Abstract
Objective
Identification of a particular cause of meningoencephalitis can be challenging owing to the myriad bacteria, viruses, fungi, and parasites that can produce overlapping clinical phenotypes, frequently delaying diagnosis and therapy. Metagenomic deep sequencing (MDS) approaches to infectious disease diagnostics are known for their ability to identify unusual or novel viruses and thus are well suited for investigating possible etiologies of meningoencephalitis.
Methods
We present the case of a 74‐year‐old woman with endophthalmitis followed by meningoencephalitis. MDS of her cerebrospinal fluid (CSF) was performed to identify an infectious agent.
Results
Sequences aligning to Balamuthia mandrillaris ribosomal RNA genes were identified in the CSF by MDS. Polymerase chain reaction subsequently confirmed the presence of B. mandrillaris in CSF, brain tissue, and vitreous fluid from the patient's infected eye. B. mandrillaris serology and immunohistochemistry for free‐living amoebas on the brain biopsy tissue were positive.
Interpretation
The diagnosis was made using MDS after the patient had been hospitalized for several weeks and subjected to costly and invasive testing. MDS is a powerful diagnostic tool with the potential for rapid and unbiased pathogen identification leading to early therapeutic targeting. Ann Neurol 2015;78:Ann Neurol 2015;78:679–696
Meningoencephalitis presents significant diagnostic challenges owing, in part, to the large number of possible underlying causes. These include infectious, inflammatory, paraneoplastic, and toxic‐metabolic etiologies, many of which can present with similar clinical phenotypes. Furthermore, confirming an infectious etiology can be challenging, given that many pathogen‐specific serological and polymerase chain reaction (PCR) assays are expensive, time‐consuming, and have poor operating characteristics. Culture‐based approaches, though they can often detect multiple species, are also limited, given that many infectious agents require special culture conditions and prolonged incubation times. Standard culture protocols are also not commercially available for many organisms.
Metagenomics is the study of genetic material recovered from an environmental sample, including bodily fluids and tissues. Rapid advances in sequencing technology and bioinformatics have made an unbiased deep sequencing approach to metagenomics an increasingly feasible method for rapidly interrogating all the genetic material contained in a sample. We previously reported on the ability of metagenomic deep sequencing (MDS) of cerebrospinal fluid (CSF) to identify the cause of infectious meningoencephalitis in real time, leading to a dramatic improvement in a patient's medical condition.1 Here, we describe a case of meningoencephalitis in a 74‐year‐old woman that illustrates the ability of an MDS‐based approach to identify an amoebic cause of meningoencephalitis. Our patient had had an extensive diagnostic workup, including two brain biopsies, but a final diagnosis was not reached before she died. Shortly before her death, a sample of CSF was submitted for MDS, which led to the identification of Balamuthia mandrillaris, a rare and almost uniformly fatal cause of meningoencephalitis. This diagnosis was later confirmed by the Centers for Disease Control and Prevention (CDC), the California Department of Public Health (CA DPH), and analysis of additional immunohistochemical stains and additional sections of brain biopsy tissue. MDS provides an efficient and unbiased diagnostic approach to identify culprit central nervous system (CNS) infections and may generate large cost savings when used to improve diagnosis rates in patients with costly conditions such as meningoencephalitis.2 Early treatment guided by rapid and accurate diagnosis may also improve outcomes in cases such as these in which delayed diagnosis has likely contributed to historically poor outcomes.
Case Report
A 74‐year‐old Chinese woman with a history of rheumatoid arthritis treated with hydroxychloroquine sulfate presented to our hospital with altered mental status. She had immigrated to the United States from China approximately 8 years previously and had been independent in all her activities. She did not use tobacco, alcohol, or illicit drugs.
In the 4 weeks preceding hospitalization at our institution, she was admitted three times to two other facilities. On each occasion, she was evaluated for intermittent fevers and mildly altered mental status. On the second admission, she was diagnosed with endophthalmitis and treated empirically with intravitreal vancomycin, ceftazidime, and voriconazole, though vitreal and blood cultures eventually showed no growth, and PCRs on the vitreous fluid for herpes simplex virus, cytomegalovirus, and toxoplasmosis ultimately proved negative. In addition, she was started on therapy for Toxoplasma gondii in the setting of a positive serum immunoglobulin M (IgM) titer (see Table). Although her vision did not improve, she was briefly discharged, but then brought back to the hospital for a third admission when a cervical spine magnetic resonance imaging (MRI) study performed to evaluate spinal tenderness was incidentally noted to show a T2 hyperintensity in the pons. A subsequent brain MRI showed small areas of restricted diffusion in multiple vascular territories consistent with infarcts (see Fig A). A stroke workup, including a transesophageal echocardiogram, was unrevealing except for paroxysmal atrial fibrillation. The patient was discharged a third time, now on warfarin for presumed cardioembolic strokes.
Table 1.
Infectious Disease Diagnostic Test Results
| Test | Site | First Hosp | Second Hosp | Third Hosp | Fourth Hosp | After Diagnosis |
|---|---|---|---|---|---|---|
| T. gondii IgG Ab | Serum | Negative | Negative | Negative | ||
| T. gondii IgM Ab | Serum | 11.4 IU/mL (<7.9 IU/mL) | Negative | |||
| T. gondii IgM ELISA | Serum | Negative | ||||
| (1‐3)‐Beta‐D‐glucan | Serum | Negative | ||||
| AFB stain and culture | Tracheal aspirate | Negative | Negative (×5) | |||
| T. gondii PCR | Vitreous | Negative | ||||
| HSV‐1, ‐2 PCR | Vitreous | Negative | ||||
| CMV PCR | Vitreous | Negative | ||||
| VZV PCR | Vitreous | Negative | ||||
| T. gondii PCR | Vitreous | Negative | Negative | |||
| Fungal culture | Vitreous | Negative | ||||
| Fungal culture | Blood | Negative (×2) | ||||
| Bacterial culture | Blood | Negative (×2) | Negative (4) | Negative (×9) | ||
| HIV‐1 Ab | Serum | Negative | Negative | Negative | ||
| HIV‐1 viral load | Serum | Negative | ||||
| Hepatitis B surface Ag | Serum | Negative | ||||
| Hepatitis B core IgM Ab | Serum | Negative | ||||
| Hepatitis C Ab | Serum | Negative | ||||
| Gram stain | CSF | Negative (×3) | ||||
| VZV PCR and IgG | CSF | Negative | ||||
| VDRL | CSF | Negative | ||||
| Cryptococcal antigen | CSF | Negative (×2) | ||||
| AFB culture | CSF | Negative (×2) | ||||
| Bacterial culture | CSF | Negative (×3) | ||||
| Fungal culture | CSF | Negative (×2) | ||||
| T. gondii PCR | CSF | Negative | ||||
| T. gondii IgG Ab | CSF | Negative | ||||
| M. tuberculosis PCR | CSF | Negative | ||||
| Bacterial culture | Urine | >10K E. coli | Negative | Negative | Negative (×4); Mixed Gram + bacteria (×1) | |
| Histoplasma Ag | Urine | Negative | ||||
| C. immitis Ab | Serum | Negative | ||||
| Bacterial culture | Tracheal aspirate | Oral flora (×2) | ||||
| Gram Stain | Tracheal aspirate | Negative (×1); Oral flora (×1) | ||||
| T. gondii PCR | Blood | Negative | ||||
| RPR | Serum | Negative | Negative | |||
| Bacterial culture | Sputum, induced | Negative | Negative | Negative | Negative | |
| Gram stain | Sputum, induced | Negative | ||||
| First pathology | Brain | Mild gliosis | ||||
| Second pathology | Brain | Vasculitis, encephalitis | ||||
| Bacterial culture | Brain | Negative (×2) | ||||
| Fungal culture | Brain | Negative (×2) | ||||
| AFB culture | Brain | Negative (×2) | ||||
| Gram stain | Brain | Negative (×2) | ||||
| B. mandrillaris IgG (CA DPH) | Serum | Borderline positive (1:32) | ||||
| A. castellanii PCR (CDC) | CSF | Negative | ||||
| B. mandrillaris PCR (CA DPH) | Brain | Positive | ||||
| B. mandrillaris PCR (CDC) | Brain | Positive | ||||
| B. mandrillaris IHC (CA DPH) | Brain | Positive | ||||
| B. mandrillaris IHC (CDC) | Brain | Positive | ||||
| B. mandrillaris 18s rRNA PCR (CDC) | CSF | Positive | ||||
| B. mandrillaris 18s rRNA PCR (DeRisi Lab) | CSF | Positive | ||||
| B. mandrillaris 18s rRNA PCR (DeRisi Lab) | Vitreous | Positive (raw fluid) | ||||
| B. mandrillaris 18s rRNA PCR (CDC) | Vitreous | Negative (extracted DNA) |
Hosp = hospitalization; Ab = antibody; ELISA = enzyme‐linked immunosorbent assay; AFB = acid fast bacillus; PCR = polymerase chain reaction; HSV = herpes simplex virus; CMV = cytomegalovirus; VZV = varicella‐zoster virus; HIV‐1 = human immunodeficiency virus; Ag = antigen; VDRL = venereal disease research laboratory; RPR = rapid plasma reagin; CA DPH = California Department of Public Health; CDC = Centers for Disease Control; IHC = immunohistochemistry; rRNA = ribosomal RNA.
Figure 1.
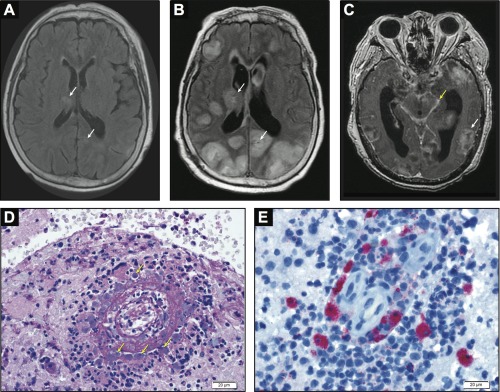
Neuroimaging and neuropathology. (A) Axial T2‐weighted/fluid‐attenuated inversion recovery (FLAIR) brain magnetic resonance imaging (MRI) demonstrating multiple hyperintensities (arrows), which correspond to areas of restricted diffusion on diffusion weighted imaging (not shown). (B) Axial T2‐weighted/FLAIR brain MRI demonstrating progression in the size and number of hyperintensities (arrows) visualized in the MRI in (A). (C) Postcontrast axial T1‐weighted brain MRI demonstrating multiple ring enhancing lesions (white arrow) and basilar meningitis (yellow arrow) as well as severe hydrocephalus. (D) Periodic acid‐Schiff stained section of brain parenchyma shows necrotizing vasculitis, chronic inflammatory cells, and a population of amoebic trophozoites that closely resemble macrophages (arrows); 400×. The majority of the tissue showed a robust necrotizing vasculitis with mixed inflammation, including macrophages and numerous eosinophils. (E) Immunohistochemistry for free‐living amoebas highlights numerous B. mandrillaris trophozoites; 400 × (courtesy of M.K.K., CDC).
Although she had been ruled out for acid‐fast bacilli during her second hospitalization, the patient presented to our institution 2 days later for an evaluation at the city's tuberculosis (TB) clinic given her recent history of pneumonia and her demographic risk factors. At the TB clinic, it was found that her neurological status had significantly deteriorated since her discharge 2 days previously. She was transferred directly to the emergency department where her vital signs were: temperature, 103°F; heart rate, 111 beats per minute; blood pressure, 123/81mm Hg; and oxygen saturation, 93% on room air. She moved all four extremities spontaneously, but was unable to follow commands or verbalize, resulting in a Glasgow Coma Scale score of 6. At this point, she was intubated for airway protection and admitted to the intensive care unit (ICU).
Initial laboratory evaluation revealed a white blood cell count of 12.6 × 103 cells/μL, hemoglobin of 13.7g/dL, serum sodium of 129mEq/L, and serum glucose of 135mg/dL. CSF analysis demonstrated 13 red blood cells/mm3, 347 white blood cells/mm3 (57% neutrophils, 11% monocytes, 31% lymphocytes, and 1% eosinophils), glucose 30mg/dL, and protein 71mg/dL. Brain MRI demonstrated innumerable ring enhancing and diffusion‐restricted supra‐ and infratentorial lesions that were felt to be most consistent with abscesses as well as severe obstructive hydrocephalus.
Based on her initial clinical and radiographic presentation, she was initiated on broad spectrum antimicrobial and anti‐toxoplasmosis coverage including vancomycin, cefepime, metronidazole, pyrimethamine, sulfadiazine, and leucovorin. Blood, urine, and CSF bacterial and fungal cultures were eventually negative, as were CSF tests for neurosyphilis, TB, and Cryptococcus (see Table). Both CSF and serological tests for toxoplasmosis were negative. Despite broad antibiotic coverage, she continued to deteriorate with progressive loss of motor activity and brainstem reflexes over the following 16 days. On hospital day 10, a repeat lumbar puncture showed worsening pleocytosis with a more prominent eosinophilia (see Table). Given these findings, broad‐spectrum antibiotics were stopped on day 10, and she was initiated on anti‐TB and antifungal therapy. A brain biopsy performed on day 4 was nondiagnostic, revealing only mild gliosis and no inflammatory infiltrate or microorganisms. A repeat biopsy 1 week later (day 11) demonstrated a severe necrotizing vasculitis of unclear etiology, with a prominent component of eosinophils, chronic inflammatory cells, and neutrophils, but without a significant granulomatous component. Repeat brain MRI on hospital day 14 demonstrated an increase in the number and size of her brain lesions as well as interval development of basilar meningitis (see Fig B, C). Her worsening cerebral edema was refractory to management with hyperosmolar therapy, as well as to the extraventricular drain that was placed during her second brain biopsy. The patient ultimately became hemodynamically unstable, likely owing to elevated intracranial pressure, and required multiple vasopressors. Her family opted for comfort care measures to be instituted, and the patient was extubated and expired on day 17 of her hospitalization.
Materials and Methods
On day 11 of the patient's hospitalization, a sample of CSF was submitted for unbiased MDS under a research protocol for the identification of potential pathogens approved by the institutional review boards of San Francisco General Hospital (SFGH) and the University of California San Francisco (UCSF). Samples were processed for MDS analysis, as previously described.1 Briefly, total RNA was extracted from CSF. Samples were randomly amplified to double‐stranded cDNA using the NuGEN Ovation v.2 kit (NuGEN, San Carlos, CA) for low nucleic acid content samples, and MDS libraries were constructed using the Nextera protocol (Illumina, San Diego, CA), as previously described.1 Library size and concentration were determined using the Blue Pippin (Sage Science, Beverly, MA) and Kapa Universal quantitative PCR kit (Kapa Biosystems, Woburn, MA), respectively. Samples were sequenced on an Illumina HiSeq 2500 instrument using 135/135‐base‐pair (bp) paired‐end sequencing. The paired‐end sequences were analyzed for pathogens using a rapid computational pipeline for pathogen detection, as described below.
Results
Identification of Balamuthia Sequences in CSF
The MDS of total RNA from the patient sample yielded 19,642,962 paired‐end reads representing 5.2 gigabases total. The data were analyzed using a rapid computational pipeline developed at our institution to classify MDS reads and identify potential pathogens by comparison to the entire National Center for Biotechnology Information (NCBI) nucleotide (nt) reference database. Briefly, paired‐end reads were quality filtered using PriceSeqFilter, a component of the PRICE assembler, followed by alignment to the human reference genome (hg19) using bowtie2 v2.2.4.3, 4 Unaligned reads that were at least 95% identical were compressed by cd‐hit (v4.6.1), resulting in a total of 33,093 remaining reads, representing approximately 0.1% of the original data.5, 6 These reads were then used as queries to search the NCBI nt database using gsnapl (v2014‐10‐16).7 Over 97% of these reads yielded gsnapl alignments to vertebrate sequences (human reads not filtered by bowtie), as well as known contaminants found in laboratory reagents.
After filtering, our analysis identified B. mandrillaris as the most highly represented organism, with 81 reads mapping to the three published B. mandrillaris genes in the NCBI repository. Twenty‐nine other individually barcoded patient CSF samples (including a water control) were included in the same sequencing run, and none contained reads that aligned to Balamuthia, nor had Balamuthia reads ever been detected in this laboratory previously. The complete pipeline run time for this analysis was 1 hour 5 minutes.
These results informed a retrospective bioinformatic analysis of the original data set wherein the raw MDS reads were directly aligned to the three available B. mandrillaris gene sequences. The alignment yielded ∼3,500 read pairs mapping concordantly to the B. mandrillaris genes, representing only 0.018% of the original data set. A random sample (30 reads) of the aligned sequences was used to seed an assembly using the targeted assembler PRICE (v1.2), ultimately generating an 806‐nt contig corresponding to the B. mandrillaris 16S ribosomal RNA (rRNA) gene (GenBank accession number KP990616).3
Confirmatory Testing for B. mandrillaris
The presence of B. mandrillaris was confirmed by multiple methods. Using a published CDC PCR protocol, the 18s rRNA gene from B. mandrillaris was detected in the patient's formalin fixed brain biopsy tissue from the second brain biopsy, CSF, and vitreous fluid.8 The PCR products were Sanger sequenced (Quintara Biosciences, Albany, CA) and confirmed to be sequences from the Balamuthia 18s rRNA gene. Based on the findings of the CSF MDS study, material from the patient's second brain biopsy was re‐evaluated with additional level sections, which revealed organisms consistent with amoeba within vessels and extending into the neural parenchyma (see Fig D). The Infectious Diseases Pathology Branch at the CDC performed confirmatory immunohistochemistry for free‐living amoeba on the brain biopsy tissue (see Fig E), and the CA DPH determined that the patient had a borderline positive serology for B. mandrillaris (see Table).9
Discussion
We present the case of a woman with several weeks of progressive meningoencephalitis that eventually proved fatal despite a 3‐week ICU stay, various antimicrobial regimens, multiple serological and CSF studies, and two brain biopsies. This extensive testing did not reveal the cause of her syndrome; however, MDS of her CSF demonstrated the etiological agent, B. mandrillaris. This finding was subsequently confirmed by PCR and by immunohistochemistry of the brain biopsy tissue. To our knowledge, this is the first case in which unbiased MDS has proven useful for identifying an amoebic infection in the context of a critical illness. The identification of B. mandrillaris was particularly notable given that sequences for only three of the likely ∼15,000 B. mandrillaris genes are publicly available in the NCBI database utilized by our bioinformatics pipeline for pathogen identification. Clearly, additional genomic information for this enigmatic pathogen would be useful in the context of future sequence‐based diagnostic assays, and for the purpose of identifying new therapeutic targets.
Since it was first isolated in 1986, B. mandrillaris has been recognized as a rare cause of amoebic encephalitis in humans, with approximately 200 cases reported worldwide.10, 11 The amoeba is distributed worldwide in soil, fresh, and saltwater sources and causes disease in both immunocompetent and ‐compromised hosts.10 In fact, heavy soil exposure has been cited as a possible risk factor for developing a B. mandrillaris infection.12 Although our patient had a small garden behind her house, she was not known to be an avid gardener. She was also not heavily immunosuppressed for her rheumatoid arthritis. The infection commonly manifests with nonspecific symptoms, including fever, headache, nausea, vomiting, seizures, and focal neurological signs.13, 14, 15, 16 As in the case presented here, imaging may reveal the presence of nodular lesions involving gray and white matter, with or without ring enhancement; disease progression commonly leads to vasogenic edema and mass effect.10
The period of incubation at the site of primary infection is variable, given that it may precede by months the development of encephalitis. From the initial site, the organisms migrate to the CNS by the bloodstream, where they proliferate, inducing a granulomatous inflammation that progresses for a period of weeks. Amoebic encephalitis can show various histological patterns, and in this case no granulomatous component was observed in the brain biopsy specimen; Balamuthia in particular is known to present with a range of inflammatory patterns.17, 18 To our knowledge, this is the first time that an ocular infection with B. mandrillaris has been described. An unsuccessful attempt was made to culture B. mandrillaris from the CSF, but no attempt at amoebic culture was made from the remaining vitreous fluid given its small volume (i.e., 30uL) and because it had been frozen for an extended period of time in a –70°C freezer. The vitreous fluid was collected in the hospital under sterile conditions when the patient first presented with endophthalmitis. All standard precautions were taken in the laboratory to prevent cross‐contamination. A negative PCR water control conducted simultaneously was negative for B. mandrillaris. The positive vitreous fluid PCR result was repeated in triplicate and confirmed by Sanger sequencing.
There are only 10 reported cases of patients surviving B. mandrillaris mengioencephalitis, in part because of extensive disease at the time of diagnosis and a lack of well‐developed treatment protocols. Pentamidine, clarithromycin, fluconazole, and sulfadiazine have been used in patients who have survived the infection.19, 20 Recently, miltefosine, in combination with some of these other agents, has shown some promise and is approved by the U.S. Food and Drug Administration as an investigational treatment of granulomatous encephalitis caused by B. mandrillaris.21
Diagnosis of B. mandrillaris has traditionally relied on its identification in tissue samples. CSF analysis usually reveals a mononuclear pleocytosis, high protein levels, and normal‐to‐low glucose concentrations.22, 23 Culturing Balamuthia amoebae is very challenging, requiring a feeder tissue monolayer, such as monkey kidney cells. Growth typically takes several weeks before the amoebae start to proliferate.24 PCR and antibody detection assays have been developed as well; however, the diagnosis is not frequently entertained and extant assays are not widely available.8, 9
As an alternative to conventional assays, MDS represents an unbiased and rapid diagnostic tool, especially for vexing cases, such as the one presented here. One criticism of an MDS‐based approach is the perceived high cost of the assay. However, we believe this technique represents a significant cost savings compared to the lengthy hospitalizations, diagnostic tests, and empiric therapies frequently associated with cases of idiopathic encephalitis.2 For example, during our patient's final hospitalization, we estimate the reimbursement from insurance, which, after adjustments, was only 18% of charges billed, to be over $140,000. We calculated reimbursement for her antimicrobials alone to total $8,000, the ICU stay over $50,000, her laboratory testing over $14,000, and brain biopsies $30,000. Meanwhile, we estimate the cost of MDS for a single sample to total approximately $2,000 for materials and labor. Given current trends in genomic technology development, this cost will likely continue to drop significantly. Furthermore, though turnaround time for MDS in this case was 6 days, this was largely owing to staffing for this experimental protocol. We have already shown that with adequate staffing, this protocol can be completed in under 48 hours.1 This represents a significant advantage compared to many of the culture‐dependent assays mentioned above. With continued decreases in cost and time, and corresponding increases in sequencing depth and advances in sequencing library preparation techniques, it is foreseeable that MDS could be deployed as a frontline diagnostic tool for patients with difficult‐to‐diagnose infectious syndromes such as encephalitis. Early diagnosis through MDS may eventually supplement or replace the numerous and costly diagnostic assays and procedures currently employed and, more important, potentially improve patient outcomes. Given that our patient's eye infection preceded her meningoencephalitis, it is tantalizing to envision how early detection of B. mandrillaris in the vitreous fluid by MDS may have created a treatment window that could have headed off the devastating and lethal CNS manifestations of her infection.
Authorship
M.R.W. and J.L.D. conceived of and coordinated the study. M.R.W. generated the MDS libraries and J.L.D., M.R.W., B.B., and B.D.O. analyzed the MDS data. J.L.D. developed the computational pipeline for ultrarapid pathogen identification from MDS data. M.R.W., B.D.O., and I.K.M.A. performed the confirmatory PCR studies. N.M.S., N.S.S., M.J.R., and J.M.G. cared for the patient and compiled clinical and pathology data. M.D.W., A.B., and M.K.K. performed the neuropathology studies. T.H.D. performed the amoeba serology and culture. J.L.D. contributed reagents/materials/analysis tools and prepared figures. H.A.S. coordinated sample collection, patient consent, and the health care cost analysis. M.R.W., N.M.S., M.J.R., J.M.G., and J.L.D. wrote the manuscript. M.R.W. and N.M.S. contributed equally to the article.
Potential Conflicts of Interest
Nothing to report.
Acknowledgment
This work was supported by the American Brain Foundation Clinical Research Training Fellowship (to M.R.W.); Howard Hughes Medical Institute (to J.L.D.); National Center for Advancing Translational Sciences of the NIH (KL2TR000143; to M.R.W. and J.M.G.). The latter's contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The findings and conclusions are those of the authors and do not necessarily represent the official position of the U.S. Department of Health and Human Services.
We would like to thank Jessica Lund and Eric Chow at the UCSF Center for Advanced Technology for their expertise and assistance operating the Illumina sequencer.
References
- 1. Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next‐generation sequencing. N Engl J Med 2014;370:2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vora NM, Holman RC, Mehal JM, et al. Burden of encephalitis‐associated hospitalizations in the United States, 1998–2010. Neurology 2014;82:443–451. [DOI] [PubMed] [Google Scholar]
- 3. Ruby JG, Bellare P, Derisi JL. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 (Bethesda) 2013;3:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Langmead B, Salzberg SL. Fast gapped‐read alignment with Bowtie 2. Nature Methods 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu L, Niu B, Zhu Z, Wu S, Li W. CD‐HIT: accelerated for clustering the next‐generation sequencing data. Bioinformatics 2012;28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W, Godzik A. Cd‐hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006;22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 7. Wu TD, Nacu S. Fast and SNP‐tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qvarnstrom Y, Visvesvara GS, Sriram R, da Silva AJ. Multiplex real‐time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri . J Clin Microbiol 2006;44:3589–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuster FL, Yagi S, Wilkins PP, et al. Balamuthia mandrillaris, agent of amebic encephalitis: detection of serum antibodies and antigenic similarity of isolates by enzyme immunoassay. J Eukaryot Microbiol 2008;55:313–320. [DOI] [PubMed] [Google Scholar]
- 10. Lorenzo‐Morales J, Cabello‐Vilchez AM, Martin‐Navarro CM, et al. Is Balamuthia mandrillaris a public health concern worldwide? Trends Parasitol 2013;29:483–488. [DOI] [PubMed] [Google Scholar]
- 11. Visvesvara GS, Martinez AJ, Schuster FL, et al. Leptomyxid ameba, a new agent of amebic meningoencephalitis in humans and animals. J Clin Microbiol 1990;28:2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schuster FL, Glaser C, Honarmand S, Maguire JH, Visvesvara GS. Balamuthia amebic encephalitis risk, Hispanic Americans. Emerg Infect Dis 2004;10:1510–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griesemer DA, Barton LL, Reese CM, et al. Amebic meningoencephalitis caused by Balamuthia mandrillaris . Pediatr Neurol 1994;10:249–254. [DOI] [PubMed] [Google Scholar]
- 14. Rowen JL, Doerr CA, Vogel H, Baker CJ. Balamuthia mandrillaris: a newly recognized agent for amebic meningoencephalitis. Pediatr Infect Dis J 1995;14:705–710. [PubMed] [Google Scholar]
- 15. Denney CF, Iragui VJ, Uber‐Zak LD, et al. Amebic meningoencephalitis caused by Balamuthia mandrillaris: case report and review. Clin Infect Dis 1997;25:1354–1358. [DOI] [PubMed] [Google Scholar]
- 16. Reed RP, Cooke‐Yarborough CM, Jaquiery AL, et al. Fatal granulomatous amoebic encephalitis caused by Balamuthia mandrillaris . Med J Aust 1997;167:82–84. [DOI] [PubMed] [Google Scholar]
- 17. Schuster FL, Yagi S, Gavali S, et al. Under the radar: balamuthia amebic encephalitis. Clin Infect Dis 2009;48:879–887. [DOI] [PubMed] [Google Scholar]
- 18. Guarner J, Bartlett J, Shieh WJ, et al. Histopathologic spectrum and immunohistochemical diagnosis of amebic meningoencephalitis. Mod Pathol 2007;20:1230–1237. [DOI] [PubMed] [Google Scholar]
- 19. Schuster FL, Visvesvara GS. Axenic growth and drug sensitivity studies of Balamuthia mandrillaris, an agent of amebic meningoencephalitis in humans and other animals. J Clin Microbiol 1996;34:385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deetz TR, Sawyer MH, Billman G, Schuster FL, Visvesvara GS. Successful treatment of Balamuthia amoebic encephalitis: presentation of 2 cases. Clin Infect Dis 2003;37:1304–1312. [DOI] [PubMed] [Google Scholar]
- 21. Martinez DY, Seas C, Bravo F, et al. Successful treatment of Balamuthia mandrillaris amoebic infection with extensive neurological and cutaneous involvement. Clin Infect Dis 2010;51:e7–e11. [DOI] [PubMed] [Google Scholar]
- 22. Jung S, Schelper RL, Visvesvara GS, Chang HT. Balamuthia mandrillaris. meningoencephalitis in an immunocompetent patient: an unusual clinical course and a favorable outcome. Arch Pathol Lab Med 2004;128:466–468. [DOI] [PubMed] [Google Scholar]
- 23. Kodet R, Nohynkova E, Tichy M, Soukup J, Visvesvara GS. Amebic encephalitis caused by Balamuthia mandrillaris in a Czech child: description of the first case from Europe. Pathol Res Pract 1998;194:423–429. [DOI] [PubMed] [Google Scholar]
- 24. Schuster FL. Cultivation of pathogenic and opportunistic free‐living amebas. Clin Microbiol Rev 2002;15:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]


