Abstract
In this review, lipid A, from its discovery to recent findings, is presented as a drug target and therapeutic molecule. First, the biosynthetic pathway for lipid A, the Raetz pathway, serves as a good drug target for antibiotic development. Several assay methods used to screen for inhibitors of lipid A synthesis will be presented, and some of the promising lead compounds will be described. Second, utilization of lipid A biosynthetic pathways by various bacterial species can generate modified lipid A molecules with therapeutic value.
Keywords: Lipid A, Endotoxin, Drug target, Antibacterial, Adjuvant
INTRODUCTION
Lipids have not been a very popular research topic for some time, because their study imposes more technical challenges than other biological molecules, such as proteins, nucleic acids, and carbohydrates. While a lot of questions remain to be answered, it is true that remarkable advances have been recently achieved in the lipid field. In this review, lipid A, from its discovery to recent findings, is presented as a drug target with therapeutic value.
Discovery of lipid A
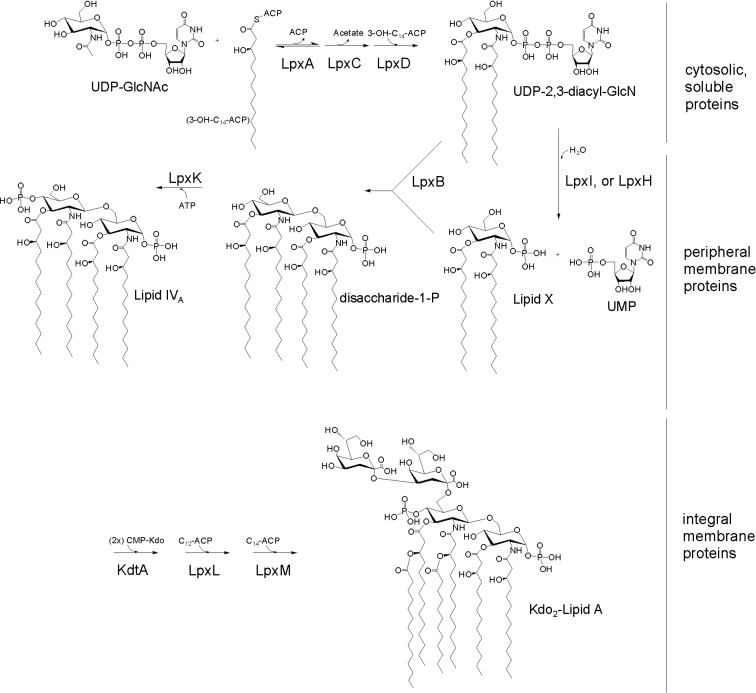
Lipid A is present mainly in Gram-negative bacteria; however, a recent study indicates it might exist in plants. It functions as a hydrophobic anchor for lipopolysaccharide (LPS) attached to the outer membrane, and it appears to be essential for the growth of cells and in LPS-free mutants. The term ‘lipid A’ was first coined by Westphal and Luderitz as the lipid rich hydrolytic fragment of LPS, and lipid B was defined as the other, more readily separated portion of phospholipids in Escherichia coli (E. coli) (Brade, 1999). The molecular identities of both lipid species are now well known, and lipid B is better known as phosphatidylethanolamine. When lipid A was first separated from the bacterial cell envelope, neither its exact molecular identity nor its biosynthetic pathway were known. In the late 1970s, when the structure of lipid A was not yet known, lipid X, a biosynthetic intermediate of lipid A, was identified from an E. coli mutant lacking phosphatidylglycerol (Nishijima and Raetz, 1979). While it was determined that lipid X contains acylated glucosamine (GlcN) and a phosphoryl group, the exact structure had yet to be defined. With the structure of lipid A not available, lipid X could not be shown to be related to lipid A. In 1983, the exact structure of lipid A was determined with the help of mass spectrometry and NMR spectroscopy (Imoto et al., 1983; Strain et al., 1983). Then, the structure of lipid X was revisited, and it was determined that lipid X is an intermediate in lipid A biosynthesis (Takayama et al., 1983). The starting compounds of lipid A biosynthesis are N-acetylglucosamine linked to uridine diphosphate (UDP-GlcNAc), and the acyl chain linked to acyl carrier protein (acyl-acyl carrier protein; acyl-ACP). After acylation of UDP-GlcNAc mediated by UDP-GlcNAc acyltransferase or lpxA (Crowell et al., 1986), eight reactions ensue to synthesize Kdo2-lipid A (Raetz and Whitfield, 2002; Raetz et al., 2007). This pathway is called the Raetz pathway (Fig. 1) as explained below.
Fig. 1.
The Raetz pathway. The biosynthetic pathway for Kdo2-lipid A (E. coli) is shown. The pathway is redrawn from (Raetz et al., 2007) for simplicity, and to indicate the location of each reaction.
Function of lipid A in Gram-negative bacteria
Obviously, the function of lipid A in Gram-negative bacteria is to anchor LPSs to the outer membrane of the cell. Generally, lipid A is considered essential for survival. Some laboratory-grown bacterial strains lack LPS, but not lipid A. The outer membrane of Francisella novicida (F. novicida) contains a lot of lipid A not linked to LPS (Wang et al., 2004). However, there are some results reporting the absence of lipid A in viable Gram-negative bacteria. Moffatt and colleagues reported a mutant of Acinetobacter baumannii in which lipid A biosynthesis is absent. These mutants were isolated from colistin-containing media used to induce colistin resistance. All the isolated mutants showed mutations in the first two enzymes of the Raetz pathway, and LPS was not present (Moffatt et al., 2011). In the case of Neisseria meningitidis, cells can grow slowly without lipid A (Steeghs et al., 1998).With the exception of the above, lipid A appears to be essential for the survival of Gram-negative bacteria. In laboratory experiments, inhibition of lipid A in a LPS-deficient mutant results in the inhibition of growth, implying that lipid A functions as more than a lipid anchor for LPS. Other biological roles of lipid A and the identities of its biosynthetic intermediates are still unclear.
Lipid A as an endotoxin
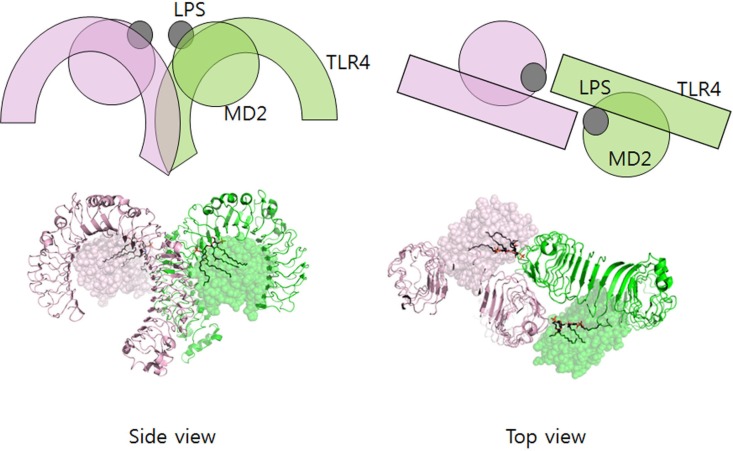
As picomolar levels of lipid A are sufficient to induce inflammation in the mammalian immune system by triggering TLR4/MD activation (Beutler and Cerami, 1988), lipid A itself can be considered an endotoxin. We can separate the immunogenicity of LPS from its toxicity; its immunogenicity depends on polysaccharide components and its toxicity is due to the lipid A moiety. While inflammation is necessary to clear up infection, overreaction of the immune system can be fatal, and Gram-negative septic shock can occur. The structure of the LPS bound to the TLR4/MD2 complex (Fig. 2) explains the formation of receptor multimers upon binding of LPS (Park et al., 2009; Ohto et al., 2012). TLR4 and MD2 exist as dimers (TLR4/MD2), and LPS binding to MD2 results in the formation of the TLR4/MD2/LPS complex (2:2:2 complex). The lipid chains in LPS are located in the hydrophobic pocket of MD2, and two phosphate groups in lipid A engage in favorable interactions with positively-charged residues in TLR4 and MD2, inducing complex formation. In human cells, the presence of two phosphate and acyloxyacyl groups is necessary to activate fully TLR4-MD-2 (Rietschel et al., 1994). Species-specificities exist among lipid A and its analogs. For example, lipid IVA, a tetra-acylated precursor of lipid A, is a weak agonist of TLR-4-MD-2 activity in mouse, while it shows antagonistic activity in human (Montminy et al., 2006). It may be possible to utilize the different lipid A analogs as therapeutics molecules. This will be discussed later.
Fig. 2.
LPS binding induces multimerization of TLR4/MD2 complexes. Model and structure (PDB ID: 3VQ2) based on (Ohto et al., 2012).
BIOSYNTHETIC PATHWAY OF LIPID A AS A DRUG TARGET
The raetz pathway
As described above, UDP-GlcNAc and acyl-ACP, in the case of E. coli, are substrates for the first step of lipid A biosynthesis. LpxA, the first enzyme in the Raetz pathway, is an acyltransferase mediating the acylation of UDP-GlcNAc and yielding UDP-3-(O)-acyl-GlcNAc. This reaction occurs in the cytoplasm, and is followed by the deacetylation of the GlcNAc moiety, mediated by lpxC (Jackman et al., 1999). This reaction yields UDP-3-(O)-acyl-GlcN as a product. As the equilibrium in the lpxA reaction is toward the substrate, the lpxC reaction is the actual step of commitment to the pathway. Once UDP-3-(O)-acyl-GlcN is formed, another acylation reaction occurs, mediated by lpxD (Vuorio and Vaara, 1995), to yield UDP-2,3-(O)-diacyl-GlcN. These three reactions occur in the cytoplasm, and the enzymes responsible are soluble proteins.
One molecule of UDP-2,3-(O)-diacyl-GlcN, and one molecule of lipid X, formed by the removal of UMP moiety from UDP-2,3-(O)-diacyl-GlcN, undergo lipid A disaccharide formation. The formation of lipid X is mediated by either lpxH (Babinski et al., 2002a; Babinski et al., 2002b) or lpxI (Metzger and Raetz, 2010), depending on the bacterial species, and lipid A disaccharide formation is mediated by lpxB (Crowell et al., 1986). These three enzymes are peripheral membrane proteins, and it appears that the reactions occur in the vicinity of the membrane.
Lipid A disaccharide undergoes further modifications. First, the phosphorylation of lipid A disaccharide results in the formation of lipid IVA, a tetra-acylated lipid A species. This reaction is mediated by lpxK (Garrett et al., 1997), a phosphorylase, or a kinase. Second, two Kdo residues are attached to lipid IVA, yielding Kdo2-IVA, and the reaction is mediated by KdtA (White et al., 1997), a Kdo transferase. The second and third reactions, mediated by lpxL (Carty et al., 1999) and lpxM (Clementz et al., 1997), are acylation reactions. Tetra-acylated Kdo2-IVA is transformed into Kdo2-lipid A, the final product in the Raetz pathway. LpxL transfers lauroyl residues while lpxM transfers a myristoyl residue to Kdo2-IVA. Two reactions occur in a sequential manner: the lpxL reaction occurs before the lpxM reaction, and the lpxM reaction does not occur in the lpxL mutant (Vorachek-Warren et al., 2002). The three enzymes that modify lipid A disaccharide are integral membrane proteins, and the reactions probably occur on the inner surface of the inner membrane.
In the above paragraphs, nine constitutive enzyme reactions in the Raetz pathway were described succinctly, using mainly E. coli as a model system. However, variations exist between bacterial species. First, lpxAs from different bacteria species show different selectivities for acyl chains and sugars. Acyl chain length can be different, either short (C10, Pseudomonas aeruginosa; P. aeruginosa (Dotson et al., 1998; Wyckoff et al., 1998), or long (Helicobacter pylori (Lee and Suh, 2003); C18, F. novicida (Wang et al., 2006b), and the lack of the 3-hydroxyl group makes a difference, as we observe myristoyl, not the 3-hydroxymyristoyl group, in the Chlamydia trachomatis lpxA reaction (Sweet et al., 2001). Sugar selectivity is observed in the case of Leptospira interrogans lpxA, as an amino sugar, UDP-2-acetamido-3-amino-2,3-dideoxy-α-D-glucopyranose, is used, resulting in the formation of amide bonds between the acyl chain and the sugar (Robins et al., 2009). Second, the formation of lipid X from UDP-diacyl-GlcN is mediated by either lpxH or lpxI, depending on the bacterial species. LpxH exists in about two-thirds of Gram-negative bacteria, while lpxI is found in the other one-third (Metzger and Raetz, 2010). LpxH and lpxI are unrelated to each other in their DNA sequences, and the enzyme mechanisms are different. These differences are important considerations in the development of inhibitors against lipid A biosynthesis.
Lipid A biosynthesis inhibitors
The biosynthetic pathway for lipid A can serve as good drug target for antibiotic development, as lipid A is essential in most Gram-negative bacteria. There have been attempts to screen for inhibitors of lipid A biosynthetic enzymes. One way is to measure the incorporation of radioactive galactose into LPS (Austin et al., 1990) to screen for an inhibitor of LPS biosynthesis. One compound, L-573,655 (Table 1), was selected during a screen of compounds by a research group at Merck Research Laboratories (Onishi et al., 1996). After selection, all nine enzymes of lipid A biosynthesis were assayed individually for sensitivity to inhibition by L-573,655, which turned out to inhibit lpxC, the second enzyme of the Raetz pathway. The inhibition constant for L-573,655 was determined to be 24 μM, and 200 analogs were synthesized to bring the inhibition constant up to ∼50 nM by L-161,240 (Table 1). While L-161,240 was an inhibitor of E. coli lpxC, the inhibition was not universal: L-161,240 could not inhibit lpxC from other species, such as P. aeruginosa or Aquifex aeolicus (A. aeolicus). Later on, hydroxamate-containing inhibitors, including TU-514 (Table 1), were developed, which turned out to be effective against both E. coli and A. aeolicus enzymes (Jackman et al., 2000). More hydroxamate-containing compounds were developed yielding several inhibitors, including CHIR-090 (Table 1) with antibacterial activity comparable to that of ciprofloxacin. CHIR-090 and other potent inhibitors have threonyl-hydroxamate in their head groups (McClerren et al., 2005). Based on knowledge of the interactions between LpxC and threonyl-hydroxamate-containing inhibitor, interactions of binding pocket and inhibitor were further increased by attaching bulky groups to threonyl-hydroxamate, as seen in the case of LPC-051 (Liang et al., 2013).
Table 1.
Structures of selected hydroxamic acids with LpxC inhibitory activity
| Compound | Structure | Ki value against E. coli LpxC | Reference |
|---|---|---|---|
| L-573,655 |
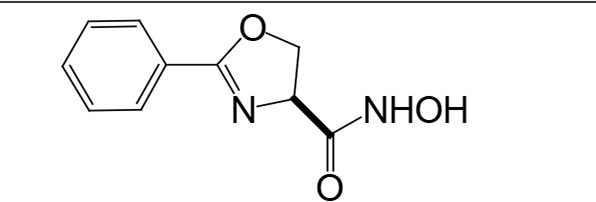
|
24 μM | Onishi et al., 1996 |
| L-161,240 |
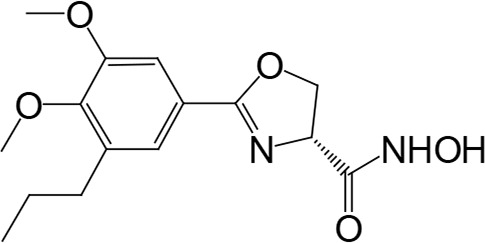
|
∼50 nM | Onishi et al., 1996 |
| TU-514 |
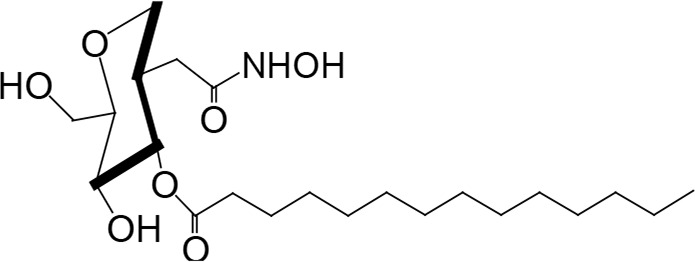
|
∼650 nM | Jackman et al., 2000 |
| CHIR-090 |
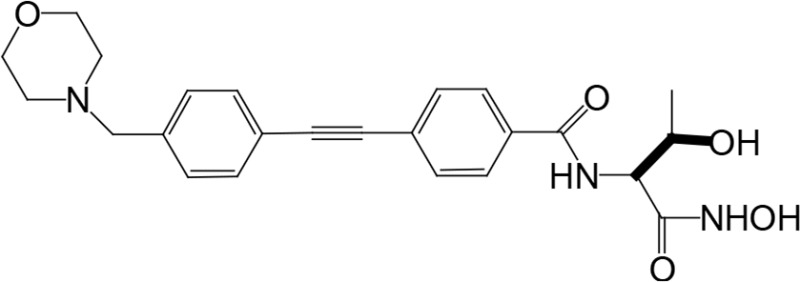
|
∼2 nM | McClerren et al., 2005 |
| LPC-051 |
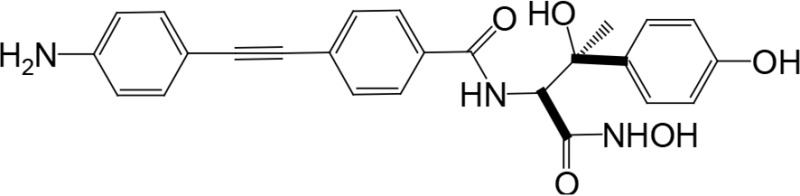
|
∼0.024 nM | Liang et al., 2013 |
As stated above, the first three enzymes of the Raetz pathway are soluble proteins, while others are non-soluble. The non-soluble ones are either peripheral membrane proteins or integral membrane proteins associated with the membrane. The development of enzyme assay systems is challenging for non-soluble proteins. Even for soluble proteins, the enzyme reactions in the Raetz pathway are hard to reconstitute in test tubes. Assays usually involve the use of radioactive isotope-labeled substrates to measure the enzyme activities, and most of the materials are not commercially available. For example, the lpxA enzyme assay requires lpxA, acylated ACPs, and radio-labeled UDP-GlcNAc, but none of these are commercially available. Therefore enzyme activity assays have not been used in high-throughput screening for inhibitors against lipid A biosynthesis. When Pirrung and colleagues developed lpxC inhibitors, they used a disk diffusion assay to test 70 oxazoline hydroxamate compounds (Pirrung et al., 2003).
Recently, a fluorescence based enzyme assay was developed for the lpxA reaction. This assay is based on the formation of free thiol from ACPs (Jenkins and Dotson, 2012): free thiols are exposed after the acyl group is transferred from acyl-ACP to UDP-GlcNAc, and this can be monitored by linking thiol exposition to other enzymatic reactions. While a previous method used a discontinuous radioassay based on thin layer chromatography (Galloway and Raetz, 1990), the new assay is a continuous assay that monitors multiple enzyme reactions on a single plate. The new method would allow higher throughput screening of large compound libraries. While most inhibitors of lipid A biosynthesis target lpxC, it does not mean lpxC is the only target. It was shown, using a phage display screening assay, that lpxA is a good antibacterial target (Benson et al., 2003), and all the enzymes in the Raetz pathway are potential targets for antibacterial agents against Gram-negative bacteria.
LIPID A ANALOGS WITH THERAPEUTIC VALUE
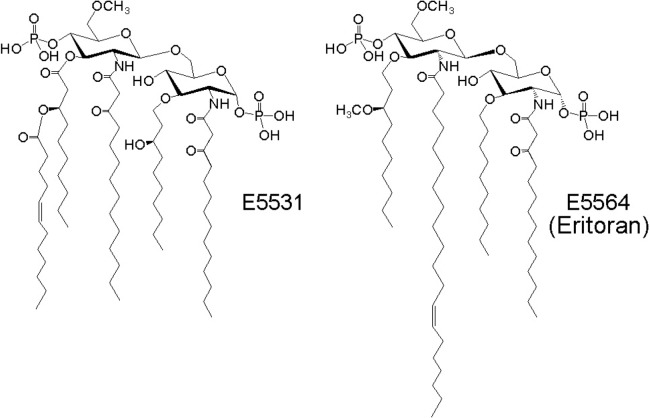
Lipid A itself is an endotoxin, and thus has no therapeutic value. However, biosynthetic intermediates and modified lipid A could have clinical applications. Lipid IVA is an antagonist of the TLR4 receptor in human cells, while it is an agonist in murine cells. It was initially hoped that lipid IVA might be useful as a therapeutic agent for the treatment of Gram-negative septicemia (Golenbock et al., 1991). However, because of the lack of stability of lipid IVA upon storage, the development of synthetic analogs was pursued instead, among which E5531 and E5564 were the most notable outcomes (Christ et al., 1995) (Fig. 3). While it is less encouraging that a recent clinical trial of E5564 (eritoran) did not reduce the mortality of septic patients (Opal et al., 2013), there are other possible clinical uses for this compound. For example, E5564 protected mice from lethal influenza infection (Shirey et al., 2013), presenting Toll-like receptors as a new drug target for antiviral therapy (Patel et al., 2014). Lipid X was once considered as an antagonist (Danner et al., 1987); however, later it was shown not to have affinity for the TLR4 receptor (Pohlman et al., 1988).
Fig. 3.
Structures of E5531 and E5564 as lipid IVA analogs.
In the above paragraph, synthetic analogs of lipid A were shown to have therapeutic value. It is also possible to prepare lipid A analogs by combining different biosynthetic enzymes from different bacterial species. As seen in the Kdo2-lipid A molecule from E. coli (Fig. 1), there are two phosphate groups, and the existence of these two phosphate groups is very important for TLR4/MD2 activation (Rietschel et al., 1994). While lipid A itself is too toxic when there are two phosphate groups present, monophospholipid A (MPLA) (Persing et al., 2002) can be prepared and used as an adjuvant, as MPLA partially activates TLR4/MD2. Interestingly, there are some bacterial species, such as Francisella tularensis, which produce lipid A devoid of phosphate groups. These species contain extra enzymes, including LpxE (Wang et al., 2004) and LpxF (Wang et al., 2006a), that function as phosphatases to get rid of the phosphate groups in lipid A. The lpxE encoding gene was inserted into a Salmonella strain to make MPLA-producing bacteria, in the hope of developing live oral vaccines (Kong et al., 2011; Wang et al., 2013). These bacterial strains could be used to deliver the antigen of choice, and to elicit moderate immune responses via MPLA, which would function as both an antigen presenter and an adjuvant.
It should also be possible to utilize biosynthetic enzymes from different species to prepare lipid A analogs in vitro. For example, PagL (Trent et al., 2001) and LpxR (Reynolds et al., 2006) exist in Salmonella species and function as lipases. These enzymes function in E. coli, and modified lipid A species are produced by cultured bacterial cells (Gibbons et al., 2000; Reynolds et al., 2006). It is expected that the repertoire of lipid A modifying enzymes from various bacterial species will allow the production of lipid A analogs with beneficial activities.
Acknowledgments
This study was supported by the Catholic University of Daegu Research Grant in 2013.
REFERENCES
- Austin EA, Graves JF, Hite LA, Parker CT, Schnaitman CA. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J Bacteriol. 1990;172:5312–5325. doi: 10.1128/jb.172.9.5312-5325.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babinski K, Kanjilal S, Raetz C. Accumulation of the lipid A precursor UDP-2,3-diacylglucosamine in an Escherichia coli mutant lacking the lpxH gene. J Biol Chem. 2002a;277:25947–25956. doi: 10.1074/jbc.M204068200. [DOI] [PubMed] [Google Scholar]
- Babinski K, Ribeiro A, Raetz C. The Escherichia coli gene encoding the UDP-2,3-diacylglucosamine pyrophosphatase of lipid A biosynthesis. J Biol Chem. 2002b;277:25937–25946. doi: 10.1074/jbc.M204067200. [DOI] [PubMed] [Google Scholar]
- Benson RE, Gottlin EB, Christensen DJ, Hamilton PT. Intracellular expression of peptide fusions for demonstration of protein essentiality in bacteria. Antimicrob Chemother. 2003;47:2875–2881. doi: 10.1128/AAC.47.9.2875-2881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Cerami A. Tumor necrosis, cachexia, shock, and inflammation: a common mediator. Annu Rev Biochem. 1988;57:505–518. doi: 10.1146/annurev.bi.57.070188.002445. [DOI] [PubMed] [Google Scholar]
- Brade H. Endotoxin in health and disease. Marcel Dekker; New York: 1999. [Google Scholar]
- Carty S, Sreekumar K, Raetz C. Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction At 12 degrees C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J Biol Chem. 1999;274:9677–9685. doi: 10.1074/jbc.274.14.9677. [DOI] [PubMed] [Google Scholar]
- Christ WJ, Asano O, Robidoux AL, Perez M, Wang Y, Dubuc GR, Gavin WE, Hawkins LD, McGuinness PD, Mullarkey MA, et al. E5531, a pure endotoxin antagonist of high potency. Science. 1995;268:80–83. doi: 10.1126/science.7701344. [DOI] [PubMed] [Google Scholar]
- Clementz T, Zhou Z, Raetz C. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J Biol Chem. 1997;272:10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- Crowell DN, Anderson MS, Raetz CR. Molecular cloning of the genes for lipid A disaccharide synthase and UDP-N-acetylglucosamine acyltransferase in Escherichia coli. J Bacteriol. 1986;168:152–159. doi: 10.1128/jb.168.1.152-159.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner RL, Joiner KA, Parrillo JE. Inhibition of endotoxin-induced priming of human neutrophils by lipid X and 3-Aza-lipid X. J Clin Invest. 1987;80:605–612. doi: 10.1172/JCI113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson G, Kaltashov I, Cotter R, Raetz C. Expression cloning of a Pseudomonas gene encoding a hydroxydecanoyl-acyl carrier protein-dependent UDP-GlcNAc acyltransferase. J Bacteriol. 1998;180:330–337. doi: 10.1128/jb.180.2.330-337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway SM, Raetz CR. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J Biol Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- Garrett T, Kadrmas J, Raetz C. Identification of the gene encoding the Escherichia coli lipid A 4′-kinase. Facile phosphorylation of endotoxin analogs with recombinant LpxK. J Biol Chem. 1997;272:21855–21864. doi: 10.1074/jbc.272.35.21855. [DOI] [PubMed] [Google Scholar]
- Gibbons H, Lin S, Cotter R, Raetz C. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem. 2000;275:32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J Biol Chem. 1991;266:19490–19498. [PubMed] [Google Scholar]
- Imoto M, Kusumoto S, Shiba T, Naoki H, Iwashita T, Rietschel ET, Wollenweber HW, Galanos C, Lüderitz O. Chemical structure of E. coli Lipid A: Linkage site of acyl groups in the disaccharide backbone. Tetrahedron Lett. 1983;24:4017–4020. doi: 10.1016/S0040-4039(00)88251-9. [DOI] [Google Scholar]
- Jackman J, Fierke C, Tumey L, Pirrung M, Uchiyama T, Tahir S, Hindsgaul O, Raetz C. Antibacterial agents that target lipid A biosynthesis in gram-negative bacteria. Inhibition of diverse UDP-3-O-(r-3-hydroxymyristoyl)-n-acetylglucosamine deacetylases by substrate analogs containing zinc binding motifs. J Biol Chem. 2000;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- Jackman J, Raetz C, Fierke C. UDP-3-O-(R-3-hydroxy-myristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme. Biochemistry. 1999;38:1902–1911. doi: 10.1021/bi982339s. [DOI] [PubMed] [Google Scholar]
- Jenkins RJ, Dotson GD. A continuous fluorescent enzyme assay for early steps of lipid A biosynthesis. Anal Biochem. 2012;425:21–27. doi: 10.1016/j.ab.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Six DA, Roland KL, Liu Q, Gu L, Reynolds CM, Wang X, Raetz CR, Curtiss R., 3rd Salmonella synthesizing 1-dephosphorylated [corrected] lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J Immunol. 2011;187:412–423. doi: 10.4049/jimmunol.1100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BI, Suh SW. Crystal structure of UDP-N-acetyl-glucosamine acyltransferase from Helicobacter pylori. Proteins. 2003;53:772–774. doi: 10.1002/prot.10436. [DOI] [PubMed] [Google Scholar]
- Liang X, Lee CJ, Zhao J, Toone EJ, Zhou P. Synthesis, structure, and antibiotic activity of aryl-substituted LpxC inhibitors. J Med Chem. 2013;56:6954–6966. doi: 10.1021/jm4007774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerren A, Endsley S, Bowman J, Andersen N, Guan Z, Rudolph J, Raetz C. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry. 2005;44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger LE, 4th, Raetz CR. An alternative route for UDP-diacylglucosamine hydrolysis in bacterial lipid A biosynthesis. Biochemistry. 2010;49:6715–6726. doi: 10.1021/bi1008744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3022–3024. doi: 10.1128/AAC.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat immunol. 2006;7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- Nishijima M, Raetz CR. Membrane lipid biogenesis in Escherichia coli: identification of genetic loci for phosphatidylglycerophosphate synthetase and construction of mutants lacking phosphatidylglycerol. J Biol Chem. 1979;254:7837–7844. [PubMed] [Google Scholar]
- Ohto U, Fukase K, Miyake K, Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc Natl Acad Sci USA. 2012;109:7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CR. Antibacterial agents that inhibit lipid A biosynthesis. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- Opal SM, Laterre PF, Francois B, LaRosa SP, Angus DC, Mira JP, Wittebole X, Dugernier T, Perrotin D, Tidswell M, Jauregui L, Krell K, Pachl J, Takahashi T, Peckelsen C, Cordasco E, Chang CS, Oeyen S, Aikawa N, Maruyama T, Schein R, Kalil AC, Van Nuffelen M, Lynn M, Rossignol DP, Gogate J, Roberts MB, Wheeler JL, Vincent JL, Group AS. Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis: the ACCESS randomized trial. JAMA. 2013;309:1154–1162. doi: 10.1001/jama.2013.2194. [DOI] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Patel MC, Shirey KA, Pletneva LM, Boukhvalova MS, Garzino-Demo A, Vogel SN, Blanco JC. Novel drugs targeting Toll-like receptors for antiviral therapy. Future Virol. 2014;9:811–829. doi: 10.2217/fvl.14.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persing DH, Coler RN, Lacy MJ, Johnson DA, Baldridge JR, Hershberg RM, Reed SG. Taking toll: lipid A mimetics as adjuvants and immunomodulators. Trends Microbiol. 2002;10:S32–37. doi: 10.1016/S0966-842X(02)02426-5. [DOI] [PubMed] [Google Scholar]
- Pirrung MC, Tumey LN, McClerren AL, Raetz CR. High-throughput catch-and-release synthesis of oxazoline hydroxamates. Structure-activity relationships in novel inhibitors of Escherichia coli LpxC: in vitro enzyme inhibition and antibacterial properties. J Am Chem Soc. 2003;125:1575–1586. doi: 10.1021/ja0209114. [DOI] [PubMed] [Google Scholar]
- Pohlman TH, Winn RK, Callahan KS, Maier RV, Harlan JM. A glycolipid precursor of bacterial lipopolysaccharide (lipid X) lacks activity against endothelial cells in vitro and is not toxic in vivo. J Surg Res. 1988;45:228–237. doi: 10.1016/0022-4804(88)90069-8. [DOI] [PubMed] [Google Scholar]
- Raetz C, Reynolds C, Trent M, Bishop R. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C, Ribeiro A, McGrath S, Cotter R, Raetz C, Trent M. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zahringer U, Seydel U, Di Padova F, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- Robins LI, Williams AH, Raetz CR. Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA. Biochemistry. 2009;48:6191–6201. doi: 10.1021/bi900629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey KA, Lai W, Scott AJ, Lipsky M, Mistry P, Pletneva LM, Karp CL, McAlees J, Gioannini TL, Weiss J, Chen WH, Ernst RK, Rossignol DP, Gusovsky F, Blanco JC, Vogel SN. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013;497:498–502. doi: 10.1038/nature12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeghs L, den Hartog R, den Boer A, Zomer B, Roholl P, van der Ley P. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- Strain SM, Fesik SW, Armitage IM. Structure and metal-binding properties of lipopolysaccharides from heptoseless mutants of Escherichia coli studied by 13C and 31P nuclear magnetic resonance. J Biol Chem. 1983;258:13466–13477. [PubMed] [Google Scholar]
- Sweet C, Lin S, Cotter R, Raetz C. A Chlamydia trachomatis UDP-N-acetylglucosamine acyltransferase selective for myristoyl-acyl carrier protein. Expression in Escherichia coli and formation of hybrid lipid A species. J Biol Chem. 2001;276:19565–19574. doi: 10.1074/jbc.M101868200. [DOI] [PubMed] [Google Scholar]
- Takayama K, Qureshi N, Mascagni P, Nashed MA, Anderson L, Raetz CR. Fatty acyl derivatives of glucosamine 1-phosphate in Escherichia coli and their relation to lipid A. Complete structure of A diacyl GlcN-1-P found in a phosphatidylglycerol-deficient mutant. J Biol Chem. 1983;258:7379–7385. [PubMed] [Google Scholar]
- Trent M, Ribeiro A, Doerrler W, Lin S, Cotter R, Raetz C. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J Biol Chem. 2001;276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- Vorachek-Warren M, Ramirez S, Cotter R, Raetz C. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J Biol Chem. 2002;277:14194–14205. doi: 10.1074/jbc.M200409200. [DOI] [PubMed] [Google Scholar]
- Vuorio R, Vaara M. Comparison of the phenotypes of the lpxA and lpxD mutants of Escherichia coli. FEMS Microbiol Lett. 1995;134:227–232. doi: 10.1111/j.1574-6968.1995.tb07942.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Kong Q, Curtiss R., 3rd New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb Pathog. 2013;58:17–28. doi: 10.1016/j.micpath.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Karbarz M, McGrath S, Cotter R, Raetz C. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of francisella novicida LpxE expressed in Escherichia coli. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McGrath S, Cotter R, Raetz C. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J Biol Chem. 2006a;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro A, Guan Z, McGrath S, Cotter R, Raetz C. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006b;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Kaltashov I, Cotter R, Raetz C. A mono-functional 3-deoxy-D-manno-octulosonic acid (Kdo) transferase and a Kdo kinase in extracts of Haemophilus influenzae. J Biol Chem. 1997;272:16555–16563. doi: 10.1074/jbc.272.26.16555. [DOI] [PubMed] [Google Scholar]
- Wyckoff T, Lin S, Cotter R, Dotson G, Raetz C. Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J Biol Chem. 1998;273:32369–32372. doi: 10.1074/jbc.273.49.32369. [DOI] [PubMed] [Google Scholar]