Abstract
Protein-ligand binding accompanies changes in the surrounding electrostatic environments of the two binding partners and may lead to changes in protonation upon binding. In cases where the complex formation results in a net transfer of protons, the binding process is pH-dependent. However, conventional free energy computations or molecular docking protocols typically employ fixed protonation states for the titratable groups in both binding partners set a priori, which are identical for the free and bound states. In this review, we draw attention to these important yet largely ignored binding-induced protonation changes in protein-ligand association by outlining physical origins and prevalence of the protonation changes upon binding. Following a summary of various theoretical methods for pKa prediction, we discuss the theoretical framework to examine the pH dependence of protein-ligand binding processes.
Introduction
Complex formation between a protein and small molecules comprises one of the most fundamental reactions in biology. The association may not only result in conformational rearrangements, but also may induce changes in the pKa values of titratable groups on either binding partner due to the altered electrostatic environment upon binding.1 While the binding-induced structural changes have been explored extensively,2 however, the shifts in the pKa, or protonation state, of the titratable groups that accompany the binding have been less well explored. In addition, the changes in protonation accompanying protein-small molecule binding1,3 have been relatively rarely analyzed compared to the analogous changes in protein-protein complex formation.4-8 However, as an estimated 60-80% of orally administered drugs are weak acids or bases, the protonation states of bound ligands can also be tuned by cellular pH and electrostatic environment of their protein binding partners.9,10 In cases where protein-ligand binding accompanies a net transfer of protons to either binding partner, the binding process is pH-dependent, i.e., the observed binding free energy is a function of pH.
However, conventional biomolecular modeling or free energy computations involving the complex formation typically employ fixed protonation states for the titratable groups in both binding partners set a priori, which are identical for the free and bound states. Clearly, in cases where ligand binding accompanies a net proton transfer to the system, this assumption ignores the possibility of protonation states changing while the chemical environments change upon binding. Consequently, when the true ensemble of conformations consists of various electrostatic environments, the use of a single, fixed protonation state may hinder the accurate description of the ensemble and can lead to significant errors.1
In this review, we outline the physical origins and prevalence of changes in protonation that accompanies the protein-small molecule complex formation; a recent review by Onufriev and Alexov very thoroughly analyzed the binding-induced pKa shifts for protein-protein, protein-small molecule, and protein-nucleic acid binding.1 We further review the theoretical methods for predicting the pKa values of titratable groups in protein-ligand binding. Finally, we address the computational protocol for obtaining pH-dependent binding free energies for the associations accompanying a net proton transfer and provide a brief outlook on the field.
Physical Origins and Prevalence of Binding-induced Protonation Changes
The observed pKa value of a titratable residue is generally obtained as a sum of the residue's intrinsic pKa in solution, pKaint, and the shift in pKa arising from the surrounding environment, ΔpKa:11,12
| (1) |
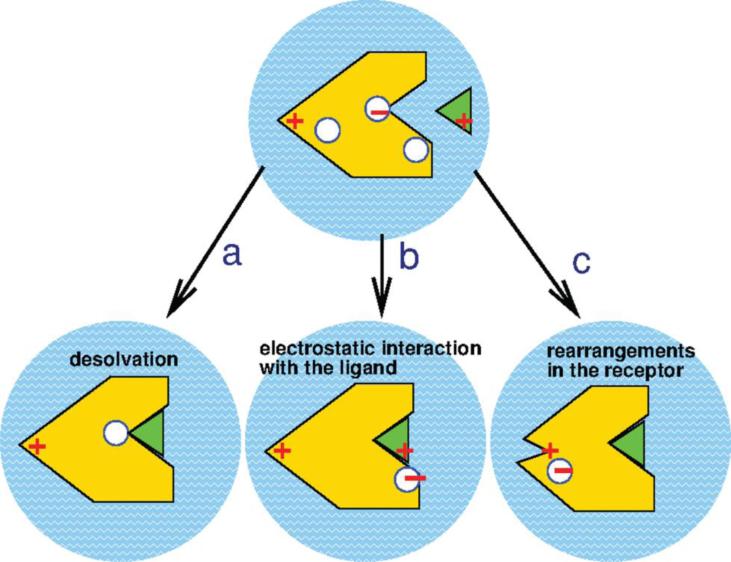
The shift in pKa of titratable residues from its pKaint is typically due to the (a) desolvation penalty; (b) direct electrostatic interaction between the protein and bound ligand; and (c) charge rearrangement following the structural change upon binding (Figure 1). The desolvation, also referred to as the Born contribution, typically favors uncharged residue because the solvent-protein interaction at the binding site is replaced by the bound ligand. The last two causes of ΔpKa together comprise the “background” contribution to the ΔpKa and include changes in hydrogen bonding pattern and interactions involving other polar or charged groups. These background effects may propagate to the titratable groups distant from the immediate binding interface due to the structural and electrostatic rearrangement, resulting in the so-called allosteric effect on the pKa shift.
Figure 1.
Three major mechanisms behind the shift in pKa. (a) Desolvation. (b) Direct protein-ligand electrostatic interaction. (c) Charge rearrangement upon structural change. The figure is adopted from Onufriev and Alexov (2013).1
A recent computational survey by Aguilar et al. showed the prevalence of binding-induced protonation changes for protein-small molecule complexes as well as protein-protein and protein-nucleic acid complexes by performing continuum electrostatic calculations on both free and bound forms of either binding partner.13 In 60% of protein-small molecule complexes considered, it was shown that at least one titratable residue completely changed its protonation state at neutral pH upon binding. It was found that all three of the above-mentioned causes of ΔpKa play roles in the changes in protonation, where the direct electrostatic effects, i.e., (a) and (b), are short-ranged and the strongest, while the charge rearrangement upon structural reorganization, i.e. (c) affects more distant residues, propagating as far as 24 Å from the binding interface.13
While the work by Aguilar et al. comprises the largest scale survey for protein-small molecule complexes to date, individual instances of binding-induced shifts in pKa were reported as well both experimentally14-23 and computationally.3,24-28 From Table 1, it is apparent that the pKa shifts of varying magnitudes are observed for the titratable residues in protein upon binding of inhibitor. In addition, several works noted the changes in pKa of titratable groups in the bound ligand upon binding to receptor, highlighting that the binding-induced changes in pKa are not limited to the protein partner.29-31
Table 1.
Experimentally observed shifts in pKa (ΔpKa) in protein-small molecule binding.
| Protein | Ligand | Titratable group | ΔpKa | PDB ID of free protein | PDB ID of complex |
|---|---|---|---|---|---|
| HIV-1 protease | Pepstatin | Asp25 | >+2.214 | 3HVP | 1HPX |
| Asp25’ | <−1.514 | 3HVP | 1HPX | ||
| HIV-1 protease | DMP-32 | Asp25/Asp25’ | +2.1915 | 1HHP | 1QBS |
| Plasmepsin II | Pepstatin | Asp34/Asp214 | +0.116 | 1PFZ | 1SME |
| His164 | +1.516 | 1PFZ | 1SME | ||
| Chymotrypsin | N-Acetyl-L-Pbhe-CF3 | His57 | +3.317 | 6GCH | 6GCH |
| Xylanase | 2FXb | Glu172 | −2.518 | 1BVV | 1BVV |
| Hydroxynitrile lyase | Thiocyanate | His235 | +5.519 | 2YAS | 2YAS |
| TSG-6 | Hyaluronan | His4 | +0.520 | 1O7B | 1O7C |
| Galectin-1 | β-Galactoside | His44/His52 | +0.321 | 1W6N | 2ZKN |
Theoretical Methods for pKa Prediction
The majority of computational pKa prediction algorithms employ continuum electrostatics models for their simplicity and speed. In the continuum electrostatics, the bulk aqueous environment is represented as an infinite, high-dielectric continuum and the protein is modeled as a low-dielectric, polarizable cavity with fixed point charges.11,32,33 The continuum approach includes Poisson-Boltzmann method34,35 and more simplified generalized Born (GB) approximation,36,37 whose underlying theories are well described in the references provided. The continuum approaches allow enhanced conformational sampling through an instantaneous dielectric response of the solute partial charges to the change in solvent dielectric and shows reliable results in computational pKa prediction.11,12,25,38-42
Structural flexibility of protein4,36,43-46 and dielectric heterogeneity47,48 have been further taken into account for more realistic description of protein conformational variability in pKa computation. The protein-dipole Langevin dipole (PDLD) model of Warshel and coworkers represents protein as polarizable dipole while the nearby solvent is described as Langevin dipoles.49 Also, quantum mechanics/molecular mechanics (QM/MM) method has been coupled with free energy computations for more rigorous pKa computations.50,51
More recent advances in the computational pKa prediction protocols couple conformational and protonation sampling by applying molecular dynamics (MD) technique that employs pH as an external thermodynamic variable. In these constant pH MD (CpHMD) methods, conformations of the system are sampled with MD and the electrostatics are periodically analyzed using various methods including continuum electrostatics, MD, or Monte Carlo (MC). Various flavors of the CpHMD techniques have been developed, which are often classified as either continuous or discrete models depending on the treatment of titratable protons in the simulation. The former considers protonation state as a continuous titration parameter that propagates with the atomic coordinates of the system.52-54 The use of mean-field approximation in the earlier implementation of the continuous CpHMD methods, however, allowed the unphysical fractional protonation states. Therefore, Lee et al. developed a novel continuous titration approach to avoid fractional protonation states by coupling λ-dynamics and applying an artificial titration barrier.55
On the other hand, in the discrete protonation state model, the titratable site is considered either protonated or deprotonated, avoiding the intermediate charge state.56-58 The stochastic titration model of Baptista et al. comprises of short segments of MD simulations periodically interrupted by MC for electrostatic energy evaluation using PB.56 Mongan et al. further applied the GB implicit solvent in the MC sampling scheme to the stochastic method of Baptista et al. for enhanced computational and sampling efficiency.59 On the other hand, Bürgi et al. avoided the continuum approach by computing electrostatic free energy by more rigorous yet expensive thermodynamic integration.60 Recent improvements on both continuous and discrete protonation models of the CpHMD methods include the application of enhanced sampling techniques such as replica exchange61-64 or accelerated MD65 in order to increase the convergence. To date, CpHMD methods have been successfully applied to predict pKa values of titratable groups in proteins28,53-56,59-65 and nucleic acids,66-68 as well as to explain the acid-base catalysis by RNase A69 and to understand the mechanisms behind the pH-dependent conformational changes critical to the function of proteins such as nitrophorin,70 rhodopsin,71 and BACE-1.28
Computation of pH-dependent binding free energies for protein-ligand complex
As stated above, simulations of protein-ligand systems are typically preceded by the assignment of fixed protonation states to titratable groups on the two binding partners. Further, docking studies often employ empirical prediction algorithms to assign fixed protonation states to the free ligands being docked. These approaches, however, fail to account for changes in protonation that may follow from the altered electrostatic environment surrounding the two binding partners upon complex formation. For instance, the pKa values of the titratable groups reported in Table 1 fall into the physiological pH range between 4 and 8, indicating that the changes in protonation upon binding can occur in biologically relevant conditions. Clearly, in cases where ligand binding is linked to the (un)binding of protons, the error in assigning incorrect protonation states in free energy computations without correcting for the pH dependence of the binding free energy can give errors greater in magnitude than the errors from typical free energy computations.72
Recently, groups of Jensen and Alexov examined the pH dependence of protein-protein binding6,8 through an application of the binding polynomial formalism devised by Wyman73 and used by Tanford to describe protein folding/unfolding.74 Based on the binding polynomial, the pH dependence of the binding free energy can be expressed as:
| (2) |
where the last two terms are corrections to the reference binding affinity, ΔG°ref, in which the binding-induced protonation change is ignored. The pKaC and pKaF are the pKa value of the complexed and free states of the receptor, respectively. This formalism for obtaining ΔG as a function of pH can further be applied to cases where multiple ligand and receptor groups titrate in the pH range considered, assuming that proton binding occurs independently. In other words, Eq. 2 can only be applied when all titratable groups are uncoupled from each other.
As protein active sites often contain multiple titratable groups whose protonation states are coupled to perform a given function, it will sometimes be wrong to assume that all titratable groups remain uncoupled upon ligand binding. In such cases, the use of a relation devised by Wyman75 provides a thermodynamic relation for proton-linked ligand binding where titratable sites may interact:
| (3) |
where ZPL, ZP, and ZL are the total charges for protein-ligand complex, protein, and ligand, respectively. This approach has been used to account for the pH dependence of protein-protein6,8 and protein-ligand binding,3,28 as well as protein stability.74,76
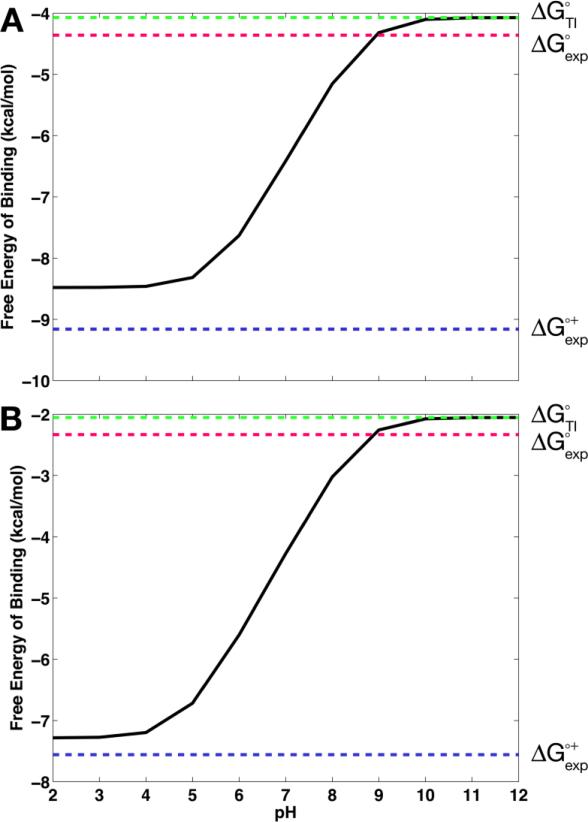
Within the binding framework, these expressions have often been used in conjunction with computational pKa predictions such as PROPKA77 or MCCE,46 as in Mason and Jensen6 and Mitra et al.8 However, due to the limited conformational sampling in such pKa computation algorithms, the coupling of conformational and protonation equilibria has not been fully accounted for. In addition, while the binding polynomial-based method has been applied to protein-protein6,8 and protein-nucleic acid binding, there had been no standard protocol to rigorously account for proton-linked ligand binding.67,78,79 Therefore, following the theoretical foundations of these studies, we recently applied the binding polynomial formalism with the CpHMD framework to compute the pH-dependent binding free energies.80 In this computational protocol, the correction terms in Eqs. 2 and 3 are obtained from the CpHMD simulations while ΔG°ref can be taken either from experiment or from thermodynamic integration (TI) computations, where the latter provides a full computational prediction of the pH dependence of binding processes. When applied to binding of small molecules to the cucurbit[7]uril (CB[7]) host, this CpHMD-based free energy method accurately obtained the pH-dependent binding free energy profiles (Figure 2).80 For instance, in Figure 2, the binding free energy profiles of CB[7] binding to benzimidazole and fuberidazole are shown, computed with the ΔG°ref obtained from the TI computation, i.e., ΔG°TI. While the CpHMD/TI computation of pH-dependent binding free energies is prone to greater error than the use of ΔG°ref from experiment (ΔG°exp), the authors found that the errors in assigning incorrect protonation states in free energy computations without correcting for the pH dependence of the binding free energy can give errors in excess of the errors from the typical free energy computations.80 The method has been further applied to several inhibitor-bound structures of BACE-1 in order to capture the pH dependence of protein-ligand systems and highlighted the significance of correctly accounting for the binding-induced protonation changes in free energy computations.28
Figure 2.
Binding free energies of cucurbit[7]uril host to the guests, (A) benzimidazole and (B) fuberidazole. ΔG°TI is the absolute reference binding free energy obtained from thermodynamic integration computations for the reference deprotonated state of the guest; ΔG°exp is the experimental binding free energy for the reference deprotonated guest; and ΔG°+exp is the binding free energy for the protonated guest derived from ΔG°exp. The figure is adopted from Kim et al. (2015).80
Outlook
We have presented a brief review of pH dependence of protein-ligand complex formation. While the prevalence of binding-induced changes in pKa or protonation state has been well appreciated especially due to the recent progress in this field, it is true that these phenomena are not always accounted for in the majority of free energy computations or computational drug discovery. The use of the expressions applying the binding polynomial formalism to address the pH dependence of binding free energies is promising in these fields, and the application of the CpHMD technique particularly enables the coupling of conformational and protonation equilibria. Though not specifically addressed, similar philosophies may also be applicable to the scoring functions in docking protocols.
Several challenges, however, still remain. The lack of binding free energies experimentally measured at various pH levels, preferably for large protein-ligand datasets, imposes a challenge to incorporating the pH-dependent effects into computational studies; availability of the experimental reference binding free energies will be of great importance to pushing the free energy computation field forward. Also, the accuracy of current pKa prediction algorithms strongly affects the quality the studies on the pH dependence. The pKa cooperative, a collaborative effort to advance structure-based calculation of pKa and electrostatics, noted that significant progress is still needed to improve the results from the current computational pKa predictions.81 Besides the performance of the pKa computation algorithms, the pKa values obtained from such computations are known to be sensitive to the details of the input structure.82 While the advances in X-ray and NMR techniques for protein structural determination are clearly encouraging, the lack of availability of high-resolution structures of both protein monomer and protein-ligand complex in many cases still hinders the progress in the study of binding-induced protonation changes. Despite these challenges, however, we strongly believe that the growing attention to this field of study is promising.
Acknowledgements
This work was supported by the NSF (MCB-1020765), NIH (NIH GM31749), Howard Hughes Medical Institute, National Biomedical Computation Resource (NBCR), and NSF supercomputer centers.
References
- 1.Onufriev AV, Alexov E. Q Rev Biophys. 2013;46:181. doi: 10.1017/S0033583513000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mobley DL, Dill KA. Structure. 2009;17:489. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexov E. Proteins. 2004;56:572. doi: 10.1002/prot.20107. [DOI] [PubMed] [Google Scholar]
- 4.Georgescu RE, Alexov E, Gunner MR. Biophys J. 2002;83:1731. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kundrotas PJ, Alexov E. Biophys J. 2006;91:1724. doi: 10.1529/biophysj.106.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason AC, Jensen JH. Proteins. 2008;71:81. doi: 10.1002/prot.21657. [DOI] [PubMed] [Google Scholar]
- 7.Jensen JH. Curr Pharm Biotechnol. 2008;9:96. doi: 10.2174/138920108783955146. [DOI] [PubMed] [Google Scholar]
- 8.Mitra RC, Zhang Z, Alexov E. Proteins. 2011;79:925. doi: 10.1002/prot.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manallack DT. SAR QSAR Environ Res. 2009;20:611. doi: 10.1080/10629360903438313. [DOI] [PubMed] [Google Scholar]
- 10.Manallack DT, Prankerd RJ, Yuriev E, Oprea TI, Chalmers DK. Chem Soc Rev. 2013;42:485. doi: 10.1039/c2cs35348b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashford D, Karplus M. Biochemistry. 1990;29:10219. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 12.Yang A-S, Gunner MR, Sampogna R, Sharp K, Honig B. Proteins. 1993;15:252. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- 13.Aguilar B, Anandakrishnan R, Ruscio JZ, Onufriev AV. Biophys J. 2010;98:872. doi: 10.1016/j.bpj.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith R, Brereton IM, Chai RY, Kent SB. Nat Struct Mol Biol. 1996;3:946. doi: 10.1038/nsb1196-946. [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki T, Nicholson LK, Wingfield P, Stahl SJ, Kaufman JD, Eyermann CJ, Hodge NC, Lam PY, Torchia DA. J Am Chem Soc. 1994;116:10791. [Google Scholar]
- 16.Xie D, Gulnik S, Collins L, Gustchina E, Suvorov L, Erickson JW. Biochemistry. 1997;36:16166. doi: 10.1021/bi971550l. [DOI] [PubMed] [Google Scholar]
- 17.Cassidy CS, Lin J, Frey PA. Biochemistry. 1997;36:4576. doi: 10.1021/bi962013o. [DOI] [PubMed] [Google Scholar]
- 18.Joshi MD, Sidhu G, Nielsen JE, Brayer GD, Withers SG, McIntosh LP. Biochemistry. 2001;40:10115. [PubMed] [Google Scholar]
- 19.Stranzl GR, Gruber K, Steinkellner G, Zangger K, Schwab H, Kratky C. J Biol Chem. 2004;279:3699. doi: 10.1074/jbc.M306814200. [DOI] [PubMed] [Google Scholar]
- 20.Blundell CD, Mahoney DJ, Cordell MR, Almond A, Kahmann JD, Perczel A, Taylor JD, Campbell ID, Day AJ. J Biol Chem. 2007;282:12976. doi: 10.1074/jbc.M611713200. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu H, Takeuchi K, Takeuchi H. Biochemistry. 2013;52:2371. doi: 10.1021/bi4001112. [DOI] [PubMed] [Google Scholar]
- 22.Kumarasiri M, Zhang W, Shi Q, Fisher JF, Mobashery S. Proteins. 2013;82:1348. doi: 10.1002/prot.24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruylants G, Wintjens R, Looze Y, Redfield C, Bartik K. Eur Biophys J. 2007;37:11. doi: 10.1007/s00249-007-0148-0. [DOI] [PubMed] [Google Scholar]
- 24.Luo R, Head MS, Moult J, Gilson MK. J Am Chem Soc. 1998;120:6138. [Google Scholar]
- 25.Trylska J, Antosiewicz J, Geller M, Hodge CN, Klabe RM, Head MS, Gilson MK. Prot Sci. 1999;8:180. doi: 10.1110/ps.8.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czodrowski P, Sotriffer CA, Klebe G. J Mol Biol. 2007;367:1347. doi: 10.1016/j.jmb.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Witham S, Zhang Z, Li L, Hodsdon M, Alexov E. Commun Comput Phys. 2013;13:207. doi: 10.4208/cicp.170911.131011s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MO, Blachly PG, McCammon JA. PLoS Comp Biol. 2015 doi: 10.1371/journal.pcbi.1004341. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szakácz Z, Béni S, Varga Z, Örfi L, Kéri G, Noszál B. J Med Chem. 2005;48:249. doi: 10.1021/jm049546c. [DOI] [PubMed] [Google Scholar]
- 30.Tóth G, Hosztafi S, Kovács Z, Noszál B. J Pharm Biomed Anal. 2012;61:156. doi: 10.1016/j.jpba.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Tóth G, Baska F, Schretner A, Rácz Á, Noszál B. Eur Biophys J. 2013;42:721. doi: 10.1007/s00249-013-0921-1. [DOI] [PubMed] [Google Scholar]
- 32.Bashford D. Front Biosci. 2004;9:1082. doi: 10.2741/1187. [DOI] [PubMed] [Google Scholar]
- 33.Simonson T. Rep Prog Phys. 2003;66:737. [Google Scholar]
- 34.Baker N. Methods Enzymol. 2004;383:94. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
- 35.Baker N. Curr Opin Struct Biol. 2005;15:137. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127. [Google Scholar]
- 37.Onufriev AV. In: Modeling Solvent Environments: Applications to Simulations of Biomolecules. Feig M, editor. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2010. [Google Scholar]
- 38.Lim C, Bashford D, Karplus M. J Phys Chem. 1991;95:5610. [Google Scholar]
- 39.Antosiewicz J, McCammon JA, Gilson MK. J Mol Biol. 1994;238:415. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 40.Chipman DM. J Phys Chem A. 2002;106:7413. [Google Scholar]
- 41.Klicic JJ, Friesner RA, Liu SY, Guida WC. J Phys Chem A. 2002;106:1327. [Google Scholar]
- 42.Gordon JC, Myers JB, Folta T, Soja V, Heath LS, Onufriev AV. Nucleic Acids Res. 2005;33:W368. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexov E. Proteins. 2003;50:94. doi: 10.1002/prot.10265. [DOI] [PubMed] [Google Scholar]
- 44.Pokala N, Handel TM. Protein Sci. 2004;13:925. doi: 10.1110/ps.03486104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Archontis G, Simonson T. Biophys J. 2005;88:3888. doi: 10.1529/biophysj.104.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexov E, Gunner MR. Biophys J. 1997;74:2075. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehler EL, Guarnieri F. Biophys J. 1999;77:3. doi: 10.1016/S0006-3495(99)76868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wisz MS, Hellinga HW. Proteins. 2003;51:360. doi: 10.1002/prot.10332. [DOI] [PubMed] [Google Scholar]
- 49.Sham YY, Chu ZT, Warshel A. J Phys Chem B. 1997;101:4458. [Google Scholar]
- 50.Jensen JH, Li H, Robertson AD, Molina PA. J Phys Chem A. 2005;109:6634. doi: 10.1021/jp051922x. [DOI] [PubMed] [Google Scholar]
- 51.Reccardi D, Schaefer P, Yang Y, Yu HB, Ghosh N, Prat-Resina X, Konig P, Li GH, Xu DG, Guo H, Elstner M, Cui Q. J Phys Chem B. 2006;110:6458. doi: 10.1021/jp056361o. [DOI] [PubMed] [Google Scholar]
- 52.Mertz JE, Pettitt BM. Supercomputer Appl High Perform Comput. 1994;8:47. [Google Scholar]
- 53.Baptista AM, Martel PJ, Petersen SB. Proteins. 1997;27:523. [PubMed] [Google Scholar]
- 54.Börjesson U, Hünenberger PH. J Phys Chem B. 2004;108:13551. [Google Scholar]
- 55.Lee MS, Salsbury FR, Brooks CL., III Proteins. 2004;56:738. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 56.Baptista AM, Teixeira VH, Soares CM. J Chem Phys. 2002;117:4184. [Google Scholar]
- 57.Dlugosz M, Antosiewicz JM. Chem Phys. 2004;302:161. [Google Scholar]
- 58.Dlugosz M, Antosiewicz JM, Robertson AD. Phys Rev E. 2004;69 doi: 10.1103/PhysRevE.69.021915. [DOI] [PubMed] [Google Scholar]
- 59.Mongan J, Case DA, McCammon JA. J Comput Chem. 2004;25:2038. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 60.Bürgi R, Kollman PA, van Gunsteren WF. Proteins. 2002;47:469. doi: 10.1002/prot.10046. [DOI] [PubMed] [Google Scholar]
- 61.Khandogin J, Chen JH, Brooks CL., III Proc Natl Acad Sci USA. 2006;103:18546. doi: 10.1073/pnas.0605216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Itoh SG, Damjanović A, Brooks BR. Proteins. 2011;79:3420. doi: 10.1002/prot.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swails JM, Roitberg AE. J Chem Theory Comput. 2012;8:4393. doi: 10.1021/ct300512h. [DOI] [PubMed] [Google Scholar]
- 64.Swails JM, York DM, Roitberg AE. J Chem Theory Comput. 2014;10:1341. doi: 10.1021/ct401042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams SL, de Oliveira CAF, McCammon JA. J Chem Theory Comput. 2010;6:560. doi: 10.1021/ct9005294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goh GB, Knight JL, Brooks CL., III J Chem Theory Comput. 2012;8:36. doi: 10.1021/ct2006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goh GB, Knight JL, Brooks CL., III J Chem Theory Comput. 2013;9:935. doi: 10.1021/ct300942z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goh GB, Knight JL, Brooks CL., III J Phys Chem Lett. 2013;4:760. doi: 10.1021/jz400078d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dissanayake T, Swails JM, Harris ME, Roitberg AE, York DM. Biochemistry. 2015;54:1307. doi: 10.1021/bi5012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Di Russo NV, Estrin DA, Marti MA, Roitberg AE. PLoS Comp Biol. 2012;8 doi: 10.1371/journal.pcbi.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laricheva EN, Arora K, Knight JL, Brooks CL., III J Am Chem Soc. 2013;135:10906. doi: 10.1021/ja4042687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilson MK, Zhou H-X. Annu Rev Biophys Biomol Struct. 2007;36:21. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 73.Wyman J. Adv Protein Chem. 1948;4:407. doi: 10.1016/s0065-3233(08)60011-x. [DOI] [PubMed] [Google Scholar]
- 74.Tanford C. Adv Protein Chem. 1970;24:1. [PubMed] [Google Scholar]
- 75.Wyman J. J Mol Biol. 1965;11:631. doi: 10.1016/s0022-2836(65)80017-1. [DOI] [PubMed] [Google Scholar]
- 76.Yang A-S, Honig B. J Mol Biol. 1993;231:459. doi: 10.1006/jmbi.1993.1294. [DOI] [PubMed] [Google Scholar]
- 77.Li H, Robertson AD, Jensen JH. Proteins. 2005;61:704. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 78.Misra VK, Hecht JL, Yang A-S, Honig B. Biophys J. 1998;75:2262. doi: 10.1016/S0006-3495(98)77671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moody EM, Lecompte JTJ, Belivacqua PC. RNA. 2005;11:157. [Google Scholar]
- 80.Kim MO, Blachly PG, Kaus JW, McCammon JA. J Phys Chem B. 2015;119:861. doi: 10.1021/jp505777n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nielsen JE, Gunner MR, García-Moreno B. Proteins. 2011;79:3249. doi: 10.1002/prot.23194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nielsen JE, McCammon JA. Prot Sci. 2003;12:313. doi: 10.1110/ps.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]