+CD44+-inhibitory efficacy of 5-fluorouracil under hyperinsulinemic conditions. Results identify molecular networks shared by MET and bioavailable soy food components, which potentially may be harnessed to increase drug efficacy in diabetic and non-diabetic patients with CRC.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-015-0499-6) contains supplementary material, which is available to authorized users.
Keywords: Metformin, Soy, Lunasin, Genistein, Colon cancer, Stem-like cell
Introduction
Colorectal cancer (CRC) remains a major contributor to cancer morbidity and mortality worldwide and is the third leading cause of cancer deaths in the USA (Siegel et al. 2015). While the molecular mechanisms by which CRC begins and then progresses to metastatic disease are better understood relative to some other cancer types (Tomasetti et al. 2014; Linnekamp et al. 2015), current treatments such as with the first-line chemotherapeutic drug 5-fluorouracil (5-FU) are largely ineffective once the tumor becomes invasive, metastatic and/or chemo-resistant (Paldino et al. 2014). Hence, preventing CRC initiation, progression and recurrence and defining bona fide targets for effective therapies are paramount for reducing CRC-related mortality.
Cancer stem cells (CSC) have emerged as attractive therapeutic targets to inhibit tumor initiation, chemo-resistance, and relapse in many cancer types (Paldino et al. 2014; Zeuner et al. 2014; White and Lowry 2015). According to the CSC hypothesis, a small population of normal stem cells requisite for maintaining tissue homeostasis/renewal can initiate a tumor upon acquiring one or more driver mutations (Chandler 2010). Moreover, a recent study (Tomasetti and Vogelstein 2015) has suggested that the lifetime risk of many different types of cancer is strongly correlated with the total number of divisions of these normal self-renewing cells, and that random mutations arising during DNA replication are likely causative for the emergence of CSC. Interestingly, among the different epithelial stem cells analyzed in that study, those derived from the colon were shown to undergo one of the highest numbers of cell divisions. Inhibiting the expansion of normal SC as well as CSC may thus constitute a useful approach for CRC prevention and treatment.
Diet is a well-accepted risk factor for CRC, the latter potentially mediated by bioactive and bioavailable components in foods that can influence colon epithelial cell growth and survival (Yan et al. 2010; Spector et al. 2013). Previous work in our laboratory demonstrated that dietary intake of soy (Xiao et al. 2008) and whey (Xiao et al. 2006) protein-based diets favorably influenced insulin-signaling components and reduced intestinal tumor incidence in rats exposed to the intestinal carcinogen azoxymethane. Further, we showed in the context of a high-fat diet that a soy protein-based dietary regimen suppressed intestinal epithelial cell proliferation and systemic insulin levels in mice (Al-Dwairi et al. 2012, 2014). The association between colonic intestinal proliferation and insulin is noteworthy, given that the anti-diabetic drug metformin (1,1-dimethylbiguanide hydrochloride; MET) has recently attracted much interest as a potential anticancer medication. Several epidemiological studies reported an inverse correlation between MET intake (at standard doses of 1500–2250 mg/day) and CRC incidence and related mortality in diabetic patients (Zhang et al. 2013; Sehdev et al. 2015). Further, MET at a low dose (250 mg/day) was found to reduce colon epithelial cell proliferation and numbers of aberrant crypt foci in non-diabetic patients (Dowling et al. 2011). Moreover, combinations of MET with conventional chemotherapeutic agents 5-fluorouracil and oxaliplatin were reported to decrease recurrence of colon tumors in a mouse xenograft model (Nangia-Makker et al. 2014). Nevertheless, the potential interactions of MET and bioactive dietary components with known anti-tumor activities for CRC prevention and treatment, and specifically in the context of targeting CSC, have not been delineated. The importance of the latter is underscored by recent reports that dietary factors including the soy isoflavone genistein (Gen) (Montales et al. 2012; Rahal et al. 2013), polyphenolic compounds present in blueberries (Montales et al. 2012), curcumin (Kakarala et al. 2010), and resveratrol (Kao et al. 2009), all of which have been reported to possess anti-metabolic/anti-diabetic properties (Gilbert and Liu 2013; Valsecchi 2013; Yeh et al. 2014; Rouse et al. 2014), can inhibit expansion of basal stem-like mammary cells with tumorigenic potential.
Here, we evaluated the effects of MET and several bioavailable and bioactive molecules present in soy protein isolate, namely the isoflavone Gen, peptide lunasin (Lun), and proteins β-conglycinin (β-con) and glycinin (Gly), alone and in combination, on the survival of CSCs present in the human colon cancer cell line HCT116 (Yeung et al. 2010). We examined potential chemo-preventive mechanisms of these components in CSCs and determined whether the combination of MET, Gen, and Lun with 5-FU could increase CRC cell death relative to 5-FU alone. Our results show that MET, Gen, and Lun together confer enhanced inhibitory effects on CSC expansion and suggest that the targeting of CSC with these molecules should be further explored for the clinical management of CRC.
Materials and methods
Cell culture and treatments
The human colon cancer cell lines HT29 and HCT116 (American Type Culture Collection; ATCC, Manassas, VA) were propagated in McCoy’s medium (ATCC) supplemented with 10 % fetal bovine serum (FBS; GIBCO, Carlsbad, CA) and 5 % antibiotic–antimycotic solution (ABAM; GIBCO) in a humidified incubator (5 % CO2:95 % air) at 37 °C. Cells were seeded in six-well plates at an initial density of 2 × 105 per well, and treated (in culture medium) with metformin (MET 60 µM; Sigma-Aldrich, St. Louis, MO), lunasin (Lun 2 µM; American Peptide Co., Sunnyside, CA), β-conglycinin (β-con 3 µM; provided by Dr. Ferreira, Federal University of Bahia, Brazil), glycinin (Gly 3 µM; provided by Dr. Ferreira), and genistein (Gen 2 µM; Sigma-Aldrich), alone or in combination. β-con and Gly were isolated and purified as described below. Metformin, Lun, β-con, and Gly were dissolved in phosphate-buffered saline (PBS; GIBCO), whereas Gen was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich). In other experiments, cells were treated with insulin (2 µM; Sigma), PTEN inhibitor SF1670 (2 µM; Echelon Biosciences, Salt Lake, UT), and 5-fluorouracil (5-FU 50 µM; Teva Parenteral Medicines, Irvine, CA). Treated cells were collected at select time points for subsequent analyses.
Extraction and isolation of β-conglycinin and glycinin
Soybean flour (60 mesh) was defatted with hexane [1:8 (wt/vol) flour-to-solvent ratio] by rocking at room temperature for 12 h and then re-extracted in the same solvent [1:6 (wt/vol)]. The extract was evaporated to dryness at room temperature and used for isolation of β-conglycinin and glycinin as previously described (Ferreira et al. 2010). The β-conglycinin and glycinin proteins were further purified by size exclusion chromatography on a Sepharose CL-6B column (1.0 × 100 cm) equilibrated with 0.05 M potassium phosphate, pH 7.5 containing 0.5 M NaCl and 0.01 % NaN3 at a column flow rate of 5.8 ml/tube. Column eluates were monitored at 280 nm, and protein-containing fractions were dialyzed and lyophilized. Column fractionation produced one major peak for each of the isolated proteins. SDS-PAGE confirmed the specific molecular weight and indicated over 95 % purity for β-con and Gly proteins (data not shown).
Cell viability, cell cycle distribution, and apoptosis
Cells were seeded in six-well plates at an initial density of 2 × 105 cells per well and incubated overnight prior to treatment. Cells were collected 48 h post-treatment, and the quantity of viable cells was determined by the trypan blue exclusion method using the Vi-Cell cell viability analyzer (Beckman Coulter Inc., Atlanta, GA). For cell cycle analysis, cells were serum-starved for 24 h after plating in 0.5 % charcoal-stripped FBS (CS-FBS; GIBCO), and 24 h later were treated with medium supplemented with 2.5 % CS-FBS and the described treatments. The analysis of cell cycle parameters followed previously described protocols (Rahal and Simmen 2010). At least 1 × 105 cells were stained with propidium iodide (PI; Sigma-Aldrich), and the proportion of cells in Sub G1, G1, S, and G2 phases was analyzed with the Becton–Dickinson LSRFortessa Flow Cytometer and associated software (BD Biosciences, San Jose, CA). The percent of apoptotic cells 48 h post-treatment was evaluated by Annexin V staining (Trevigen, Gaithersburg, MD), followed by analysis in the Becton–Dickinson LSRFortessa Flow Cytometer.
Flow cytometry
Single-cell suspensions were evaluated for relative frequency of CD133+CD44+-stained cells by flow cytometry and the use of PE-conjugated anti-CD133 (Miltenyi Biotec Incorporated, San Diego, CA) and FITC-conjugated anti-CD44 (BD Pharmingen, San Jose, CA), as previously described (Chen et al. 2011). Cells were sorted on a FACS Aria Cell Sorting Flow Cytometer (BD Biosciences); Supplementary Figure S1 shows the gating strategy used for FACS analysis. Dead cells were excluded using 4′,6-diamidino-2-phenylindole (DAPI; 1 µg/ml; Sigma-Aldrich). CD133+CD44+-positive cells were collected for colonosphere formation assay and gene expression analysis (below).
Colonosphere formation
To examine treatment effects on colonosphere formation, unsorted HCT116 cells and FACS-sorted CD133+CD44+ cell fractions were seeded at a density of 2 × 103 cells per well in ultra-low attachment plates (Corning Inc., Corning, NY) in plating medium [phenol red-free MEM supplemented with B27 media (Invitrogen); 20 ng/ml human basic fibroblast growth factor (Invitrogen, Carlsbad, CA), 20 ng/ml epidermal growth factor (Invitrogen), 10 μg/ml heparin (Sigma-Aldrich), 1 % ABAM, and 100 μg/ml gentamicin (Sigma-Aldrich)] (Montales et al. 2012). Treatments were MET (60 µM), soy bioactive factors (3 µM each for β-con and Gly; 2 µM each for Lun and Gen), and insulin (2 µM), singly and with the indicated combinations. To investigate whether MET limited colonosphere-forming ability, HCT116 cells were treated after initial plating with MET or vehicle, collected 24 h later and then grown as single-cell suspensions under low attachment-plating conditions. For PTEN inhibitor studies, unsorted cells (2 × 105 per well) were plated in six-well plates, allowed to attach overnight, and then treated with the PTEN inhibitor SF1670 (2 µM) for 24 h, prior to collection and re-plating in ultra-low attachment plates. Colonosphere number was counted 5 days after the initial (one-time) treatment using an inverted microscope. To evaluate self-renewal, primary colonospheres (P1) were collected after counting, dissociated into single cells, filtered using a 40 μM sieve, and re-plated in ultra-low attachment plates. Colonospheres formed at secondary passage (P2) were counted at 7 days post-plating and expressed as a percentage of the total number of initially plated cells. Treatment effects were determined from at least three independent experiments, with 3–4 replicates per experiment.
RNA isolation and quantification
Total RNA was extracted from unsorted and FACS-sorted cells and from colonospheres using TRIzol reagent (Invitrogen). cDNA synthesis and real-time quantitative PCR (QPCR) were performed following published methods (Montales et al. 2012). Primers (Supplemental Table 1) were designed using Primer Express (Applied Biosystems, Foster City, CA) and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Gene expression was determined by QPCR using Bio-Rad iTaq SYBR Green Supermix (Bio-Rad; Hercules, CA). Transcript levels for target genes were calibrated to a standard curve generated using pooled cDNAs and normalized to a factor that was derived from the geometric mean of expression for β-actin (ACTB) and cyclophilin A (CYPA) genes, using GeNorm excel file software as previously described (Vandesompele et al. 2002).
Western blot analysis
Whole extracts from treated cells (60 µM MET or vehicle; 24 h) were harvested in RIPA buffer containing protease inhibitor cocktail (Santa Cruz Biotechnology; Santa Cruz, CA). Protein concentrations were determined using the bicinchoninic acid protein assay kit (Pierce Biotechnology; Rockford, IL). Equal amounts of lysate protein (50 μg) were subjected to immunoblotting as described previously (Al-Dwairi et al. 2012). Primary antibodies used were anti-PTEN (1:1000; Cell Signaling, Danvers, MA), anti-FASN (1:1000; Abcam, Cambridge, MA), and anti-β-actin (1:10,000; Sigma-Aldrich). Blots were stripped with Restore Western blot stripping buffer (Pierce Biotechnology) prior to incubation with another primary antibody. Immunoreactive proteins were visualized with Amersham ECL Plus kit (GE Health Care Life Sciences, Piscataway, NJ). Digital images were captured with a FlourChem HD2 Imager system (Alpha Innotech, San Leandro, CA), and densitometry was performed using Alpha View software (Cell Biosciences; Santa Clara, CA). Levels of immunoreactive PTEN and FASN proteins were normalized to that for β-actin.
Data analysis
All experiments were performed at least twice, with each experiment conducted in triplicate or quadruplicate. Data are presented as the mean ± SEM; except for that in Fig. 5b which are LS mean ± SEM. Statistical significance between diet groups was evaluated by Student’s t test or one-way analysis of variance using Sigma Stat version 3.5 for Windows. Data in Fig. 5b were subjected to a two-way ANOVA with ‘experiment’ being considered as a ‘random’ factor, and pairwise multiple comparison procedures (Holm–Sidak method) were used to ascribe statistically significant differences between treatment groups. A P value <0.05 was considered to be statistically significant, with tendency for significance at 0.05 < P < 0.1.
Fig. 5.
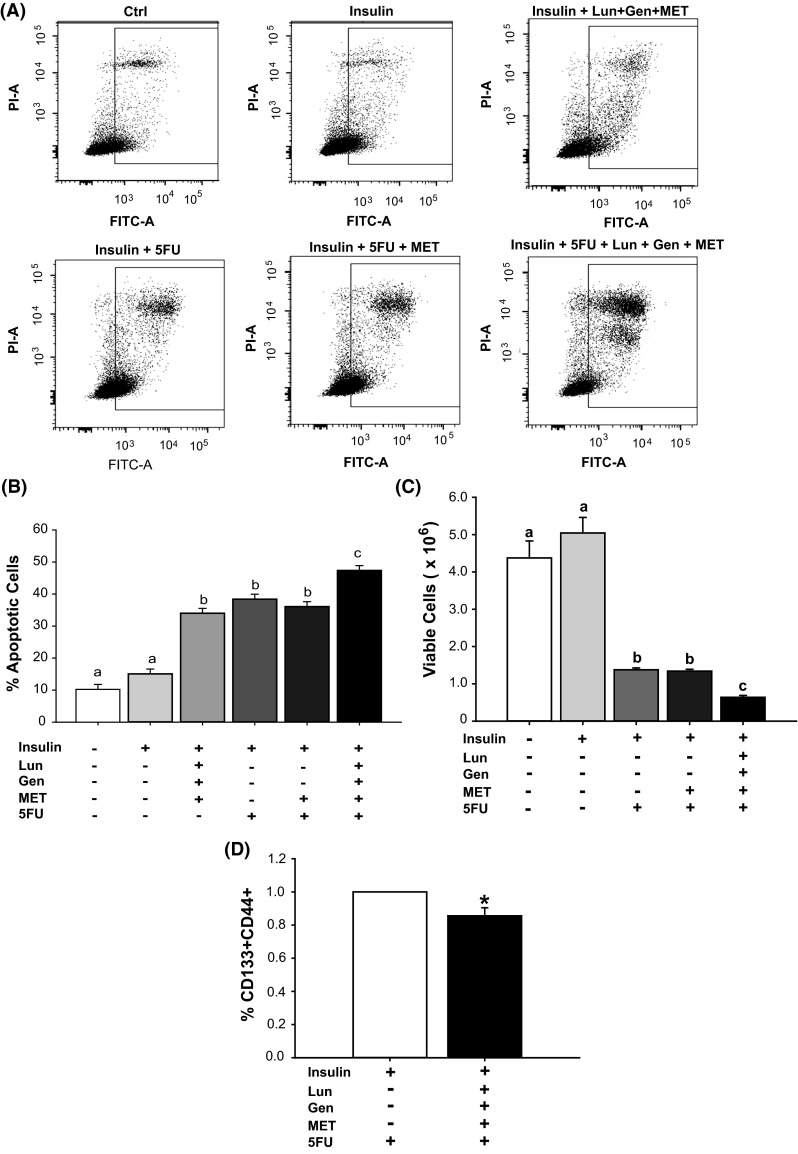
MET plus Lun plus Gen augmented effects of 5-fluorouracil on apoptosis, viability, and frequency of stem cell-like subpopulation. a Representative plots of apoptotic cells (Annexin V FITC- and PI-positive) after treatment with insulin (2 μM); insulin + Lun + Gen + MET; insulin + 5FU; insulin + 5FU + MET (60 μM); or insulin + 5FU + MET + soy bioactives (Lun + Gen; 2 μM each). b Apoptosis induced by 5FU in combination Lun + Gen + MET. Values (Least-square mean ± SEM) are from two independent experiments, with each experiment carried out in triplicate. Means with distinct letters differed at P < 0.05. c HCT116 cells under the hyperinsulinemic condition were pre-treated with MET or MET + Lun + Gen for 48 h prior to receiving 5 FU. MET + Lun + Gen augmented the 5FU-mediated decrease in cell viability. Mean values with distinct letters differed at P < 0.05; two experiments, with each carried out in triplicates. d HCT116 cells under hyperinsulinemic conditions were treated with 5FU alone or 5FU after pre-treatment with MET + Lun + Gen. The percentage of CD133+CD44+ subpopulation in treated cells was evaluated by FACS. Pre-treatment with the three factors decreased the percentage of CSC-like cells in 5FU-treated cells. Data are mean ± SEM, relative to that of insulin-treated cells (two experiments, each performed in triplicates). *P < 0.05
Results
HT29 and HCT116 colonospheres
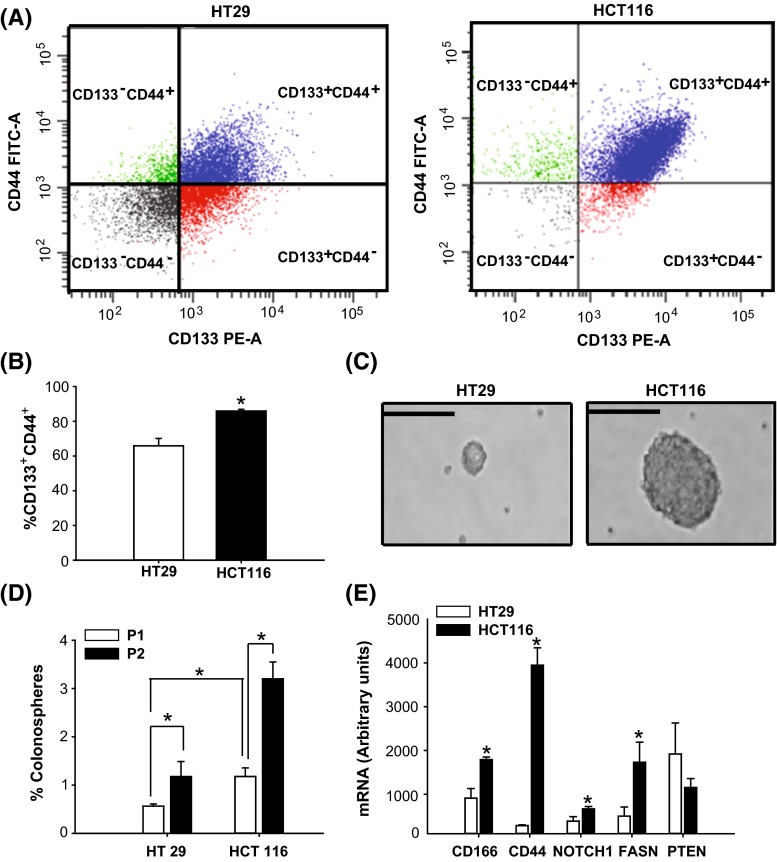
The human colon cancer cell lines HT29 and HCT116 contain a sub-population of cancer stem cell-like cells (CSC), which are distinguished by their expression of cell surface antigens CD133 and CD44 (Sahlberg et al. 2014) and their ability to initiate tumors in nude mice (Yeung et al. 2010; Chen et al. 2011). The relative abundance of the CD133+CD44+ subpopulation between these cell lines was determined by FACS. HCT116 cells showed a higher percentage of CD133+CD44+ subpopulation (~80 %) when compared to HT29 cells (~60 %) (Fig. 1a, b). Further, colonospheres formed from CD133+CD44+ fractions of HCT116 were larger in diameter (Fig. 1c) and were expressed at higher frequencies in both initial (P1; 1 %) and secondary (P2; 3 %) passages (Fig. 1d) than those of HT29 (P1; 0.5 %; P2; 1 %) cells. The increased frequencies in the CD133+CD44+ epithelial subpopulation in P2 relative to P1 for each cell line indicated self-renewal competence. Transcript levels for the CSC marker genes CD166,CD44, and NOTCH1 and for the key lipogenic enzyme, fatty acid synthase (FASN), were higher in the CD133+CD44+ subpopulation from HCT116 than from HT29 cells (Fig. 1e). By contrast, PTEN mRNA abundance was comparable between the two cell subpopulations (Fig. 1e). These collective results provided a rationale for our selection of the HCT116 cell line as in vitro model to test the effects of MET and soy-derived bioactive molecules on human colon cancer.
Fig. 1.
Comparison of human colon cancer cell lines HCT29 and HCT116 for content of cancer stem-like cells. a Single-cell suspensions of HT29 and HCT116 cells were subjected to flow cytometry to assess the relative expression of CD133+CD44+ colon cancer stem-like cells. Representative FACS plots of cells from HT29 (left) or HCT116 (right). b HCT116 cells exhibited a greater percentage of the CD133+CD44+-enriched subpopulation of stem-like subpopulation compared to HT29 cells. Data are mean ± SEM; *P < 0.05 for HCT116 compared to HT29. c CD133+CD44+-stained cells were sorted, collected, and plated for colonosphere formation. Representative pictures of primary colonospheres formed from sorted HT29 (left) and HCT116 (right) cells grown in suspension culture for 5 days. Magnification = 40×, scale bar 50 µm. d CD133+CD44+ epithelial subpopulation (from FACS of HCT116 cells) had greater colonosphere formation compared to that from HT29 cells. Passage 1 (P1) spheres were dissociated to single cells and grown in suspension culture to form secondary spheres (Passage 2; P2). Counting of colonospheres was done at day 5 (for P1) and day 7 (for P2). Data (mean ± SEM) represent the percent of colonospheres formed from 2.5 × 103 plated cells (P1) and from 1 × 103 plated cells (P2), respectively, from two independent experiments, with each experiment carried out in quadruplicates. e Transcript levels of stem cell markers (CD166, CD44), and key genes involved in the maintenance of cancer stem/progenitor cells (NOTCH 1, FASN, PTEN) were quantified by QPCR in FACS-sorted CD133+CD44+–HCT116 and CD133+CD44+–HT29 cells. Transcripts were normalized to a factor derived from the geometric mean of expression values for β-actin ACTB) and cyclophilin A (CYPA). Data (mean ± SEM; n = 5 samples/cell type) were obtained from two independent experiments. *P < 0.05 for HCT116 compared to HT29
MET attenuated cell viability and colonosphere formation, induced cell apoptosis and altered PTEN and FASN expression of HCT116 cells
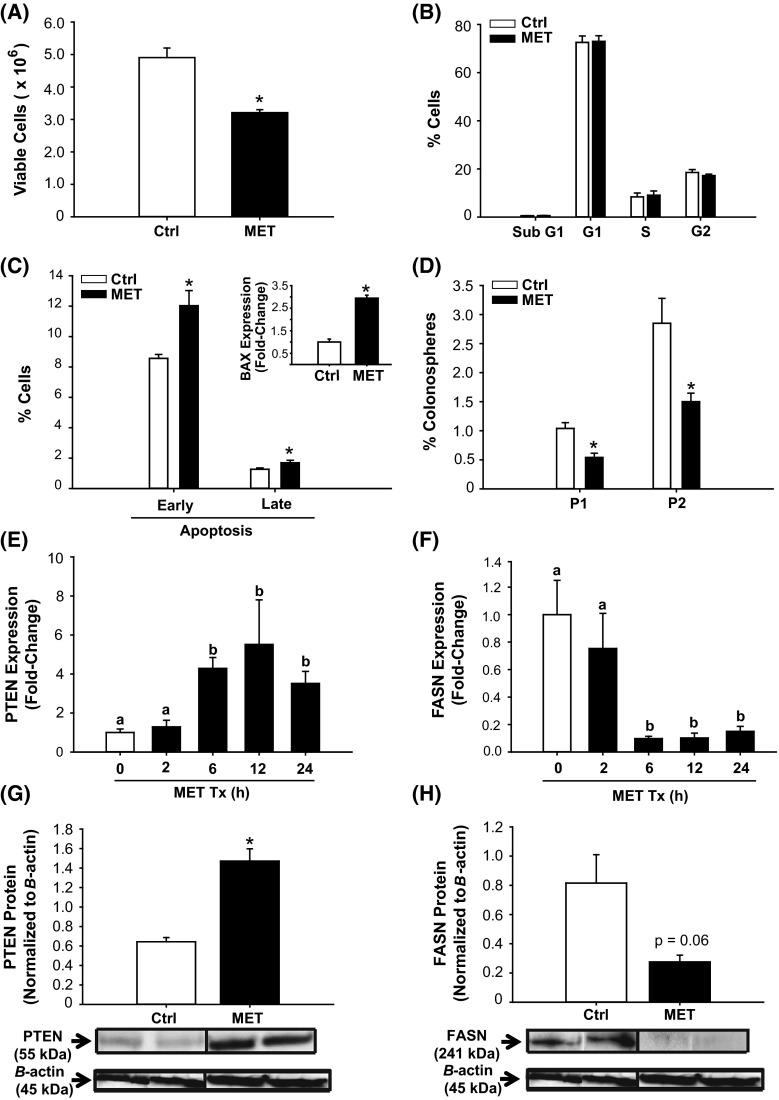
Clinical studies suggest that MET treatment of diabetic patients reduces their risk of and mortality from CRC (Zhang et al. 2013; Sehdev et al. 2015). The effects of MET on colon cancer proliferation and survival were investigated in HCT116 cells using MET at a near-physiological concentration of 60 µM (readily achievable in humans with daily doses of oral MET) (Ochs 2014). MET decreased cell viability (Fig. 2a) in the absence of changes in cell cycle distribution (Fig. 2b). MET increased the percentage of cells in early and late phases of apoptosis (Fig. 2c), and induced the expression of the pro-apoptotic gene BAX (Fig. 2c, inset) relative to control (non-treated) cells. Cells treated with MET had lower percentage of colonospheres formed at both P1 and P2 (a ~ 50 % decrease) when compared to control cells; however, both control and MET-treated cells retained the property of self-renewal (P2 > P1) (Fig. 2d).
Fig. 2.
Metformin (MET) altered growth and survival parameters and modified PTEN and FASN expression of HCT116 cells. a HCT116 cell viability after treatment with 60 μM MET for 48 h. b Treatment with MET for 48 h had no effect on cell cycle distribution, as determined by propidium iodide (PI) staining and subsequent FACS. c MET increased the number of apoptotic cells relative to control. Proportion of apoptotic cells was determined by Annexin V staining and FACS (“Materials and methods”), and the induction in apoptosis was associated with an increase in expression of the pro-apoptotic BAX gene (inset). d MET lowered P1 and P2 colonosphere formation of HCT116 cells. Results (a–d) are mean ± SEM; *P < 0.05 relative to control, with n = 2–3 independent experiments and each experiment performed in quadruplicates. e, f Transcript levels of PTEN and FASN genes at specific time points were quantified (QPCR) in control and MET-treated cells. Data are mean ± SEM of fold-change in normalized expression (n = 5 samples/group), relative to control. g, h Representative Western blots of whole cell extracts after treatment with MET. PTEN and FASN protein levels were compared to control (vehicle-treated cells). Each lane represents an individual treatment sample and contained 50 μg of total protein. Immunoreactive bands were quantified by densitometry; expression values (mean ± SEM) were normalized to those of the loading control β-actin and are presented as histograms. *P < 0.05 (relative to control)
We next quantified the temporal expression of the tumor suppressor PTEN and the lipogenic enzyme FASN in control and MET-treated cells. MET increased PTEN mRNA levels (Fig. 2e) and decreased FASN mRNA abundance (Fig. 2f) in a time-dependent manner. Interestingly, MET effects on PTEN and FASN gene expression were maximal at 6 h and persisted to 24 h (Fig. 2e, f) and 48 h (data not shown). The increase in PTEN and decrease in FASN mRNA abundance at 24 h with MET were confirmed at the level of their respective proteins (Fig. 2g, h), demonstrating concordance of mRNA and protein expression.
Soy bioactive components alter HCT116 cell viability, apoptosis, and colonosphere formation
To examine effects of soy bioactive components with potential anti-carcinogenic activities (Yan et al. 2010; Dia and Gonzales de Mejia 2011) on the cell cycle, HCT116 cells were treated with Gen, Lun, β-con, and Gly at concentrations (2–3 μM) that, in the case of Gen, approximated that found for sera of regular soy food consumers (Iwasaki et al. 2008). Treatment with the soy-derived components decreased cell viability although to differing extents, with the efficacy of Gen > Gly > β-con = Lun (Fig. 3a). There was an associated, although not a proportionate, decrease in cyclin D1 (CCND1) gene expression with loss of cell viability (Lun > Gen = Gly = β-con) (Fig. 3b). Gen, β-con, and Gly, but not Lun inhibited cell cycle progression relative to control cells (Fig. 3c). Specifically, Gen induced G1-S arrest, whereas β-con and Gly induced G2-M arrest (Fig. 3c). With apoptosis as the measured endpoint, Gen and Lun demonstrated enhancement (Gen > Lun), with β-con and Gly showing no comparable activities over control cells (Fig. 3d).
Fig. 3.
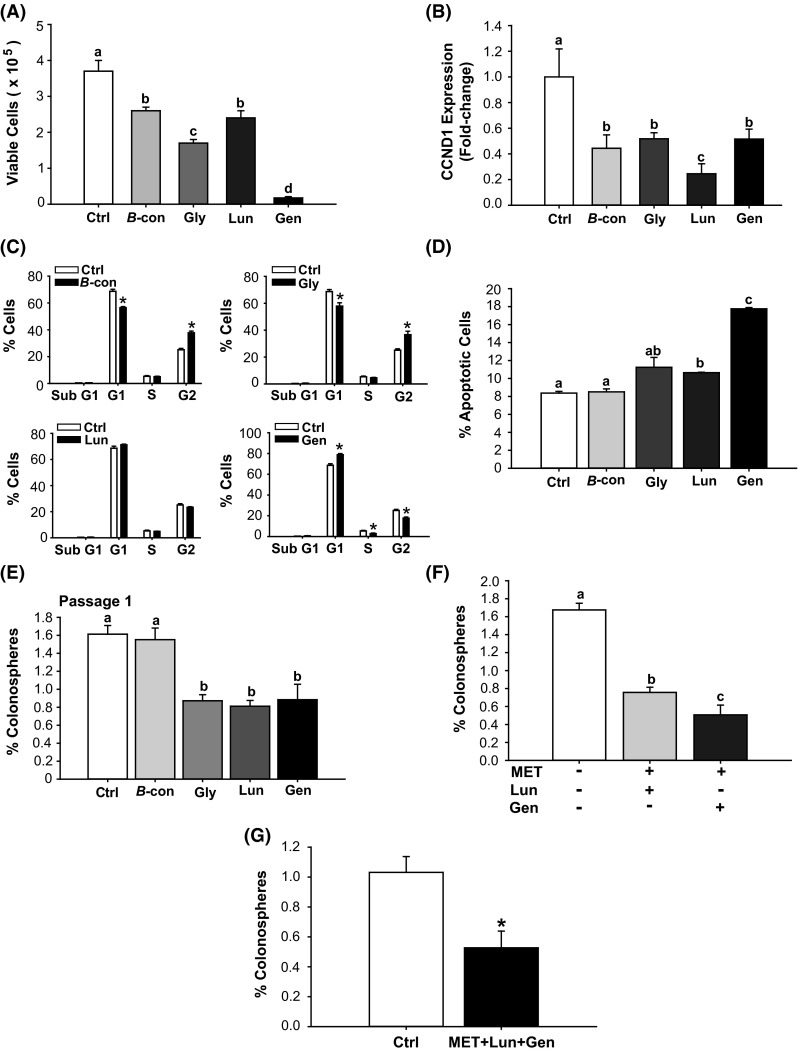
Bioactive soy factors are anti-proliferative, pro-apoptotic and colonosphere inhibitory in HCT116 cells. a HCT 116 cells were treated with the indicated bioactive factors (Gen 2 µM, Lun 2 µM, β-con 3 µM, and Gly 3 µM), and cell viability was measured 48 h later. b Transcript levels of cyclin D1 (CCND1) were determined 48 h after treatment with individual soy factors. Data (mean ± SEM) represent fold-change in normalized expression relative to control; bars with distinct letters differed from control values (P < 0.05). c Cell cycle distribution, determined by fluorescence-activated cell sorting, showed lower percentage of cells in the S and G2 phases and greater percentage of cells in G1, after Gen treatment. β-con and Gly induced lower percentages of cells in G1 and greater percentage of cells in G2. Lun had no effect on cell cycle parameters. Values are means ± SEMs from two independent experiments with each performed in quadruplicate. *P < 0.05 relative to control for each stage of cell cycle. d HCT116 cells were treated with 2 µM (Gen, Lun) or 3 µM (β-con, Gly) of soybean factor and 48 h later, were analyzed for the percentage of apoptotic cells. Lun and Gen induced cell apoptosis (Gen > Lun). Means with distinct letters differed at P < 0.05. e HCT116 cells were treated with soy factors (described above) or vehicle, which were added to plating medium in low attachment plates. Gly, Lun, and Gen diminished the percentage of P1 colonospheres. Means with different letters differed at P < 0.05. Data (mean ± SEM) are from three independent experiments, with each carried out in quadruplicate. f HCT116 cells were treated with MET + Lun or MET + Gen and evaluated for percentage of P1 colonosphere formation. Means with distinct letters differed at P < 0.05, (two independent experiments; each performed in quadruplicate). g Co-addition of MET + Gen + Lun repressed P1 colonosphere formation by 50 %. Data are mean ± SEM, two independent experiments each performed in quadruplicate. *P < 0.05 between groups
We next examined whether the above soy factors affected colonosphere formation in HCT116 cells. Gen, Lun, and Gly each suppressed colonosphere formation by ~50 %, while β-con was ineffective at the concentration tested (Fig. 3e). Co-treatment with MET and Lun reduced colonosphere formation to a slightly lesser extent (by ~60 %) than did co-treatment with MET and Gen (by ~76 %) (Fig. 3f). Interestingly, co-treatment with all three components failed to reduce colonosphere formation further than that elicited with MET in the presence of either Lun or Gen (Fig. 3g).
MET, Lun, and Gen limit the cancer stem cell-like cell subpopulation, in part, through PTEN
The changes in growth and survival elicited by Gen and by Lun (Fig. 3) were accompanied by decreased FASN and increased PTEN mRNA levels in P1 colonospheres (Fig. 4a). By contrast, P1 colonospheres from β-con- and Gly-treated cells failed to manifest changes in PTEN and FASN gene expression (Fig. 4a). The tumor suppressor PTEN is a common target for inactivation in cancer, including CRC (Rahal and Simmen 2010; Sawai et al. 2008; Molinari and Frattini 2013). We next evaluated whether induction of PTEN is responsible, in part, for the inhibitory effects of MET, Gen, and Lun on colonosphere formation. HCT116 cells were pre-treated with the PTEN inhibitor SF1670 (2 μM) for 24 h (untreated HCT116 cells served as control); treated cells were subsequently plated under non-adherent conditions with added MET (60 µM), Lun (2 µm), or Gen (2 µM). SF1670 binds to the PTEN active site, resulting in elevated phosphatidylinositol (3,4,5) triphosphate signaling (Rosivatz et al. 2006). We found that inhibition of PTEN activity increased colonosphere formation relative to control cells (Fig. 4b). MET, Lun, or Gen alone reduced the colonosphere-promoting effect of SF1670, with inhibition for Gen = Lun > MET (Fig. 4b).
Fig. 4.
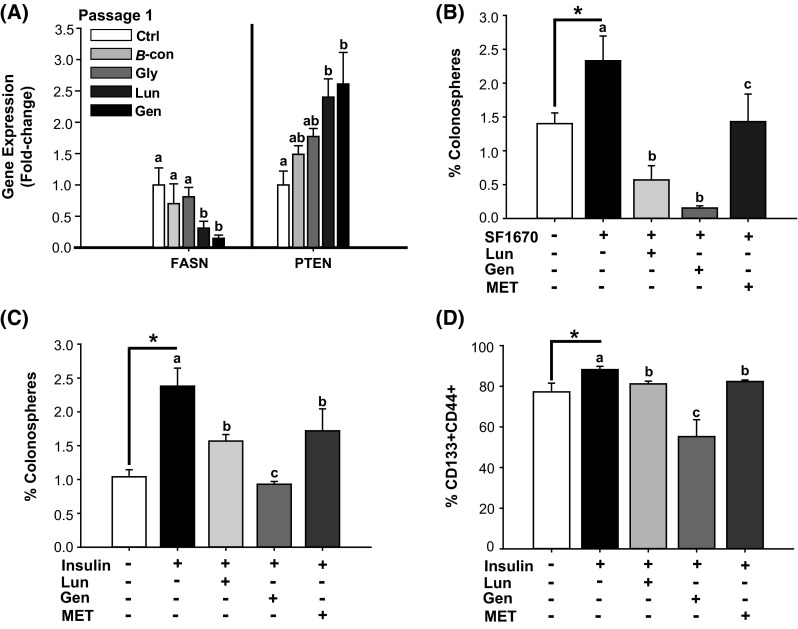
MET and soy factors inhibit colonosphere formation in part through regulation of FASN and PTEN gene expression. a Lun and Gen decreased FASN mRNA abundance and induced PTEN transcript levels in P1 colonospheres formed from unsorted HCT116 cells. Means with distinct letters differed at P < 0.05, relative to untreated cells (n = 4 independent samples). b Colonosphere formation in HCT116 cells treated with Lun, Gen, or MET in the presence of the PTEN inhibitor SF1670. HCT116 cells were pre-treated with SF1670 (2 µM) for 24 h prior to plating under low attachment conditions with added soy factors (Gen, Lun) and MET. P1 colonospheres were quantified after 5 days. *P < 0.05 between control and SF1670-treated cells. Means with distinct letters differed at P < 0.05 (two independent experiments, each in quadruplicates). c Insulin added to 2 µM final concentration increased P1 colonosphere formation (*P < 0.05 between control and insulin-treated group). Treatment with Lun, Gen, or MET reduced colonosphere formation (Gen > Lun = MET) under insulin supplementation. Mean ± SEM are from two independent experiments, each performed in quadruplicate. Means with different letters differed at P < 0.05. d FACS analysis, using stem cell surface antigen markers CD133 and CD44, of attached HCT116 cells treated with Lun, Gen, or MET for 72 h in the presence of insulin (2 μM final concentration). Addition of each factor decreased the percentage of CD133+CD44+ cells, relative to insulin-treated cells. Data (mean ± SEM) are from three independent experiments, each performed in quadruplicate. *P < 0.05 between control and insulin-treated groups. Means with different letters differed at P < 0.05
Epidemiological studies suggest that hyperinsulinemia is associated with increased CRC risk (Giovannucci 2007). Moreover, insulin signaling was found to inhibit PTEN expression (Liu et al. 2014), and insulin resistance has been linked to increased FASN expression (Menendez et al. 2009), indicating that aberrant insulin signaling may underlie altered PTEN and FASN expression levels and activity in CRC. To evaluate whether Lun, Gen, and/or MET attenuate CSC frequency by opposing insulin action, we exposed HCT116 cells to an elevated level of insulin (2 µM, modeling hyperinsulinemia) and determined effects of added factors on colonosphere formation and on the CD133+CD44+ subpopulation. Insulin increased colonosphere formation (Fig. 4c) and the percentage of CD133+CD44+ cells (Fig. 4d). Lun, Gen, and MET reduced CSC formation under in vitro hyperinsulinemic condition, with Gen demonstrating greater inhibitory activity than either MET and Lun (MET = Lun) (Fig. 4c, d).
HCT116 cell killing by 5-FU plus MET, Gen, and Lun
5-Fluorouracil (5-FU) is a first-line chemotherapeutic agent for CRC treatment. To examine whether combination treatment with Lun + Gen + MET enhances the sensitivity of HCT116 cells to the cytotoxic effects of 5-FU under a hyperinsulinemic condition, cells were treated with insulin (2 µM) and 5-FU in the presence of MET alone or in combination with Lun + Gen (Fig. 5a, b). Amount of apoptosis induced by 5-FU in the presence of insulin was similar to that elicited by MET + Gen + Lun alone (in the absence of 5-FU) but was not affected by co-addition of only MET (Fig. 5a, b). However, co-addition of MET + Gen + Lun under hyperinsulinemic conditions modestly but significantly increased 5-FU effects on apoptosis (Fig. 5a, b) and cell viability (Fig. 5c) relative to the effects seen with MET alone. Since we only tested combinations of factors at a single dose for each, it is not possible to discern whether effects were additive or synergistic. In addition, exposure to MET + Gen + Lun for 72 h significantly attenuated (by ~20 %) the percentage of CD133+CD44+ cells, when compared to 5-FU alone-treated cells (Fig. 5d).
Discussion
CRC is increasingly considered to have metabolic underpinnings; hence, dietary factors that maintain metabolic homeostasis or normalize metabolic dysfunction may be beneficial to inhibiting CRC progression, metastasis, and relapse. Clinical and epidemiological studies have implicated the anti-diabetic drug metformin (MET) in reducing the risk of CRC among diabetic patients as well as improving CRC outcome in patients with diabetes (Dowling et al. 2011; Zhang et al. 2013; Nangia-Makker et al. 2014; Sehdev et al. 2015). Additionally, in diabetic patients with CRC, the decrease in mortality with MET was inversely associated with the frequency of CD133+ subpopulation with CSC properties (Zhang et al. 2013). Nevertheless, MET effects on colon cancer remain controversial (Sui et al. 2014). Moreover, MET elicits unfavorable side effects in some individuals, precluding its use at typically prescribed doses for diabetic patients. Thus, factors that could increase the efficacy of MET may be advantageous for its inclusion as a therapy to address CRC prevention and/or treatment.
Previous studies in several laboratories including our own (Xiao et al. 2008; Al-Dwairi et al. 2012; Yeh et al. 2014; Isanga and Zhang 2011) have shown that soybean components when consumed can favorably affect metabolic pathways and associated genes in animal models and in humans. Moreover, in a mouse model of breast cancer and in breast cancer cells in vitro, dietary intake of soy protein isolate and treatment with Gen, respectively, decreased the frequency of the mammary basal stem cell-like subpopulation, which was associated with decreased mammary tumor incidence and oncogenic molecular profile (Montales et al. 2012; Rahal et al. 2013). Based on this previous body of work, we hypothesized that combination(s) of MET and bioactive soy factors would increase the therapeutic efficacy of the CRC drug 5-FU by targeting cancer stem cells. Our findings present a novel and potentially important role for combination MET, Lun, and Gen in not only reducing cancer cell viability and survival, but in limiting the frequency of the cancer cell subpopulation (CD133+CD44+) with self-renewal properties. We also provide evidence, using the PTEN-specific inhibitor SF1760 and insulin, to indicate that MET, Gen, and Lun actions are comparably mediated by their concurrent induction of tumor suppressor PTEN and inhibition of pro-oncogenic FASN expression. The functional significance of these results is supported by the increased efficacy of 5-FU in eliminating CSCs under high-insulin conditions, the latter mimicking aspects of type 2 diabetes and/or obesity, when co-exposed with MET, Gen, and Lun.
In these studies, we primarily utilized the human colon cancer cell line HCT116 since its highly aggressive nature and increased proportion of non-differentiating CSCs are clinically correlated with lymph node metastases and reduced survival (Yeung et al. 2010). We show that relative to the more differentiated, less aggressive colon cancer cell line HT29, HCT116 cells exhibit a greater proportion of the CSC subpopulation (designated CD133+CD44+), increased ability to form colonospheres (a measure of tumor-forming ability and self-renewal in vitro), and enhanced expression of CSC genes. Using these cells to model aggressive CRC in vitro, we observed that MET, Gen, and Lun, individually and together, limited the expansion of the CSC subpopulation, suggesting their potential value in inhibiting disease progression and conceivably, metastasis and recurrence. The higher efficacy of Gen relative to MET, at the doses used here, in reducing colonosphere formation under high-insulin conditions is a novel and an unexpected finding. Previous studies have shown a dose-related inverse association between soy consumption and CRC risk (Yan et al. 2010). Nevertheless, molecular mechanisms to explain the preventative effects of soy on colon cancer remain relatively unknown. The demonstration of the efficacy of Gen, Lun, and Gly in targeting CSCs at relatively low doses (2–3 uM) suggests their potential to reduce currently prescribed doses of CRC conventional therapies with adverse side effects. While our data point to potential combination therapies of soybean components and 5FU, the current study has not attempted the complex task of defining optimal doses for these single agents and combinations (future studies). Our findings also provide an impetus to pursue preclinical studies of these bioactive molecules in animal models of colon cancer metastasis and under conditions of hyperinsulinemia.
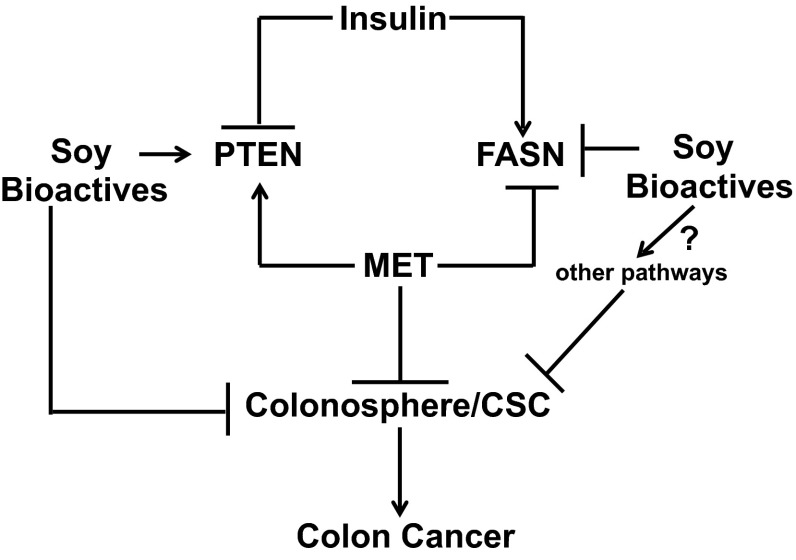
The present data identify PTEN and FASN as target genes for MET, Lun, and Gen in the CSC subpopulation (Fig. 6). The bioactive molecules did not demonstrate additive or synergistic effects on PTEN and FASN expression, possibly indicating that once the threshold induction (for PTEN) or reduction (for FASN) in these signaling pathways is achieved, further blocking of the CSC self-renewal/expansion by these molecules becomes limited. The latter idea is consistent with the notion that additional pathways independent of MET, Lun, and Gen signaling regulate CSC expansion. Our observations that MET alone was less effective than MET + Gen in inhibiting CSC expansion, however, suggest that Gen affects other pathways to further limit CSC expansion and hence, CRC (Fig. 6). The identities of these potential contributing pathways are unknown, but may involve inflammatory, cytokine/chemokine, and epithelial–mesenchymal transition signaling, each of which has been separately linked to the promotion of CSCs when aberrantly activated (Bruttel and Wischhusen 2014) and to the favorable health effects of soy when inhibited (Hsu et al. 2010).
Fig. 6.
Model that integrates the potential pathways by which MET and soy bioactives influence the frequency of cancer stem-like cell population. The pathways demonstrate the involvement of the tumor suppressor PTEN and the lipid biosynthesis-associated enzyme FASN under obesogenic/diabetic conditions. Designations (→, ⊥) indicate promotion and inhibition, respectively, of a specific pathway. The identities of other possible pathways mediated by soy bioactives are unknown (indicated by question mark)
Conclusions
The present study utilizing an in vitro model of aggressive colon cancer in a hyperinsulinemic environment, provides insight into molecular networks shared by the well-prescribed, anti-diabetic drug MET and bioactive/bioavailable soy food components, which potentially may be harnessed to increase drug efficacy in patients with CRC. Additionally, given that PTEN loss of function/expression is a hallmark of many primary and metastatic CRCs, our results suggest further exploration of the potential of combination therapy with MET and the soy factors to improve CRC outcome. Induction of tumor suppressor PTEN expression in cancer cells by dietary bioactive factors is an emerging theme (Liu et al. 2013; Pabona et al. 2013) that warrants further confirmation in vivo. In addition, our results for β-conglycinin and glycinin suggest further examination of these molecules for efficacy in CRC, in light of their high abundance and bioavailability either as intact or smaller proteins/peptides in consumed soy products. The mode(s) of action of these two latter proteins in cancer cell growth regulation remain virtually unexplored, with the exception of a previous report on β-conglycinin as being inhibitory to leukemia cell growth in vitro (Wang et al. 2008). Finally, the identification of food constituents that target CSCs underscores the potential importance of diet/nutrition in the genesis (and treatment) of primary and metastatic CRC and of other cancer types with metabolic underpinnings.
Electronic supplementary material
Acknowledgments
We thank other members of our laboratories for helpful discussions during the course of this work. We are also grateful to Lorenzo Fernandes and Eric Siegel for their assistance with statistical analysis and data presentation.
Authors’ contributions
MTM carried out the experiments and performed statistical analysis. EF and VN performed the extraction and purification of β-conglycinin and glycinin. MTM, RCMS, and FAS interpreted the data and drafted the manuscript. RCMS and FAS supervised the entire project. All authors read and approved the final manuscript.
Funding
This work was supported by NIH R01CA136493, NIH UL1TR000039, and the Huie Family Trust.
Abbreviations
- β-con
β-Conglycinin
- CSC
Cancer stem cell
- CRC
Colorectal cancer
- FASN
Fatty acid synthase
- 5-FU
5-Fluorouracil
- Gen
Genistein
- Gly
Glycinin
- Met
Metformin
- PTEN
Phosphatase and tensin homologue deleted on chromosome ten
- QPCR
Real-time quantitative polymerase chain reaction
References
- Al-Dwairi A, Pabona JM, Simmen RC, Simmen FA. Cystolic malic enzyme 1 (ME1) mediates high fat diet-induced adiposity, endocrine profile, and gastrointestinal tract proliferation-associated biomarkers in male mice. PLoS ONE. 2012;7(10):e46716. doi: 10.1371/journal.pone.0046716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dwairi A, Brown AR, Pabona JM, Van TH, Hamdam H, Mercado CP, Quick CM, Wight PA, Simmen RC, Simmen FA. Enhanced gastrointestinal expression of cytosolic malic enzyme (ME1) induces intestinal and liver lipogenic gene expression and intestinal cell proliferation in mice. PLoS ONE. 2014;9(11):e113058. doi: 10.1371/journal.pone.0113058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruttel VS, Wischhusen J. Cancer stem cell immunology: key to understanding tumorigenesis and tumor immune escape? Front Immunol. 2014;5:360. doi: 10.3389/fimmu.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JM Cancerous. Cancerous stem cells. Deviant stem cells with cancer causing misbehavior. Stem Cell Res Ther. 2010;1:13. doi: 10.1186/scrt13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KL, Pan F, Jiang H, Chen JF, Pei L, Xie FW, Liang HJ. Highly enriched CD133+CD44+ stem-like cells with CD133+CD44hi metastatic subset in HCT 116 colon cancer cells. Clinical Exp. Metastasis. 2011;28:751–763. doi: 10.1007/s10585-011-9407-7. [DOI] [PubMed] [Google Scholar]
- Dia VP, Gonzales de Mejia E. Lunasin potentiates the effect of oxaliplatin preventing outgrowth of colon cancer metastasis, binds to α5β1 integrin and suppresses FAK/ERK/NF-κB signaling. Cancer Lett. 2011;313:168–180. doi: 10.1016/j.canlet.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Goodwin P, Stambolic V. Understanding the benefit of metformin use in cancer treatment. BMC Med. 2011;9:33. doi: 10.1186/1741-7015-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira E, Silva M, Demonte A, Neves V. β-Conglycinin (7S) and glycinin (11S) exert a hypocholesterolemic effect comparable to that of fenofibrate in rats fed a high-cholesterol diet. J Funct Foods. 2010;2:275–283. doi: 10.1016/j.jff.2010.11.001. [DOI] [Google Scholar]
- Gilbert ER, Liu D. Anti-diabetic functions of soy isoflavone genistein: mechanisms underlying its effects on pancreatic β-cell function. Food Funct. 2013;4:200–212. doi: 10.1039/C2FO30199G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am J Clin Nutr. 2007;86:s836–s842. doi: 10.1093/ajcn/86.3.836S. [DOI] [PubMed] [Google Scholar]
- Hsu A, Bray TM, Ho E. Anti-inflammatory activity of soy and tea in prostate cancer prevention. Exp Biol Med. 2010;235(6):659–667. doi: 10.1258/ebm.2010.009335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isanga J, Zhang G-N. Soybean bioactive components and their implications to health—a review. Food Rev Int. 2011;24:252–276. doi: 10.1080/87559120801926351. [DOI] [Google Scholar]
- Iwasaki M, Inoue M, Otani T, Sasazuki S, Kurahashi N, Miura T, Yamamoto S, et al. Plasma isoflavone levels and subsequent risk of breast cancer among Japanese women: a nested case-control study from the Japan Public Health Center-based prospective study group. J Clin Oncol. 2008;26:1677–1683. doi: 10.1200/JCO.2007.13.9964. [DOI] [PubMed] [Google Scholar]
- Kakarala M, Brenner D, Korkaya H, Cheng C, Tazi K, Ginestier C, Liu S, Dontu G, Wicha MS. Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res Treat. 2010;122:777–785. doi: 10.1007/s10549-009-0612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CL, Huang PI, Tsai PH, Lo JF, Lee YY, Chen YJ, Chen YW, Chiou SH. Resveratrol-induced apoptosis and increased radiosensitivity in CD133-positive cells derived from atypical teratoid/rhabdoid tumor. Int J Radiat Oncol Biol Phys. 2009;74:219–228. doi: 10.1016/j.ijrobp.2008.12.035. [DOI] [PubMed] [Google Scholar]
- Linnekamp JF, Wang X, Medema JP, Vermeulen L. Colorectal cancer heterogeneity and targeted therapy: a case for molecular disease subtypes. Cancer Res. 2015;75:245–249. doi: 10.1158/0008-5472.CAN-14-2240. [DOI] [PubMed] [Google Scholar]
- Liu YL, Zhang GQ, Yang Y, Zhang CY, Fu RX, Yang YM. Genistein induces G2/M arrest in gastric cancer cells by increasing the tumor suppressor PTEN expression. Nutr Cancer. 2013;65:1034–1041. doi: 10.1080/01635581.2013.810290. [DOI] [PubMed] [Google Scholar]
- Liu J, Visser-Grieve S, Boudreau J, Yeung B, Lo S, et al. Insulin activates the insulin receptor to downregulate the PTEN tumour suppressor. Oncogene. 2014;33:3878–3885. doi: 10.1038/onc.2013.347. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Vazquez-Martin A, Ortega FJ, Fernandez-Real JM. Fatty acid synthase: association with insulin resistance, type 2 diabetes, and cancer. Clin Chem. 2009;55:425–438. doi: 10.1373/clinchem.2008.115352. [DOI] [PubMed] [Google Scholar]
- Molinari F, Frattini M. Functions and regulation of the PTEN gene in colorectal cancer. Front Oncol. 2013;3:326. doi: 10.3389/fonc.2013.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montales MT, Rahal O, Kang J, Rogers TJ, Prior R, Wu X, Simmen RC. Repression of mammosphere formation of human breast cancer cells by soy isoflavone genistein and blueberry polyphenolic acids suggests diet-mediated targeting of cancer stem-like/progenitor cells. Carcinogenesis. 2012;33:652–660. doi: 10.1093/carcin/bgr317. [DOI] [PubMed] [Google Scholar]
- Nangia-Makker P, Yu Y, Vasudevan A, Farhana L, Rajendara SG, Levi E, Majumdar AP. Metformin: a potential therapeutic agent for recurrent colon cancer. PLoS ONE. 2014;9(1):e84369. doi: 10.1371/journal.pone.0084369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs R. Metformin: a case study in the meaning of serum concentrations. J Mol Pharm Org Process Res. 2014;2:e111. doi: 10.4172/2329-9053.1000e111. [DOI] [Google Scholar]
- Pabona JM, Dave B, Su Y, Montales MT, de Lumen BO, et al. The soybean peptide lunasin promotes apoptosis of mammary epithelial cells via induction of tumor suppressor PTEN: similarities and distinct actions from soy isoflavone genistein. Genes Nutr. 2013;8:79–90. doi: 10.1007/s12263-012-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paldino E, Tesori V, Casalbore P, Gasbarrini A, Puglisi MA. Tumor initiating cells and chemoresistance: which is the best strategy to target colon cancer stem cells? Biomed Res Int. 2014;2014:859871. doi: 10.1155/2014/859871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal O, Simmen RC. PTEN and p53 cross-regulation induced by soy isoflavone genistein promotes mammary epithelial cell cycle arrest and lobuloalveolar differentiation. Carcinogenesis. 2010;31:1491–1500. doi: 10.1093/carcin/bgq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal O, Machado H, Montales MT, Pabona JM, Heard M, Nagarajan S, Simmen RC. Dietary suppression of the mammary CD29(hi)CD24(+) epithelial subpopulation and its cytokine/chemokine transcriptional signatures modifies mammary tumor risk in MMTV-Wnt1 transgenic mice. Stem Cell Res. 2013;11:1149–1162. doi: 10.1016/j.scr.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosivatz E, Matthews J, McDonald N, Mulet X, Ho KK, Lossi N, et al. A small-molecule inhibitor for phosphatase and tensin homologue deleted on chromosome 10 (PTEN) ACS Chem Biol. 2006;1:780–790. doi: 10.1021/cb600352f. [DOI] [PubMed] [Google Scholar]
- Rouse M, Younes A, Egan JM. Resveratrol and curcumin enhance pancreatic β-cell function by inhibiting phosphodiesterase activity. J Endocrinol. 2014;223:107–117. doi: 10.1530/JOE-14-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlow B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS ONE. 2014;9(4):e94621. doi: 10.1371/journal.pone.0094621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Yasuda A, Ochi N, Ma J, Matsuo Y, et al. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterol. 2008;8:56. doi: 10.1186/1471-230X-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehdev A, Shih YC, Vekhter B, Bissonnette MB, Olopade O, Polite BN. Metformin for primary colorectal cancer prevention in patients with diabetes: a case-control study in a US population. Cancer. 2015;121(7):1071–1078. doi: 10.1002/cncr.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller K, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Spector D, Anthony M, Alexander D, Arab L. Soy consumption and colorectal cancer. Nutr Cancer. 2013;47:1–12. doi: 10.1207/s15327914nc4701_1. [DOI] [PubMed] [Google Scholar]
- Sui X, Xu Y, Yang J, Fang Y, Lou H, Han W, et al. Use of metformin alone is not associated with survival outcomes of colorectal cancer cell but AMPK activator AICAR sensitizes anti-cancer effect of 5-fluorouracil through AMPK activation. PLoS ONE. 2014;9(5):e97781. doi: 10.1371/journal.pone.0097781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Marchionni L, Nowak M, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci USA. 2014;112:118–123. doi: 10.1073/pnas.1421839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi AE. The soy isoflavone genistein reverses oxidative and inflammatory state, neuropathic pain, neurotrophic and vasculature deficits in diabetes mouse model. Eur J Pharmacol. 2013;650:694–702. doi: 10.1016/j.ejphar.2010.10.060. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bringe NA, Berhow MA, de Meija Gonzalez. Beta-conglycinins among sources of bioactives in hydrolysates of different soybean varieties that inhibit leukemia cells in vitro. J Agric Food Chem. 2008;56:4012–4020. doi: 10.1021/jf8002009. [DOI] [PubMed] [Google Scholar]
- White AC, Lowry W. Refining the role for adult stem cells as cancer cells of origin. Trends Cell Biol. 2015;25:11–20. doi: 10.1016/j.tcb.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Carter JA, Linz AL, Ferguson M, Badger TM, Simmen FA. Dietary whey protein lowers serum C-peptide concentration and duodenal SREBP-1c mRNA abundance, and reduces occurrence of duodenal tumors and colon aberrant crypt foci in azoxymethane-treated male rats. J Nutr Biochem. 2006;17:626–634. doi: 10.1016/j.jnutbio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Xiao R, Su Y, Simmen RC, Simmen FA. Dietary soy protein inhibits DNA damage and cell survival of colon epithelial cells through attenuated expression of fatty acid synthase. Am J Physiol Gastrointest Liver Physiol. 2008;294:G868–G876. doi: 10.1152/ajpgi.00515.2007. [DOI] [PubMed] [Google Scholar]
- Yan L, Spitznagel EL, Bosland MC. Soy consumption and colorectal cancer risk in humans: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19:148–158. doi: 10.1158/1055-9965.EPI-09-0856. [DOI] [PubMed] [Google Scholar]
- Yeh WJ, Yang HY, Chen JR. Soy β-conglycinin retards progression of diabetic nephropathy via modulating the insulin sensitivity and angiotensin-converting enzyme activity in rats fed with high salt diet. Food Funct. 2014;5:2898–2904. doi: 10.1039/C4FO00379A. [DOI] [PubMed] [Google Scholar]
- Yeung TM, Gandhi S, Wilding J, Muschel R, Bodmer W. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci USA. 2010;107:3722–3727. doi: 10.1073/pnas.0915135107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guan M, Zheng Z, Zhang Q, Gao F, Xue Y. Effects of metformin on CD133+ colorectal cancer cells in diabetic patients. PLoS ONE. 2013;8(11):e81264. doi: 10.1371/journal.pone.0081264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.