Abstract
The field of antigen processing and presentation is likely one of the most well defined areas in immunology based on decades of intense molecular and structural studies. Many molecules contributing to antigen processing and presentation have been discovered and their mechanisms of action been largely defined, yet a major question, which lies at the very core of the field has remained hard to pin down. The question is what determines immunodominance? Immunodominance is defined as a few specific epitopes being selected to represent an antigen to the immune system and provide targets for T cells. Many studies have aimed at understanding how epitopes are selected. A range of hypotheses related to the structural features of antigens, sensitivity to proteases, epitope affinity for MHC II, T cell precursor frequency, and T cell receptor affinity for peptide/MHC II have been considered. However, because of the variety of proteins and factors involved in antigen processing and enormous complexity, finding an answer has been challenging. Here we make an effort to tease out the sequence of events in antigen processing that promote selection of immunodominant epitopes for exogenous antigens.
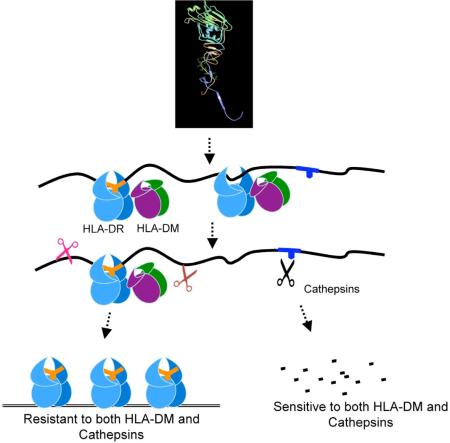
Graphic Abstract legend
DM and Cathepsins Co-Operate in Regulating Pathogen-Derived Dominant Epitope Selection
Pathogen-derived antigens bind to MHC class II as full-length proteins or large fragments, while DM facilitates the selection of the best fitting epitopes. As such, the best fitting epitopes form compact dimers with MHC II that do not dissociate by DM. Being insensitive to DM-mediated dissociation rescues those epitopes from being cleaved into peptides too short to form stable complexes with MHC II by cathepsins. Non-dominant epitopes are sensitive to DM-mediated dissociations, and to degradation by cathepsins; they dissociate from MHC II groove by DM, and are destroyed by cathepsins. As such, dominant epitopes/MHC II complexes accumulate and become relatively more abundant.
Cathepsins are shown as scissors, peptides and epitopes are depicted as part of the denatured proteins, or in short stretches of sequences that carry a MHC II P1 fitting anchor or no anchor. Small dots represent degraded peptides.
1. Introduction
In brief, antigen presentation to CD4+ T cells by APCs begins by the uptake of exogenous antigens and its coordinated transfer through a series of endosomal compartments containing a suitable denaturing environment, accessory chaperones, and cathepsins that process the antigens (Kim and Sadegh-Nasseri, 2015). Newly synthesized MHC II molecules associate with the class II invariant chain (Ii), which targets it to specialized endosomal compartments (MIIC) where the Ii is proteolysed by antigen processing proteases called cathepsins until only a fragment known as the class II-associated invariant chain peptide (CLIP), remains bound in the MHC II peptide-binding groove. Loading of exogenous peptides onto MHC II requires displacement of CLIP from the MHC groove. While CLIP can readily dissociate from some MHC II alleles on its own, other alleles require help from the accessory molecule HLA-DM in human or H2-DM in mice (DM). DM functions by inducing conformational changes in class II/CLIP complexes resulting in the release of CLIP and inducing a peptide-receptive MHC II. A peptide-receptive MHC II can quickly sample epitopes derived from exogenously acquired proteins (Chou and Sadegh-Nasseri, 2000; Natarajan et al., 1999a). DM helps in shaping epitope selection by selectively dissociating some p/MHC II complexes while leaving other complexes unaffected. Whether pMHC II complexes remain untouched or dissociate relates to the differences in conformation of pMHC II complexes somehow recognized by DM (Chou et al., 2008; Narayan et al., 2007; Narayan et al., 2009; Sadegh-Nasseri et al., 2012; Sadegh-Nasseri et al., 2010). With the ability to recognize small differences displayed by different pMHC complexes, DM is expected to be a critical player in the selection of immunodominant epitopes. Another accessory molecule in antigen processing is HLA-DO in human or H-2O in mice (DO). DO has a restricted expression; it is mainly expressed in B cells, thymic medulla, and some DC subpopulations. Understanding the contribution of DO to epitope selection has been a difficult task, as the DO knockout mice did not show a readily detectable phenotype (Liljedahl et al., 1998; Poluektov et al., 2013). Finally, antigen processing cannot be done without cathepsins, the proteases in antigen processing. Different cathepsins have been associated with the generation of epitopes. Some are cell, or tissue specific and some others are mainly found in the extracellular matrix. Many cathepsins in antigen processing function in acidic pH, although they may also function in neutral pH, but with different specificities.
An issue that has remained controversial over the years is the sequence of events in antigen capture by MHC II. While there are a number of publications reporting binding of protein antigens or large fragments to MHC II molecules (Jensen, 1995; Lee et al., 1988; Lindner and Unanue, 1996), the generally accepted view envisions that protein antigens are first cut into short peptides by cathepsins, and then binding to MHC II and DM editing follow. This latter notion is mainly based on our understanding of MHC class I antigen processing that relies on precisely cut epitopes fitting MHC molecules bearing closed ends.
2. Current Status
2.1. A Cell Free Minimalist Antigen Processing System
A recent discovery by our laboratory has offered great help in teasing out steps in antigen processing and in deciphering the order of events (Hartman et al., 2010; Kim et al., 2014). This system is composed of only five soluble purified components, HLA-DR1 (DR1), DM, and three cathepsins CatB, cat H and CatS and yet has proved to be sufficient for processing of full-length antigens and in depicting physiologically selected immunodominant epitopes. Combined with high-resolution mass-spectrometry for the analysis of the peptide mixtures eluted from MHC II molecules, the results provide unambiguous answers to the fate of the antigenic precursors and their proteolytic products. Several lines of experimental designs validated the authenticity of outcomes of this cell free system. By immunizing and analyzing the specificity of the CD4 T-cell responses in appropriate DR1 transgenic mice we confirmed accuracy of epitope predictions by the minimalist system. Moreover, in one instance, human volunteers who were vaccinated with a recombinant protein variant of Malaria Plasmodium falciparum Liver Stage Antigen-1 (LSA-1) recognized the epitopes predicted by the reductionist system as dominant. Among eight individuals who were responsive to the LSA-1 protein immunization, only two individuals responded to the predicted epitope, and it turned out that those two were the only DR1 positive individuals among the cohort. Of note, is that the testing of human samples for responsiveness to identified epitopes were done prior to HLA typing of the volunteers’ cells. Further physiological validity of our system was established by using specific pharmacological inhibitors of the cathepsins used in cellular assays. Specific inhibition of only Cathepsin B led to complete impairment of antigen processing and presentation by the antigen processing cells in vitro (Kim et al., 2014; Kim and Sadegh-Nasseri, 2015). As such, the cell free system proved to be a reliable and trustworthy tool for prediction of physiologically relevant dominant epitope(s).
2.2. Short Epitopes from Exogenous Antigens Are Destroyed by Cathepsins
The ability to add the components sequentially in any desired order allowed us to assess the temporal relationship of protein versus peptide capture by MHC II during processing. We tested the fundamental question of whether the epitopes were generated independently of the MHC or required MHC II molecules to protect them from proteolytic destruction. In other words, is it feasible to assume that the selection for immunodominant epitopes takes place among short epitopes? Our observations disproved that assumption. By sequential incubation of antigen with cathepsins first, and then with DR1 and DM, we discovered that short epitopes from foreign antigens were highly sensitive to proteolytic digestion by the cathepsins in the system. Three different pathogen-derived protein antigens were examined; recombinant HA1 proteins from A/Vietnam/1203/2004 (H5N1) strain and A/Texas/1/77 influenza stains, as well as LSA-NRC of malaria antigen. When HA1 protein of A/Texas/1/77 influenza strain containing the well-characterized dominant epitope HA(306–318), was exposed to cathepsins first, few rHA1-derived peptides were detectable by Mass spectrometry after 1h, or only a 15min exposure time, but HA(306-318) containing epitope was not found under any conditions tested. Peptides containing the immunodominant epitope were identified only when all components were mixed together, or when the order of reaction was changed such that full-length rHA1 protein was first incubated with DR1 and DM and then with the cathepsin mix. We surmised that the successful generation of immunodominant epitopes from rHA1 protein depends on capture by DR and editing by DM prior to the proteolytic fragmentation of the antigen.
The dominant epitopes of HA1-H5N1 and LSA-NRC have been described previously (Hartman et al., 2010). Similar results as above were observed for both antigens. Occasionally, however, much shorter versions of the epitopes could be found. Those results were verified by exposing premade synthetic peptides to cathepsins in the absence of DR1, followed by Mass Spectrometry analysis of the products, which revealed that cathepsins had destroyed the epitopes. Shortened sequences that had remained were too short to bind DR1 explaining why we could not find the dominant epitopes when protein antigens were exposed to cathepsins first and then subjected to DR binding and editing (Kim et al., 2014). Therefore, it was only by binding to MHC II groove and resisting to DM-mediated dissociation that the dominant epitopes were protected from destruction. Based on these observations, for successful selection of dominant epitopes, protein antigens may bind to MHC II first while at the same time DM is at work selecting the best fitting epitopes.
2.3. Full-Length Proteins Bind to MHC II Binding Groove
To test the notion that protein antigens bind to MHC II before enzymatic digestion, we utilized a gentle SDS-PAGE assay originally used to detect peptide binding to MHC class II in which samples were not boiled (Dornmair et al., 1989; Natarajan et al., 1999b). Binding of full-length rHA1 protein to DR1 was detected by its slower migrating molecular species at the correct estimated molecular mass of the complex. Importantly, in the presence of excess HA(306-318) in the reaction, the band corresponding to the DR1/protein complex was significantly reduced while a new band that matched the migration pattern of HA(306-318) peptide/DR1 complex appeared. Parallel Western blots and antibody staining confirmed that the observed complexes indeed were composed of DR1 and rHA1 protein (Kim et al., 2014). These data established that interaction with HA1 protein occurred specifically through the peptide-binding groove of DR1. Consistent with the role for DM in generating the peptide-receptive conformation of DR molecules, inclusion of DM together with DR1 and the rHA1 proteins resulted in more intensely stained bands corresponding to the protein/DR1 complexes (Sadegh-Nasseri et al., 2012; Yin et al., 2015).
If the full length proteins bind to MHC class II molecules, it should be possible to detect simultaneous binding of a stretch of protein to two MHC II alleles that have non-overlapping epitopes on the same protein. We already know that several DR4 restricted epitopes of H5N1-rHA1 do not overlap with DR1 restricted immunodominant epitope. Taking advantage of BIAcore Surface Plasmon Resonance (SPR) instrument, we immobilized preformed biotinylated DR4 in complex with denatured H5N1-rHA1 on streptavidin coated (SA) BIAcore chip and then followed by injection of receptive DR1 in the presence of DM. Binding of peptide receptive DR1 to the complexes of DR4/H5N1-rHA1 indicated that more than one MHC class II molecule is capable of binding to the H5N1-rHA1 protein. Consistent with our in vitro study, binding of a single protein to two MHC alleles has precedented earlier in cells (Castellino et al., 1998), further strengthening that a major pathway of Ag processing involves the initial binding of MHC II to large protein substrates, followed by cleavage and/or trimming of the exposed protein around this bound region. Thus; a) MHC class II molecule can bind to full-length protein antigens, b) just like peptide binding, DM enhances binding of full-length protein to DR1, and c) binding of protein to DR1 occurs through the peptide binding groove of DR1 rather than other possible sites.
2.4. Accessibility of the dominant epitopes for binding to MHC II
The availability of stretches of sequences that can readily be captured by MHC II before the antigens are cut is another parameter that could contribute to preferred epitope selection during antigen processing (Dai et al., 2001). Along these thoughts, GILT (gamma interferon-inducible lysosomal thiol reductase) has been shown to enhance antigen processing for antigens with several disulfide bonds such as HEL and HA protein of influenza (Hastings and Cresswell, 2011; Nguyen et al., 2015). The open-ended nature of the MHC class II peptide binding groove supports the ‘binding first, trimming later’ model. In our system, two out of three dominant epitopes, HA(306-318) from rHA1 and LSA(434–453) from LSA-NRC, are located at C- and N-termini of proteins respectively. Being at either end of the proteins might offer an advantage for their capture by MHC II, likely due to less structural constraints. Both epitopes form DM-resistant complexes with DR1. Accordingly, the epitopes will be protected from proteolysis and will be displayed at a higher abundance relative to other potential epitopes that are located further away from the protein termini. This might explain why many known immunodominant epitopes are located at the C or N-termini (Guillet et al., 1986; Lee et al., 1988; Nepom et al., 2001), or on flexible strands of protein antigens (Dai et al., 2001).
2.5. DM and Cathepsins Together Increase the Abundance of the MHC II/Dominant Epitopes
We find that immunodominant epitopes gain dominance by increased abundance. To achieve higher abundance, both cathepsin sensitivity and DM sensitivity play key roles. While a role for DM in shaping epitope repertoire has been previously proposed, using our system, we find that DM resistance is only part of the equation. Contribution of endosomal proteases, cathepsin B, H and S, combined with DM-mediated epitope-exchange critically influence the repertoire of peptides captured by DR molecules. Of note, is that the proteases used in our system appear to be the major cathepsins contributing to epitope generation and selection in vivo as well. We found that processing and presentation of epitopes from two proteins, HA(259-274) and type II collagen CII(280-294), were completely inhibited when specific pharmacological inhibitor of CatB (CA-074ME, a cell-permeable CatB inhibitor) was included in the system (Kim et al., 2014). The inhibition was due to defective antigen processing and not T cell responsiveness. In the presence of DM and the cathepsins, immunodominant epitopes are selected in part by being resistant to DM, hence being retained by DR molecules, which protects them from degradation by the cathepsins. Concurrently, epitopes that are sensitive to DM-mediated dissociation are dislodged from MHC II groove and are destroyed by the cathepsins. We have also demonstrated that epitopes from autoantigens are insensitive to degradation by cathepsins although they may or may not be sensitive to DM. They gain dominance by resisting degradation by cathepsins when released from the MHC II groove, which allows them to rebind. Non-dominant epitopes are sensitive to both DM and cathepsins and are destroyed (Kim et al., 2014). The important net result of all these events is that the immunodominant epitopes bound to MHC II gain relative abundance, as we have shown by quantitative MS (Kim et al., 2014) and as suggested for cellular processing (Ma et al., 1999; Rinderknecht et al., 2010).
3. Future perspectives
The cell free minimalist system described has been of significant help in teasing out the order of events in antigen processing for MHC II. It has allowed us to determine that different antigens have differential sensitivity to cathepsins and that external antigens are captured by MHC class II molecules in full-length form rather than as short peptides. This system has also allowed us to evaluate how DM contributes to immunodominance. These new concepts can certainly guide us in better prediction of most likely immunogenic epitopes from pathogens. Moreover, although not discussed here, we have also learned from our reductionist system that autoantigens might not follow the same path that pathogen-derived antigens do (Kim et al., 2014; Kim and Sadegh-Nasseri, 2015). Autoantigens are generally captured as precut peptides that happen to be resistant to further proteolysis by cathepsins. This characteristic was shared by all immunodominant epitopes that we tested from 6-8 autoantigens known for induction of anti-self reactivity. The chemical characteristics of such epitopes that make them resistant to antigen processing enzymes need further investigation. Gaining answers to this question can open new ways for better prediction of epitopes from auto-antigens that may become targets of autoreactivity.
The last but not the least factor contributing to epitope selection is the accessory molecule DO. Future experiments would include DO together with DM in the reductionist antigen processing system to address differences in repertoire generation in antigen presenting cells that do or do not express DO. Since DO is expressed in the thymic medulla where negative selection takes place, it is important to find out how its inclusion during antigen processing would change epitope selection and hence T cell repertoire.
Acknowledgements
Authors thank Dr. Isamu Z. Hartman who contributed to some of the work discussed here. This work was supported by grants from NIAID R01AI063764 and R21 AI101987 to SS-N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Castellino F, Zappacosta F, Coligan JE, Germain RN. Large protein fragments as substrates for endocytic antigen capture by MHC class II molecules. J Immunol. 1998;161:4048–57. [PubMed] [Google Scholar]
- Chou CL, Mirshahidi S, Su KW, Kim A, Narayan K, Khoruzhenko S, Xu M, Sadegh-Nasseri S. Short peptide sequences mimic HLA-DM functions. Mol Immunol. 2008;45:1935–43. doi: 10.1016/j.molimm.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai G, Steede NK, Landry SJ. Allocation of helper T-cell epitope immunodominance according to three-dimensional structure in the human immunodeficiency virus type I envelope glycoprotein gp120. J Biol Chem. 2001;276:41913–20. doi: 10.1074/jbc.M106018200. [DOI] [PubMed] [Google Scholar]
- Dornmair K, Rothenhausler B, McConnell HM. Structural intermediates in the reactions of antigenic peptides with MHC molecules. Cold Spring Harb Symp Quant Biol. 1989;54:409–16. doi: 10.1101/sqb.1989.054.01.050. Pt 1. [DOI] [PubMed] [Google Scholar]
- Guillet JG, Lai MZ, Briner TJ, Smith JA, Gefter ML. Interaction of peptide antigens and class II major histocompatibility complex antigens. Nature. 1986;324:260–2. doi: 10.1038/324260a0. [DOI] [PubMed] [Google Scholar]
- Hartman IZ, Kim A, Cotter RJ, Walter K, Dalai SK, Boronina T, Griffith W, Lanar DE, Schwenk R, Krzych U, Cole RN, Sadegh-Nasseri S. A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes. Nat Med. 2010;16:1333–1340. doi: 10.1038/nm.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings KT, Cresswell P. Disulfide reduction in the endocytic pathway: immunological functions of gamma-interferon-inducible lysosomal thiol reductase. Antioxidants & redox signaling. 2011;15:657–68. doi: 10.1089/ars.2010.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE. Antigen unfolding and disulfide reduction in antigen presenting cells. Semin Immunol. 1995;7:347–53. doi: 10.1006/smim.1995.0039. [DOI] [PubMed] [Google Scholar]
- Kim A, Hartman IZ, Poore B, Boronina T, Cole RN, Song N, Ciudad MT, Caspi RR, Jaraquemada D, Sadegh-Nasseri S. Divergent paths for the selection of immunodominant epitopes from distinct antigenic sources. Nature communications. 2014;5:5369. doi: 10.1038/ncomms6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Sadegh-Nasseri S. Determinants of immunodominance for CD4 T cells. Curr Opin Immunol. 2015;34:9–15. doi: 10.1016/j.coi.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Matsueda GR, Allen PM. T cell recognition of fibrinogen. A determinant on the A alpha-chain does not require processing. J Immunol. 1988;140:1063–8. [PubMed] [Google Scholar]
- Liljedahl M, Winqvist O, Surh CD, Wong P, Ngo K, Teyton L, Peterson PA, Brunmark A, Rudensky AY, Fung-Leung WP, Karlsson L. Altered antigen presentation in mice lacking H2-O. Immunity. 1998;8:233–43. doi: 10.1016/s1074-7613(00)80475-6. [DOI] [PubMed] [Google Scholar]
- Lindner R, Unanue ER. Distinct antigen MHC class II complexes generated by separate processing pathways. Embo J. 1996;15:6910–20. [PMC free article] [PubMed] [Google Scholar]
- Ma C, Whiteley PE, Cameron PM, Freed DC, Pressey A, Chen SL, Garni-Wagner B, Fang C, Zaller DM, Wicker LS, Blum JS. Role of APC in the selection of immunodominant T cell epitopes. J Immunol. 1999;163:6413–23. [PubMed] [Google Scholar]
- Narayan K, Chou CL, Kim A, Hartman IZ, Dalai S, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Su KW, Chou CL, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM mediates peptide exchange by interacting transiently and repeatedly with HLA-DR1. Mol Immunol. 2009;46:3157–62. doi: 10.1016/j.molimm.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan SK, Assadi M, Sadegh-Nasseri S. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J Immunol. 1999a;162:4030–6. [PubMed] [Google Scholar]
- Natarajan SK, Stern LJ, Sadegh-Nasseri S. Sodium dodecyl sulfate stability of HLA-DR1 complexes correlates with burial of hydrophobic residues in pocket 1. J Immunol. 1999b;162:3463–70. [PubMed] [Google Scholar]
- Nepom GT, Lippolis JD, White FM, Masewicz S, Marto JA, Herman A, Luckey CJ, Falk B, Shabanowitz J, Hunt DF, Engelhard VH, Nepom BS. Identification and modulation of a naturally processed T cell epitope from the diabetes-associated autoantigen human glutamic acid decarboxylase 65 (hGAD65) Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1763–8. doi: 10.1073/pnas.98.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Steede NK, Robinson JE, Landry SJ. Conformational instability governed by disulfide bonds partitions the dominant from subdominant helper T-cell responses specific for HIV-1 envelope glycoprotein gp120. Vaccine. 2015;33:2887–96. doi: 10.1016/j.vaccine.2015.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluektov YO, Kim A, Sadegh-Nasseri S. HLA-DO and Its Role in MHC Class II Antigen Presentation. Front Immunol. 2013;4:260. doi: 10.3389/fimmu.2013.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderknecht CH, Roh S, Pashine A, Belmares MP, Patil NS, Lu N, Truong P, Hou T, Macaubas C, Yoon T, Wang N, Busch R, Mellins ED. DM influences the abundance of major histocompatibility complex class II alleles with low affinity for class II-associated invariant chain peptides via multiple mechanisms. Immunology. 2010;131:18–32. doi: 10.1111/j.1365-2567.2010.03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh-Nasseri S, Chou CL, Hartman IZ, Kim A, Narayan K. How HLA-DM works: recognition of MHC II conformational heterogeneity. Front Biosci (Schol Ed) 2012;4:1325–32. doi: 10.2741/s334. [DOI] [PubMed] [Google Scholar]
- Yin L, Maben ZJ, Becerra A, Stern LJ. Evaluating the Role of HLA-DM in MHC Class II-Peptide Association Reactions. J Immunol. 2015 doi: 10.4049/jimmunol.1403190. [DOI] [PMC free article] [PubMed] [Google Scholar]