Abstract
Kinesin superfamily members play important roles in many diverse cellular processes, including cell motility, cell division, intracellular transport, and regulation of the microtubule cytoskeleton. How the properties of the family-defining motor domain of distinct kinesins are tailored to their different cellular roles remains largely unknown. Here, we employed molecular-dynamics simulations coupled with energetic calculations to infer the family-specific interactions of kinesin-1 and kinesin-3 motor domains with microtubules in different nucleotide states. We then used experimental mutagenesis and single-molecule motility assays to further assess the predicted residue-wise determinants of distinct kinesin-microtubule binding properties. Collectively, our results identify residues in the L8, L11, and α6 regions that contribute to family-specific microtubule interactions and whose mutation affects motor-microtubule complex stability and processive motility (the ability of an individual motor to take multiple steps along its microtubule filament). In particular, substitutions of prominent kinesin-3 residues with those found in kinesin-1, namely, R167S/H171D, K266D, and R346M, were found to decrease kinesin-3 processivity 10-fold and thus approach kinesin-1 levels.
Main Text
Kinesins are a large superfamily of microtubule-based motor proteins, with individual family members playing essential roles in cell division, cell motility, and intracellular trafficking. All kinesins contain a family-defining motor domain that enables nucleotide-dependent interactions with the microtubule lattice. General principles of how kinesin motor domains interact with nucleotide and microtubules have been established based on extensive biochemical and biophysical studies of kinesin-1. In particular, it has been demonstrated that alternating ATP binding and hydrolysis in each kinesin-1 motor domain leads to coordinated changes in microtubule binding affinity that enable processive motility (the ability to undergo many steps along the microtubule surface without dissociating) (1, 2). It has been assumed that this is the mechanistic paradigm for all kinesin motors. However, recent work indicates that some kinesin motors utilize their core motor domain for very different functions. For example, the kinesin-8 and kinesin-13 families depolymerize microtubules (3, 4). Even for the conventional property of processive motility, evolutionary tuning of the core motor domain has resulted in a range of family-specific processivities. For example, some kinesin-4 motors are nonprocessive (5), whereas kinesin-3 motors are superprocessive, being 10-fold more processive than kinesin-1 (6). How sequence divergence within the motor domain gave rise to these different properties remains an outstanding question in the field of cellular and molecular biology.
To identify microtubule interactions that contribute to family-specific motility properties, we utilized our recent cryo-electron microscopy structures of kinesin-3 and kinesin-1 on microtubules (7) and employed molecular-dynamics simulations and energetic calculations. We then used experimental mutagenesis and single-molecule motility assays to assess the predicted determinants of their distinct microtubule binding properties.
Kinesin-3 displays more extensive microtubule interactions than kinesin-1
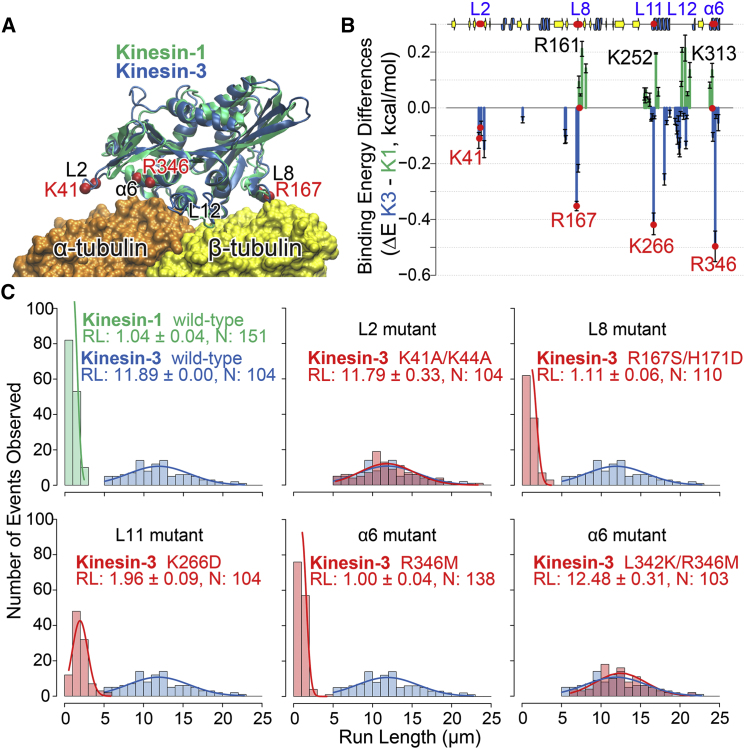
Molecular-dynamics simulations and molecular mechanics with generalized Born and surface area solvation (MM/GBSA) binding energy calculations indicate that the residues that contribute to motor-microtubule stability are clustered in the same six regions (L2, L7, L8, L11-α4, L12, and α6) on the surface of the kinesin-1 and kinesin-3 motor domains (Fig. 1, A and B, and Figs. S1–S3 in the Supporting Material). In general, kinesin-3 was found to exhibit more extensive interactions with the tubulin surface than kinesin-1 (Fig. 1 B). The L2 region makes a comparatively minor contribution to the tubulin binding of both kinesin-1 and kinesin-3 (Figs. S2 and S3), despite the presence of a family-specific seven-amino-acid insertion in this region of kinesin-3 motors (Fig. S4). The L7 region also makes comparatively minor energetic contributions to complex stability for both kinesin-1 and kinesin-3 (Figs. 1 B, S2, and S3). By contrast, the L8 region provides a larger contribution due to extensive contacts with the H12 region of β-tubulin. Particularly notable are the strong electrostatic contributions from the kinesin-3 residues R167 and R169 in all nucleotide states. Kinesin-1 lacks direct equivalents of these charged residues and instead coordinates with the microtubule via R161 in the Apo state and K166 in the ADP state (Figs. S2 and S3).
Figure 1.
Altering select family-specific tubulin interactions of kinesin-3 motors reduces processivity. (A) Refined molecular structures for kinesin-1 (green) and kinesin-3 (blue) resulting from cryo-electron microscopy (7) and subsequent molecular-dynamics simulations (see Supporting Materials and Methods for full details). (B) Differences in the residue contribution to the binding energy for kinesin-1 (green) and kinesin-3 (blue) in the ATP state. These values were determined from four replicate 40-ns molecular-dynamics simulations and subsequent energetic calculations. Note that specific interactions of the L2, L7, L8, L11, and α6 regions are predicted to enhance the binding affinity of kinesin-3 in relation to kinesin-1. (C) Processivity measurements from single-molecule motility assays of wild-type kinesin-1 (green), wild-type kinesin-3 (blue), and kinesin-3 mutants (red). The average run length (RL) and number of observations (N) are noted.
Different charge distributions and energetic contributions are also evident for the L11-α4, L12, and α6 regions. For L11-α4, both motors display extensive interactions and high binding energies with the microtubule surface. Notable are R254, K261, and K266 of kinesin-3, which display strong interactions with tubulin in the ATP and Apo states but weaker interactions in the ADP state (Figs. 1 B and S3), as well as the interactions of K237 of kinesin-1 in both the Apo and ATP states and K252 in the ADP state (Fig. S2). The L12 region also contributes strong interactions with tubulin in both motors. Particularly notable is residue R307 in kinesin-3 and the equivalent R278 in kinesin-1 (Figs. S2 and S3). Finally, strong interactions of the α6 helix with tubulin can be observed for kinesin-3 and originate from residues R346 and R350 in all nucleotide states (Figs. 1 B and S3). However, the α6 region of kinesin-1 displays a different pattern of exposed charges and makes comparatively minor energetic contributions through residues K313 and R321 (Figs. 1 B and S2).
Collectively, this analysis identified differences between kinesin-1 and kinesin-3 motors in the extent of their microtubule interactions and binding energies (see the distinct kinesin-3 and kinesin-1 peaks in Figs. 1 B and S5). We hypothesized that these differences contribute to distinct stabilities for each motor-microtubule complex that in turn determine the distinct motile properties of these motors. To test this hypothesis, we replaced kinesin-3 residues with the corresponding kinesin-1 amino acids (Figs. S4 and S5) and examined the effects on the energetics of motor-tubulin interactions computationally and on motility properties experimentally in single-molecule assays. For this analysis, we focused on the L2, L8, L11, and α6 regions, as L7 was predicted to contribute little to motor-microtubule stability (see above) and L12 was demonstrated in both Brownian dynamics simulations (8) and experimental work (9) to enhance the association kinetics of kinesin-3 motors with the microtubule rather than the processivity. Intriguingly, the results presented here indicate that the family-specific K-loop insertion in L12 of kinesin-3 motors lacks significant energetic interactions with tubulin (Fig. S3) and thus is predicted to have relatively little effect on motor-microtubule stability. L2 was included in this analysis despite its predicted minor contributions to tubulin binding energetics (Figs. 1 B, S2, and S3) due to its functional importance in other kinesin families (10).
Specific regions of the kinesin-3-microtubule interface contribute to interaction energy, velocity, and processivity
For L2, we replaced the two lysine residues at positions 41 and 44 with alanine residues to negate their electrostatic contributions to motor-microtubule stability. The resulting K41A/K44A mutant did not significantly alter the predicted binding affinity (ΔΔG 1.6 kcal/mol) and did not affect the experimentally determined velocity and run length values of the motor (2.13 ± 0.13 μm/s and 11.79 ± 0.33 μm, respectively; Figs. 1 C, S6, and S7). These results indicate that the L2 region is not a major determinant of distinct kinesin-3 motility properties.
We introduced mutations in the L8 region (R167S/H171D) to investigate the role of the predicted strong interactions of this region with the H12 region of β-tubulin. In the mutant Apo state, the missing wild-type interactions were predicted to be partially compensated for by R307 in L12, and by R346 and R350 in α6. These interactions result from a motor domain conformational rearrangement that positions α6 2.8 Å closer to the microtubule surface, leading to stronger interactions with α-tubulin H11 and H12 (Fig. S8) and an increased Apo-state binding affinity (ΔΔG −7.36 kcal/mol). In contrast, these new interactions were absent from the ATP state, which displayed an overall destabilization of the tubulin interface, resulting in a large reduction of the predicted binding affinity (ΔΔG 11.07 kcal/mol). The experimental motility assays for this mutant displayed reduced velocity and processivity values (1.32 ± 0.03 μm/s and 1.11 ± 0.06 μm; Fig. 1 C, S6, and S7). This result indicates that the identified residues in the L8 region of kinesin-3 contribute to the enhanced motility of wild-type kinesin-3.
The L11 region was also found to contribute to the enhanced stability of the kinesin-3/tubulin complex. In particular, the K266D mutation was observed to weaken tubulin interactions in both the ATP and Apo states (ΔΔG 6.66 and 7.58 kcal/mol, respectively) and resulted in a slower (1.68 ± 0.03 μm/s) and less processive (1.96 ± 0.09 μm) mutant motor (Figs. 1 C, S6, and S7). These results indicate that K266 in L11 is important for the enhanced processivity of kinesin-3.
Finally, we further investigated the potential family-specific interactions of the α6 region with tubulin via the single mutation R346M and double mutation L342K/R346M. For wild-type kinesin-3, R346 forms strong electrostatic interactions with the α-tubulin residues E415 and E421. Mutagenesis studies in budding yeast have suggested that both of these H12 α-tubulin residues are important for the interaction of kinesin proteins with microtubules (11). The R346M mutant removes the exposed positive charge on helix α6, and the double mutant L342K/R346M is predicted to retain a charge in this region complementary to that in kinesin-1 (on the next turn of the α6 helix; Fig. S1). Consistent with this energetic analysis, the R346M single mutant showed reduced velocity (1.19 ± 0.44 μm/s) and processivity (1.00 ± 0.04 μm), whereas the double mutant L342K/R346M displayed little variation in velocity (1.73 ± 0.01 μm/s) or processivity (12.48 ± 0.31 μm; Figs. 1, B and C, S6, and S7). These results highlight how analogous interactions can result from nonequivalent positions (i.e., nonaligned residues), indicating that one should consider multiple substitutions and potential epistatic effects when examining the collective determinants of enhanced kinesin-3 processivity.
In summary, using a combined computational and experimental approach, we have uncovered kinesin-3 family-specific tubulin interactions that influence motor-microtubule complex stability and motor processivity. Our results indicate that the family-specific distribution of exposed charges in L8, L11, and α6 regions result in distinct energetic interactions with the microtubule that affect kinesin motility. In particular, kinesin-3 R167 in L8, K266 in L11, and R346 in α6 contribute to the enhanced processivity of this motor in relation to kinesin-1, as their independent mutation resulted in a reduction of velocity and processivity. More broadly, these findings emphasize how processivity can be modulated by sequence differences intrinsic to individual motor domains in addition to established factors such as neck-linker composition (12, 13, 14). We suggest that our predictive approach should be widely applicable to additional families as well as to motor domain mutations linked to various diseases, including neurodegeneration and tumorigenesis. These expanded studies will generate new insights into how new motors can be custom engineered with distinctive motility properties.
Author Contributions
B.J.G., K.J.V., and C.A.M. designed the study. G.S., V.S., X.-Q.Y., B.J.G., and J.A. performed the research. G.S., B.J.G., K.J.V., and C.A.M. wrote the manuscript.
Acknowledgments
We thank Dr. Rob Cross for valuable discussions.
This work was supported by the University of Michigan, the Medical Research Council (MR/J000973/1 to J.A. and C.A.M.), and the National Institutes of Health (R01070862 to K.J.V.).
Editor: E. Michael Ostap.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Guido Scarabelli and Virupakshi Soppina contributed equally to this work.
Supporting Materials and Methods and eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00862-0.
Contributor Information
Kristen J. Verhey, Email: kjverhey@umich.edu.
Barry J. Grant, Email: bjgrant@umich.edu.
Supporting Material
References
- 1.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 2.Toprak E., Yildiz A., Selvin P.R. Why kinesin is so processive. Proc. Natl. Acad. Sci. USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanenbaum M.E., Macurek L., Medema R.H. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr. Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Hunter A.W., Caplow M., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bringmann H., Skiniotis G., Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- 6.Soppina V., Norris S.R., Verhey K.J. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atherton J., Farabella I., Moores C.A. Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. eLife. 2014;3:e03680. doi: 10.7554/eLife.03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant B.J., Gheorghe D.M., Cross R.A. Electrostatically biased binding of kinesin to microtubules. PLoS Biol. 2011;9:e1001207. doi: 10.1371/journal.pbio.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soppina V., Verhey K.J. The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors. Mol. Biol. Cell. 2014;25:2161–2170. doi: 10.1091/mbc.E14-01-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H., Fonseca C., Stumpff J. A unique kinesin-8 surface loop provides specificity for chromosome alignment. Mol. Biol. Cell. 2014;25:3319–3329. doi: 10.1091/mbc.E14-06-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchimura S., Oguchi Y., Muto E. Key residues on microtubule responsible for activation of kinesin ATPase. EMBO J. 2010;29:1167–1175. doi: 10.1038/emboj.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shastry S., Hancock W.O. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreasson J.O., Milic B., Block S.M. Examining kinesin processivity within a general gating framework. elife. 2015;4:e07403. doi: 10.7554/eLife.07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milic B., Andreasson J.O., Block S.M. Kinesin processivity is gated by phosphate release. Proc. Natl. Acad. Sci. USA. 2014;111:14136–14140. doi: 10.1073/pnas.1410943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.