Abstract
Intrinsically disordered proteins and intrinsically disordered regions are implicated in many biological functions and associated with many diseases, but their conformational characterizations are challenging. The disordered β6/β7 loop of Staphylococcus aureus sortase A is involved in the binding of both sorting signals and calcium. Calcium binding allosterically activates the enzyme, but the detailed mechanism has been unclear. Here we adapted the replica exchange with solute tempering method to sample the conformations of the β6/β7 loop, in apo form and in three liganded forms (bound with a sorting signal or calcium or both). The extensive molecular dynamics simulations yield atomic details of the disordered-to-order transition of the loop and suggest a mechanism for allosteric activation: calcium binding results in partial closure and ordering of the loop, thereby leading to preorganization of the binding pocket for the sorting signal. The approach has general applicability to the study of intrinsically disordered regions.
Introduction
Intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) are implicated in many biological functions including signaling and regulation, and associated with many diseases (1, 2, 3). The structure and function of IDPs and IDRs are receiving ever-increasing attention (4, 5). In their functional processes, many IDPs and IDRs gain well-defined structures upon binding targets or ligands, often following a dock-and-coalesce mechanism (6). Conformational characterizations of IDPs and IDRs are a key step toward understanding functions and mechanisms. Here we report the adaption of a molecular dynamics simulation method called replica exchange with solute tempering (REST), to sample the conformations of an intrinsically disordered active-site loop of an enzyme, Staphylococcus aureus sortase A (SrtA).
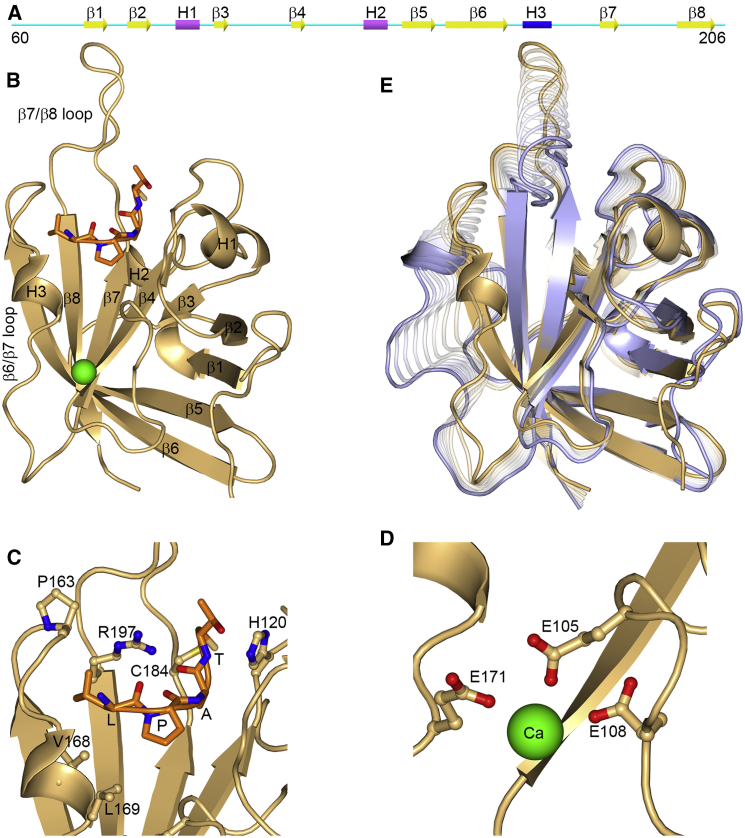
SrtA anchors to proteins that bear a cell-wall sorting signal LPXTG (X representing any amino acid). A part of this process involves cleavage of the peptide bond between the Thr and Gly residues, and subsequent formation of a thioester bond between the SrtA catalytic residue Cys184 and the sorting signal Thr residue (7). As shown by nuclear magnetic resonance (NMR) and x-ray structures, the catalytic domain of SrtA (residues 60–206) adopts an eight-strand β-barrel fold (Fig. 1) (8, 9, 10). The sorting signal sits on a floor formed by residues around the termini of the β4, β7, and β8 strands and is enclosed by residues in the N-terminal region of the long β6/β7 loop (Fig. 1, B and C) (8, 9). In the apo form, residues in this active-site loop showed exceedingly low NMR order parameters (11) as well as missing electron densities and large conformational differences between subunits within the crystallographic asymmetric unit (9), suggesting structure disorder (Fig. 1 E). Ca2+ binding was found to increase the enzymatic activity by eightfold (10). The Ca2+ binding site is formed by residues in the β3/β4 loop and in the C-terminal region of the active-site loop, and hence is distal to the active site (Fig. 1, B and D) (8, 10, 11). Ca2+ binding quenched the fast (picoseconds to nanoseconds) dynamics of the active-site loop and redistributed slow (microseconds to milliseconds) dynamics toward the active site (11), but a detailed mechanism for the allosteric activation by Ca2+ is still lacking.
Figure 1.
Structure of S. aureus sortase A. (A) Secondary structure of the catalytic domain. (B) The catalytic domain (colored in gold) bound with a sorting signal (sequence LPAT; in orange) and a Ca2+ ion (as green sphere), from model 1 of the NMR structure in PDB: 2KID (8). (C) Enlarged view of sorting signal binding site, displaying (as ball-and-stick) sorting signal-contacting residues around the C-termini of β4 and β7 and N-terminus of β8 and in the N-terminal region of the active site (i.e., β6/β7) loop. (D) Enlarged view of the Ca2+ binding site, displaying (as ball-and-stick) the three glutamates that coordinate the Ca2+ ion. (E) Superposition of the x-ray structure for apo SrtA (chain C in PDB: 1T2P,blue) (9) and the NMR structure for SrtAPep/Ca (gold). Conformational changes from apo to holo, especially prominent in the active-site loop (residues 159–177) and β7/β8 loop (residues 182–198), are illustrated by morphing calculated by the Yale Morph Server (36). The active-site loop in apo SrtA also exhibits significant structure disorder, as demonstrated by missing electron densities in chain B and large conformational differences between chains A and C (9). To see this figure in color, go online.
Conformational characterizations of IDPs and IDRs present a significant challenge for both experimental and computational approaches. Among the most useful experimental techniques, NMR spectroscopy can provide valuable but nevertheless limited data (12). Molecular dynamics simulations hold great potential, and a number of enhanced sampling methods have been applied to the study of IDPs and IDRs. For example, accelerated molecular dynamics simulations of SrtA by Kappel et al. (13) identified multiple possible binding modes for a substrate peptide. Temperature replica exchange molecular dynamics (14) has also been used to sample conformations of IDPs and IDRs (15, 16). As the system size increases, the number of replicas required for adequate sampling becomes a limiting factor. As an alternative, Hamiltonian replica exchange methods have been developed. Among them, a method called multiscale enhanced sampling (MSES) was used to study the disorder-to-order transition of the SrtA active-site loop (17). The basic idea of MSES is that an all-atom representation of the system is coupled to a coarse-grained model and replicas with different coupling strengths are allowed to exchange. The sampling efficiency is dictated by the ability of the coarse-grained simulations to capture the conformational dynamics of the all-atom model (18). MSES simulations found little effect on the conformations of the N-terminal region of the active-site loop by Ca2+ binding (17).
REST is a different form of Hamiltonian replica exchange, introduced by Liu et al. (19) and subsequently modified to improve sampling efficiency (20, 21). An implementation in GROMACS is now available (21, 22). In REST, the total potential energy of a system is divided into three components: the protein energy Upp, the protein-solvent interaction energy Upw, and the solvent energy Uww. Scaling is introduced as
and replicas are assigned a range of scaling factors with an upper bound of 1. Although all the replicas are simulated at the same temperature (T), with the scaling the effective temperature of the protein can be higher (at T/λ). By limiting the scaling to the first two energy terms, the exchange probability between two replicas is only determined by the energy of the protein and its interaction with the solvent, not by the energy of the large number of solvent molecules. Because the first two terms generally account for only a small percentage of the total energy of the system, a relatively small number of replicas is needed. REST has been used mostly for simulating folding and for conformational sampling of peptides (19, 20, 21, 22, 23), but also for conformational sampling of lipids in a bilayer (24) and of a ligand in a protein-ligand complex (25).
Normally a whole solute molecule (e.g., a peptide) is chosen for scaling. However, a region of a large solute molecule (e.g., a protein) can be just as well chosen to achieve enhanced sampling, and this choice is a perfect match for exploring the conformational ensembles of IDRs. Here we used this REST protocol to investigate the disorder-to-order transition of the SrtA active-site loop. We demonstrate significant gain in sampling efficiency over a conventional molecular dynamics (cMD) simulation and arguably also gain over the previous MSES simulations. In our REST simulation on apo SrtA, the active-site loop exhibits considerable flexibility, as evidenced by the coexistence of three conformational substates. Binding of either a sorting signal or Ca2+ or both reduces the flexibility of the active-site loop, now largely confined to a single conformational state. Importantly, the simulations show that binding of Ca2+ alone induces partial closure and ordering of the active-site loop, thereby leading to preorganization of the binding pocket for the sorting signal. That is, prebinding of Ca2+ with the β3/β4 loop accelerates the docking of the C-terminal region of the active-site loop via electrostatic attraction, and furthermore facilitates the coalescence of the N-terminal region of this loop around the sorting signal.
Materials and Methods
System and simulation setup
The starting structure for simulations on apo SrtA was model 1 of the NMR structure of Ca2+-bound SrtA (SrtACa; PDB: 1IJA), in which Ca2+ was missing due to lack of structural restraints. The starting structures for simulations on the three liganded forms were all from model 1 of the NMR structure of holo SrtA (SrtAPep/Ca; PDB: 2KID), with either the sorting signal (sequence LPAT) or the Ca2+ ion or both retained. The sorting signal was capped by acetyl and N-methyl amide groups, respectively, at the N- and C-termini. Similar to Moritsugu et al. (17), we weakly constrained the sorting signal and Ca2+ ion to their respective binding sites, by one-sided harmonic potentials. The harmonic potentials had a force constant of 0.5 kcal/mol/Å2 and started at an interatomic distance of 5 Å for the sorting signal or 3.5 Å for the Ca2+ ion. For the sorting signal, the one-sided harmonic constraints were imposed on peptide-protein heavy atom pairs within a 4-Å distance cutoff in model 1 of 2KID, excluding the active-site loop (residues 159–177) and the β7/β8 loop (residues 182–198), but including the pair between the carbonyl oxygen of the peptide C-terminal threonine and the sulfur of the protein Cys184 (to mimic the thioester bond). For the Ca2+ ion, the constraint partners were the oxygen atoms in the side chains of Glu171, Glu105, Glu108, and Asp112 and the backbone of Asn114.
We performed five separate simulations using GROMACS 4.6.3 (26) patched with the PLUMED 2.0 plug-in (22). One was a cMD simulation on apo SrtA, lasting 1600 ns. The other four were REST simulations on apo SrtA, SrtACa, sorting signal-bound SrtA (SrtAPep), and SrtAPep/Ca. Each REST simulation involved 16 replicas and each replica was simulated to 100 ns; hence the total simulation time was also 1600 ns. The force field was Amber99sb (27). The solvent between each protein system and the nearest side of the cubic simulation box was at least 10 Å thick, comprising ∼9000 TIP3P water molecules and 0.15 M neutralizing NaCl. Long-range electrostatic interactions were treated using particle-mesh Ewald (28), with a direct-space cutoff of 10 Å and a grid spacing of 1.2 Å. Temperature was maintained at 300 K by velocity rescaling. All bonds involving hydrogen atoms were restrained by LINCS (29), allowing an integration time of 2 fs.
Details of REST protocol
The 19 residues of the active-site loop were selected for enhanced sampling. In the REST simulations, replica exchanges were attempted every 2 ps (20). The values of the scaling factor λ were adjusted by trial and error to achieve roughly equal acceptance rates for exchanges between neighboring replicas. For each REST simulation, 16 replicas were used, and the λ-values finally chosen were: 1, 0.965, 0.930, 0.895, 0.860, 0.825, 0.790, 0.755, 0.721, 0.688, 0.656, 0.625, 0.595, 0.566, 0.538, and 0.511. Correspondingly, the effective temperatures (i.e., T/λ) of the active-site loop in the 16 replicas were 300, 311, 323, 335, 349, 364, 380, 397, 416, 436, 457, 480, 504, 530, 558, and 587 K. The acceptance rate was ∼0.47.
Principal component analysis
Principal component analysis (PCA) was performed on conformations collected from all the five simulations and NMR and x-ray structures of SrtA. The total collection consisted of 50,000 conformations from the cMD simulation on apo SrtA, sampled at every 32 ps; 50,000 conformations from each of the four REST simulations, sampled on the replica with λ = 1 at every 2 ps; 25 models in the NMR structure of SrtACa (PDB: 1IJA); 20 models in the NMR structure of SrtAPep/Ca (PDB: 2KID); and two chains in the x-ray structure of apo SrtA (PDB: 1T2P). Before PCA, all the conformations were superimposed on the Cα atoms of the β1 (residues 74–78), β2 (residues 83−87), β5 (residues 141–146), and β6 (residues 149–156) strands. PCA on the Cα atoms of the active-site loop used the built-in tools in GROMACS.
Chemical shift prediction
Backbone chemical shifts were predicted using the SPARTA+ program (30), on 2500 conformations from each REST simulation (sampled at every 40 ps). SPARTA+ was trained on a database of chemical shifts for structured proteins. Chemical shift predictions for the SrtA IDR could therefore be worse than for the structured regions.
Results
We carried out extensive molecular dynamics simulations to explore the conformational ensembles of the SrtA active-site loop in the apo form and the three liganded forms. A long (1600-ns) cMD simulation on apo SrtA served as a benchmark for evaluating sampling efficiency. Four 100-ns REST simulations, each involving 16 replicas in which the potential energy of the active-site loop was scaled downward to different extents to mimic heating up to 587 K, were used to compare conformational preferences among apo SrtA, SrtACa, SrtAPep, and SrtAPep/Ca.
REST versus cMD on apo SrtA
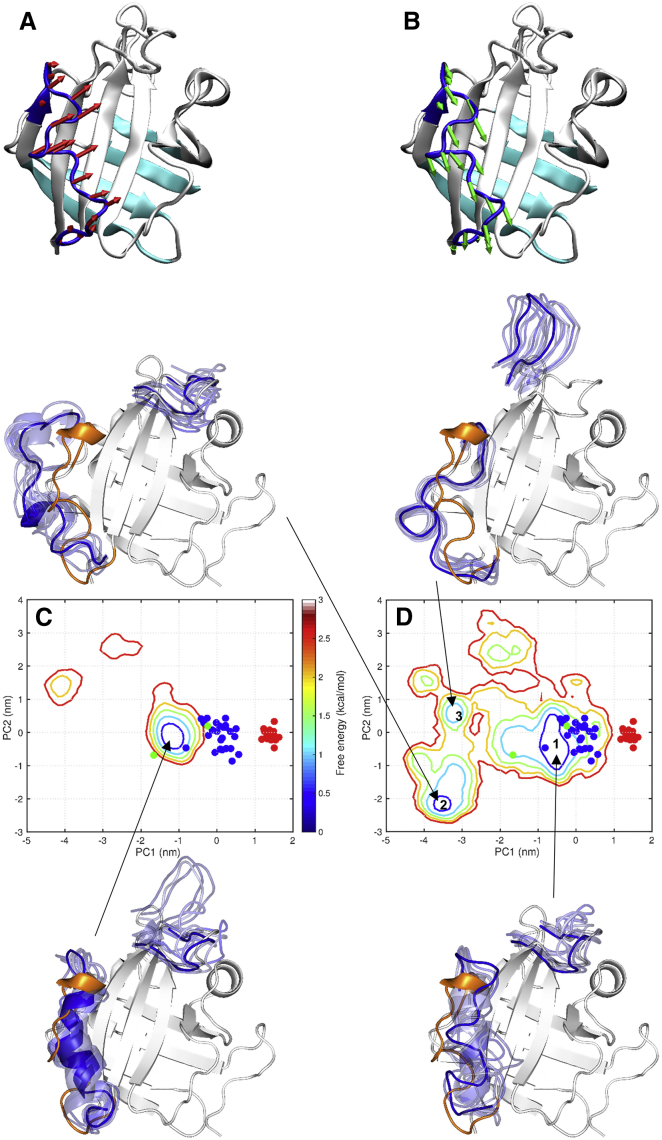
The cMD and REST simulations on apo SrtA accrued the same total simulation time (1600 ns) and started from the same structure, i.e., model 1 of the NMR structure for SrtACa (PDB: 1IJA) (10), in which Ca2+ was missing. The conformational space covered by the REST simulation is significantly broader than the cMD simulation (Fig. 2). To display the difference, we carried out PCA on the active-site loop after superposition on four β-strands (Fig. 2, A and B, cyan ribbon). The first two principal components, PC1 and PC2, capture global movements of the active-site loop (Fig. 2, A and B, arrows). These movements are largely confined to a plane parallel to the central β-sheet comprising β4, β7, and β8, and can thus be described as “gliding”. PC1 is approximately directed at the sorting signal binding site whereas PC2 is approximately directed at the Ca2+ binding site. We obtained the distributions of PC1 and PC2 in the cMD and REST simulations and then converted them into free-energy surfaces according to the Boltzmann relation. In the cMD simulation the active-site loop is mostly in a semiopen substate, defined by a narrow free-energy basin bordered by the conformations of chains A and C in the x-ray structure for apo SrtA (PDB: 1T2P; Fig. 2 C) (9). The corresponding basin in the REST simulation is significantly broader, and contains both of the two x-ray conformations for apo SrtA and the 25 NMR models for SrtACa (Fig. 2 D). Moreover, two other basins appear. Hereafter the conformational substates defined by the three free-energy basins will be numbered 1, 2, and 3, respectively. Whereas the active-site loop is semiopen in substate 1, it is wide open in the N-terminal region in substate 2, and wide open in the C-terminal region in substate 3. Below, we will use substate 1 of apo SrtA in the REST simulation as the reference for describing conformational changes in the liganded forms.
Figure 2.
Comparison of conformational ensembles of apo SrtA sampled by the cMD and REST simulations. (A and B) Conformational differences represented by the first two principal components, PC1 (red arrows) and PC2 (green arrows), on a SrtA conformation with both PC1 and PC2 near 0. Four β-strands (cyan), β1, β2, β5, and β6, were used for superposition before PCA on the active-site loop. The view is rotated by ∼45° around a vertical axis from that for Fig. 1. (C and D) Free-energy surfaces, calculated from distributions of PC1 and PC2 in the cMD and REST simulations, respectively, on apo SrtA and contoured at 0.5 kcal/mol internals. PC coordinates are also displayed for chains A and C in the x-ray structure of apo SrtA (PDB: 1T2P) (green dots); 25 models in the NMR structure of SrtACa (PDB: 1IJA) (blue dots); and 20 models in the NMR structure of SrtAPep/Ca (PDB: 2KID) (red dots). One substate from the cMD simulation and three substates from the REST simulations are each illustrated by 10 conformations within the first contour around a free-energy local minimum, with only the active-site loop and β7/β8 loop displayed (in blue) along with chain C of 1T2P (gray with active-site loop in orange). To see this figure in color, go online.
The apo x-ray structure and holo NMR structure show that residues in the active-site loop have a tendency to form a 310 helix, but not an α-helix. This tendency is captured well by the REST simulation, showing 15% of average 310 helix content for the active-site loop and little sign of α-helix. On the other hand, in the cMD simulation the active-site loop has a significant α-helix content (at 17%; see also Fig. 2 C). The propensity for a 310 helix is also seen in the REST simulations on the three liganded forms (see Fig. S1 in the Supporting Material for residue-specific 310 helix probabilities). A 310 helix is present in the active-site loop of apo SrtA from Streptococcus pyogenes (31), which has 24% sequence identity with the S. aureus ortholog.
Besides the active-site loop, the β7/β8 loop also exhibits a significant conformational change from the apo x-ray structure to the holo NMR structure, opening up as if to make room for the bound sorting signal (Fig. 1 E). In the cMD simulation, the β7/β8 loop is sporadically open (Fig. 2 C). In the REST simulation, the β7/β8 loop is stably open in substate 3 (Fig. 2 D). Here the N-terminal region of the active-site loop has the closest approach to β8, thereby mimicking an arriving sorting signal in pushing open the β7/β8 loop. The REST simulation thus was able to reveal an apparent cooperative effect between the active-site and β7/β8 loops, even though only one of them was subject to energy scaling for enhanced sampling.
Taken together, we can conclude that the REST simulation provides a much more extensive sampling of the conformational space of the active-site loop than the cMD simulation. The results below further suggest that REST may exceed MSES in sampling efficiency. The REST sampling efficiency came about because the replicas readily exchanged (Fig. S2). Regardless where along the effective temperature ladder a replica was started, it was able to traverse the entire ladder with significant probabilities during the simulations.
Disorder-to-order transition of the active-site loop upon ligand binding
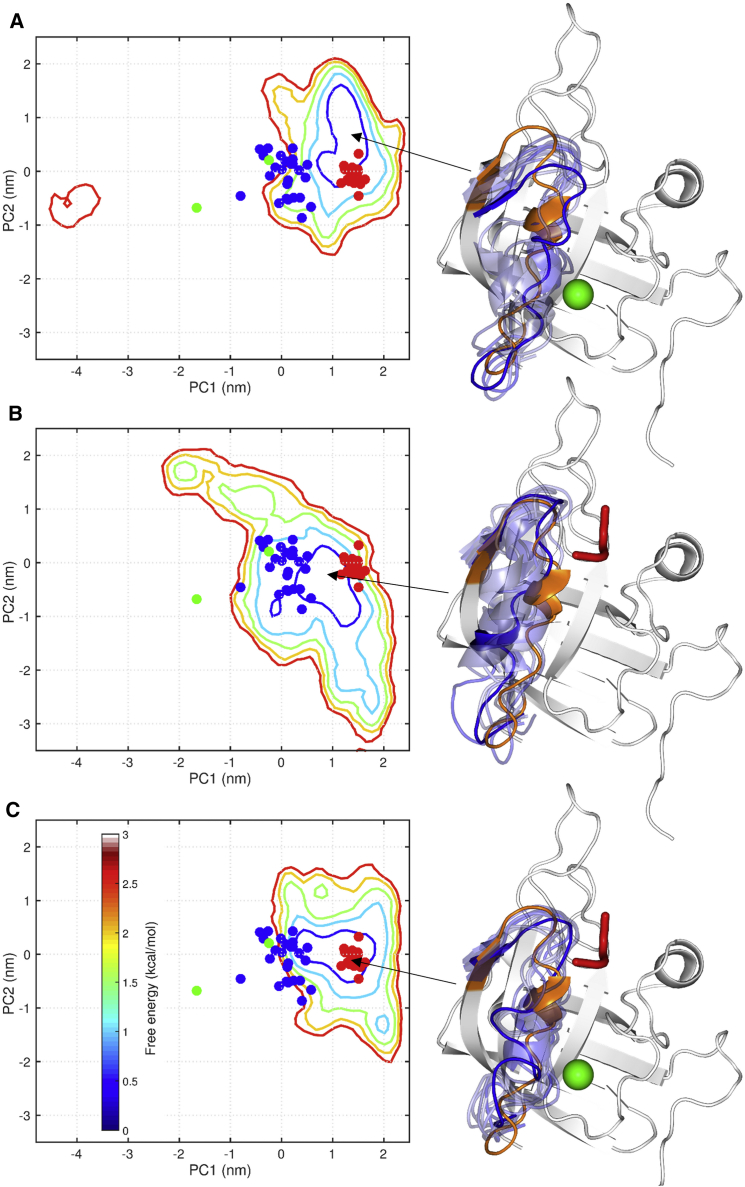
The preceding REST simulation shows that in the apo form the active-site loop can readily exchange between three conformational substates, which together cover a broad area over PC1 and PC2. Upon binding of either Ca2+ or the sorting signal, and especially upon binding both, the active-site loop significantly loses flexibility and becomes largely confined to a single conformational state (Fig. 3). Relative to substate 1 of apo SrtA (Fig. 2 D), the free-energy basins of the liganded forms all shift toward larger PC1, representing closure of the active-site loop. The merging and compaction of free-energy basins exposed by the REST simulations are the essence of the ligand-induced, disorder-to-order transition of the active-site loop.
Figure 3.
Conformational ensembles of SrtA in three liganded forms. (A) SrtACa. (B) SrtAPep. (C) SrtAPep/Ca. The information presented is the same as in Fig. 2, C and D, except that the free-energy surfaces are calculated from the REST simulations on the three liganded forms. Each free-energy basin is illustrated by 10 conformations within the first contour (at 0.5 kcal/mol) around the minimum, with only the active-site loop displayed (in blue) along with model 1 of 2KID (gray with active-site loop in orange). To see this figure in color, go online.
To test for sampling convergence, we divided the 100-ns REST simulations into two equal segments and compared the results separately calculated from the conformations in the two segments (Fig. S3). Overall, the results from the two segments are very similar to each other and to those in Figs. 2 D and 3 for the entire series of 100-ns simulations. The major free-energy basins are matched well, although the two smaller basins for apo SrtA show some differences between the two simulation segments, with basin 2 being more prominently sampled in the first segment, and basin 3 being more so in the second segment. The convergence is not surprising given that, during the 100-ns simulations, each replica readily traversed the entire temperature ladder (Fig. S2).
The holo form has the most compact free-energy basin; the 20 NMR models for SrtAPep/Ca are clustered around the free-energy minimum (Fig. 3 C). Relative to the apo substate 1, the active-site loop in the holo form is tightly closed, and maintains contact with both the sorting signal and Ca2+ (through the N-and C-terminal regions, respectively). In comparison, the free-energy basin of SrtAPep is less compact, and at the free-energy minimum the active-site loop is closed not as tightly as in the holo form, but does maintain the N-terminal interaction with the sorting signal (Fig. 3 B). The vast majority of the SrtACa conformations are found in a very compact free-energy basin where the active-site loop is closed nearly as tightly as in SrtAPep/Ca but does not reach far into the sorting signal binding site (Fig. 3 A). In a small minority of the SrtACa conformations, the active-site loop is wide open.
Calcium promotes sorting signal binding by preorganization of the binding pocket
The foregoing results provide direct support to the speculation of Naik et al. (11) that Ca2+ achieves allosteric activation by stabilizing closed conformations of the active-site loop poised for binding the sorting signal. These PCA results capture the global movements of the active-site loop. We now present detailed information on the formation of the binding pockets for Ca2+ and the sorting signal.
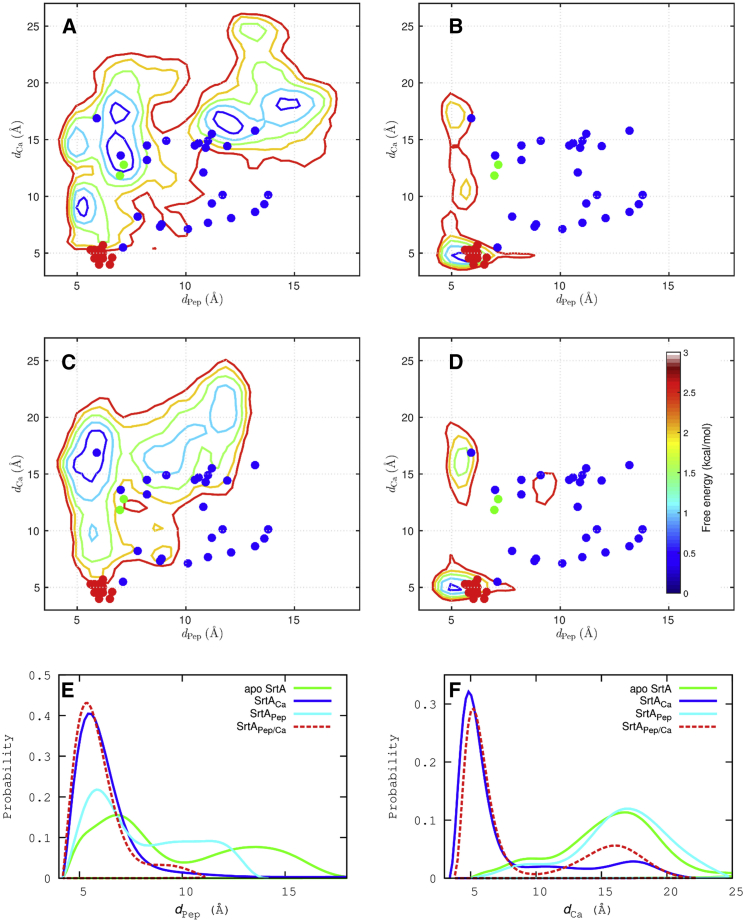
We used two distances, dCa and dPep, to indicate the formation of the two binding pockets. Their definitions are the same as those of Moritsugu et al. (17), to allow for easy comparison with the previous MSES simulations. Namely, dCa is the smaller of the Cδ-Cδ distances from Glu171 to Glu105 and Glu108 (Fig. 1 D), whereas dPep is the Cα-Cα distance from Pro163 to Arg197 (Fig. 1 C). The free-energy surfaces over dPep and dCa calculated from the REST simulations on apo SrtA and the three liganded forms are shown in Fig. 4. The free-energy surface for apo SrtA spreads over a broad area, with multiple local minima (Fig. 4 A). One cluster of minima corresponds to substate 1 defined above (featuring a semiopen active-site loop), with dPep averaging to ∼7 Å; the other cluster encompasses substates 2 and 3, with dPep ranging from 10 to 17 Å (Fig. 4, A and E). In both clusters, the Ca2+ binding pocket is not formed, as indicated by a broad distribution in dCa (Fig. 4 F). The free-energy surface for SrtAPep also covers a relatively broad area, with a major basin centered at dPep ∼ 6 Å and dCa ∼ 16 Å (Fig. 4 C). Again, the Ca2+ binding pocket is not formed, with a distribution in dCa similar to that in apo SrtA (Fig. 4 F). The absence of a well-formed Ca2+ binding pocket in both apo SrtA and SrtAPep is understandable, as its presence would mean direct contact between like-charged Glu171 and Glu105/Glu108.
Figure 4.
Status of sorting signal and Ca2+ binding pockets, monitored by dPep and dCa, respectively. (A–D) Free-energy surfaces over dPep and dCa for apo SrtA, SrtACa, SrtAPep, and SrtAPep/Ca, respectively, contoured at 0.5 kcal/mol internals. The dPep and dCa values are also displayed for conformations in 1T2P (green dots); 1IJA (blue dots); and 2KID (red dots). (E and F) Distributions of dPep and dCa, respectively. To see this figure in color, go online.
PC1 and dPep present two ways to describe the movement of the active-site loop relative to the sorting signal binding site. Not surprisingly, there is strong correlation between PC1 and dPep for apo SrtA, SrtACa, and SrtAPep, with R2 of linear regression at 0.78, 0.65, and 0.52, respectively. For SrtAPep/Ca, the correlation between PC1 and dPep becomes weak because of the restricted ranges of the sampled PC1 and dPep values in this case. Although both PC2 and dCa describe the movement of the active-site loop relative to the Ca2+ binding site, the former is defined with only Cα positions whereas the latter is also dictated by significant side-chain motions, and hence their correlation is weak for all four systems.
Moritsugu et al. (17) presented free-energy surfaces over dPep and dCa from their MSES simulations on apo SrtA and SrtAPep. In both cases, when compared to the counterparts in the REST simulations, significantly smaller areas over dPep and dCa were covered, suggesting less extensive conformational sampling. In particular, dPep extended to 12 and 8 Å for apo SrtA and SrtAPep, respectively, in the MSES simulations, but to 17 and 13 Å in the REST simulations. The conformational characterizations of the holo form are more similar in the MSES and REST simulations.
Notably, for SrtACa our free-energy surface spans very limited regions over dCa and dPep (Fig. 4 B). The compact basin over PC1 and PC2 shown in Fig. 3 A is now characterized here by both short dCa (appropriate for Ca2+ coordination; Fig. 4 E) and a distribution in dPep sharply peaked at a low value (∼5.5 Å), but with a right tail (Fig. 4 F). The short dPep in the vast majority of the conformations in this basin indicates that Ca2+ binding is accompanied by closure and ordering of the N-terminal region of the active-site loop, leading to preorganization of the binding pocket for the sorting signal. The right tail of the distribution in dPep shows that the N-terminal region (as represented by Pro163) of the active-site loop in SrtACa can transiently open but strongly prefers the closed state, through interactions with the N-terminus (as represented by Arg197) of β8. In a second, minor free-energy basin, the N-terminal region of the active-site loop interacts with the N-terminus of β8, but the C-terminal region of the active-site loop loses coordination with the bound Ca2+, with the side chain of Glu171 pointing outward and thus resulting in long dCa. Overall, the extent of closure and ordering of the active-site loop in SrtACa nearly approaches that in SrtAPep/Ca (Fig. 4, B and D–F). In contrast, the MSES simulation on SrtACa by Moritsugu et al. (17) found Ca2+-induced formation of the sorting signal binding pocket to be “limited to a small range”, as shown by a broad distribution in dPep that was only slightly left-shifted from the counterpart for apo SrtA.
The values of dPep and dCa map the two chains of the apo x-ray structure to the center of the major free-energy basin for apo SrtA (Fig. 4 A) and the 20 models of the holo NMR structure to the center of the major free-energy basin for SrtAPep/Ca (Fig. 4 D). However, only model 1 of the 25 models in the NMR structure for Ca2+-bound SrtA is located within the major free-energy basin for SrtACa (Fig. 4 B). That the other 24 models show significant scatter in both dPep and dCa is largely a reflection of the paucity of structural restraints on the active-site loop (especially side chains therein) due to resonance overlap and line-broadening (10).
The considerable increase in structure order shown in our simulation on SrtACa is consistent with the Ca2+-induced quenching of picosecond motions within the active-site loop indicated by NMR spectroscopy (10). Quenching of fast (picosecond-nanosecond) motions, along with initiation of slow (microsecond-millisecond) motions, appears to be a common phenomenon for allosteric activators (32). To obtain further validation of our simulations, we calculated Cα chemical shifts on the sampled conformations of SrtACa and apo SrtA and compared the resulting chemical shift perturbations to the data presented by Ilangovan et al. (10). As shown in Fig. S4, the calculated chemical shift perturbations are qualitatively similar to the experimental data, with large values for the β3/β4 loop and the active-site loop. Moreover, the occasional long dPep sampled by SrtACa may explain the significant line-broadening observed on some residues in the N-terminal region of the active-site loop, and the occasional long dCa may be consistent with residual picosecond motions in the C-terminal region in the presence of Ca2+ (10).
Discussion
We have adapted the REST method to carry out extensive simulations on SrtA in the apo form and in three liganded forms. Compared to conventional molecular dynamics simulation and even possibly to the MSES method, REST significantly enhances the sampling efficiency. The simulations show that the ligand-induced disorder-to-order transition of the active-site loop is characterized by merging and compaction of free-energy basins. Calcium binding leads to the preorganization of the binding pocket for sorting signals, but not vice versa.
Mechanism of allosteric activation
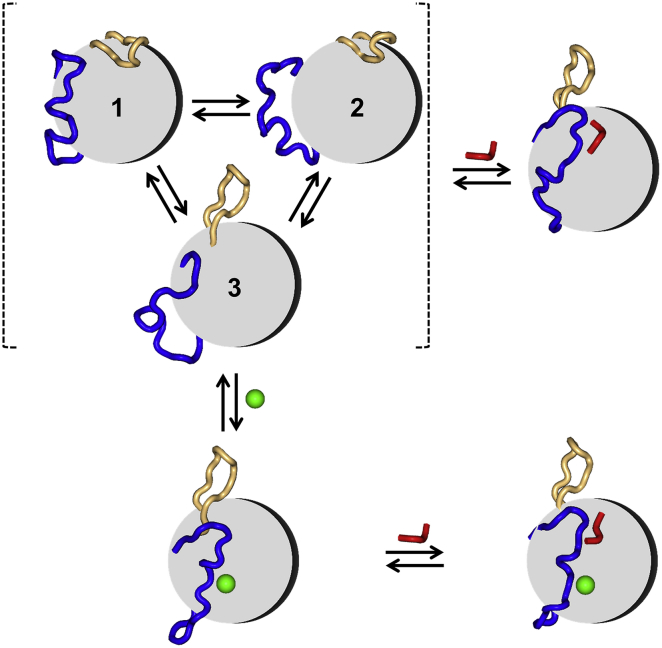
Our simulation results delineate a detailed mechanism for the allosteric activation of SrtA by Ca2+ (Fig. 5). In apo SrtA, the active-site loop undergoes conformational exchange among three substates, one that is semiopen and two that are wide open; the β7/β8 loop is closed most of the time, but tends to open when the N-terminal region of the active-site loop encroaches. When the sorting signal binds, the β7/β8 loop opens up to make room, and the active-site loop has the N-terminal region enclosing the peptide but the C-terminal region remaining open to avoid charge-charge repulsion by anionic residues including Glu105 and Glu108. When Ca2+ binds, the entire active-site loop closes up, stabilized by interactions between Glu171 and the prebound Ca2+ and between the N-terminal region and the β8 N-terminus. The latter interaction increases the tendency of the β7/β8 loop to open, preorganizing the binding pocket for the sorting signal. When the sorting signal then comes in for binding, the binding pocket just needs to make fine adjustments, including the upshift of the N-terminal region of the active-site loop.
Figure 5.
Mechanistic model for allosteric activation of SrtA by Ca2+. β-barrel of SrtA (gray), active-site loop (blue), β7/β8 loop (orange), sorting signal (red), and Ca2+(green). In the apo form, the active-site loop undergoes conformational exchange among three substates, and the β7/β8 loop is mostly closed but can transiently open. Upon binding of the sorting signal, the active-site loop closes up but retains significant mobility, partly due to repulsion of Glu171 by anionic residues such as Glu105 and Glu108. The Ca2+ binding leads to closure of the entire active-site loop and increased open probability for the β7/β8 loop. The resulting preorganization of the binding pocket for the sorting signal facilitates the latter’s tight binding. To see this figure in color, go online.
In this allosteric mechanism, the active-site loop plays a critical role. The conformational heterogeneity exhibited by the active-site loop in the apo form persists to a large degree when the sorting signal is bound without Ca2+, especially in the C-terminal region but also in the N-terminal region. Here, the N-terminal region of the active-site loop therefore does not strongly contribute to the proper positioning and/or binding stability of the sorting signal, leading to the relatively low enzymatic activity of the Ca2+-free enzyme. When Ca2+ is bound, the active-site loop, through partial closure and ordering, appears to be the main conduit for the allosteric communication from the Ca2+ binding site to the active site.
Based on our other studies on the binding of IDPs to structured targets (6), we can further speculate a possible pathway for the allosteric communication. Initially the prebound Ca2+ might, through long-range electrostatic attraction, recruit Glu171 for coordination, leading to the docking of the C-terminal region of the active-site loop. Thereafter the N-terminal region would look for interaction partners and eventually coalesce around the β8 N-terminus (and the sorting signal when present). The S. pyogenes SrtA is Ca2+-insensitive and there a cationic residue, Lys126, substitutes for the S. aureus Glu105 (31). Interestingly, Lys126 may serve a role similar to that of Ca2+. It forms a salt bridge with Asp196 (corresponding to S. aureus Glu171) on the active-site loop, and this interaction may initiate the dock-and-coalesce pathway just outlined for the Ca2+-loaded S. aureus SrtA.
If the allosteric mechanism outlined here is correct, then an E105K mutation of S. aureus SrtA may allow the protein in apo form to reach the catalytic activity of the Ca2+-loaded form of the wild-type protein, because the anticipated salt bridge with Glu171 may elicit a similar allosteric effect to Ca2+ binding. It will be interesting to test the catalytic activities of this type of mutant.
General use of REST for simulations on IDRs
Our study demonstrates that the REST method is ideally suited for efficient conformational sampling of IDRs. With SrtA solvated in explicit solvent, only 16 replicas were needed to cover an effective temperature range from 300 to nearly 600 K, at an acceptance rate of ∼0.47 for replica exchange. Using the traditional temperature replica exchange, to achieve the coverage of the same temperature at the same acceptance rate would require as many as 200 replicas (33). Our simulations also appear to be more efficient than simulations for the same SrtA systems using MSES, which like REST is also a form of Hamiltonian replica exchange. Specifically, our simulations yielded broader coverage of the conformational space for the active-site loop in apo SrtA and in SrtAPep. More importantly, our simulations presented direct evidence for the allosteric activation of SrtA by Ca2+, in the form of closure and ordering of the entire active-site loop and the consequent preorganization of the binding site for the sorting signal. In contrast, Moritsugu et al. (17) concluded that Ca2+ binding had little effect on the N-terminal region of the active-site loop. The apparently more limited sampling by MSES may be a reflection of the intrinsic ability of this method, or may be due to artificial bias in the choice of the coarse-grained model. The use of two versions of the Amber force field (Amber99sb here and Amber03 in the MSES study, differing in backbone torsion parameters and partial charges) could conceivably result in a difference in sampling efficiency. A rigorous comparison of the sampling efficiencies of REST and MSES will require extensive simulations using the same force field, best done on test systems much smaller than SrtA.
IDRs/IDPs are implicated in many biological functions and often associated with diseases. Designing drugs for disordered proteins is a challenge due to the conformational heterogeneity. A critical step is to better understand the binding mechanisms and disorder-to-order transitions of disordered proteins, particularly the motifs involved in ligand recognition. SrtA itself is an ideal drug target (34, 35). The types of conformational substates and binding and allosteric mechanisms revealed by REST simulations may guide drug discovery in the future. While for SrtA the IDR selected for enhanced sampling appears to contain most of the allosteric pathway, in other cases allosteric pathways may not be largely confined to isolated, small IDRs. Finding the balance between optimizing sampling efficiency and maximizing the coverage of the allosteric pathways will be a challenge for future studies.
Author Contributions
X.P. and H.-X.Z. designed the research; X.P. performed the research and analyzed the data; and X.P. and H.-X.Z. wrote the article.
Acknowledgments
We thank Dr. Robert Clubb for discussion.
This work was supported by National Institutes of Health grant No. GM058187.
Editor: Michael Feig.
Footnotes
Four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00921-2.
Supporting Material
References
- 1.Dunker A.K., Lawson J.D., Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 2.Babu M.M., van der Lee R., Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 5.Uversky V.N. Introduction to intrinsically disordered proteins (IDPs) Chem. Rev. 2014;114:6557–6560. doi: 10.1021/cr500288y. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H.X., Pang X., Lu C. Rate constants and mechanisms of intrinsically disordered proteins binding to structured targets. Phys. Chem. Chem. Phys. 2012;14:10466–10476. doi: 10.1039/c2cp41196b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ton-That H., Liu G., Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suree N., Liew C.K., Clubb R.T. The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J. Biol. Chem. 2009;284:24465–24477. doi: 10.1074/jbc.M109.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zong Y., Bice T.W., Narayana S.V.L. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J. Biol. Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 10.Ilangovan U., Ton-That H., Clubb R.T. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naik M.T., Suree N., Clubb R.T. Staphylococcus aureus Sortase A transpeptidase. Calcium promotes sorting signal binding by altering the mobility and structure of an active site loop. J. Biol. Chem. 2006;281:1817–1826. doi: 10.1074/jbc.M506123200. [DOI] [PubMed] [Google Scholar]
- 12.Jensen M.R., Ruigrok R.W.H., Blackledge M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr. Opin. Struct. Biol. 2013;23:426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Kappel K., Wereszczynski J., McCammon J.A. The binding mechanism, multiple binding modes, and allosteric regulation of Staphylococcus aureus Sortase A probed by molecular dynamics simulations. Protein Sci. 2012;21:1858–1871. doi: 10.1002/pro.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugita Y., Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 15.Zhang W., Ganguly D., Chen J. Residual structures, conformational fluctuations, and electrostatic interactions in the synergistic folding of two intrinsically disordered proteins. PLOS Comput. Biol. 2012;8:e1002353. doi: 10.1371/journal.pcbi.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knott M., Best R.B. A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: evidence from molecular simulations. PLOS Comput. Biol. 2012;8:e1002605. doi: 10.1371/journal.pcbi.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moritsugu K., Terada T., Kidera A. Disorder-to-order transition of an intrinsically disordered region of sortase revealed by multiscale enhanced sampling. J. Am. Chem. Soc. 2012;134:7094–7101. doi: 10.1021/ja3008402. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W.H., Chen J.H. Accelerate sampling in atomistic energy landscapes using topology-based coarse-grained models. J. Chem. Theory Comput. 2014;10:918–923. doi: 10.1021/ct500031v. [DOI] [PubMed] [Google Scholar]
- 19.Liu P., Kim B., Berne B.J. Replica exchange with solute tempering: a method for sampling biological systems in explicit water. Proc. Natl. Acad. Sci. USA. 2005;102:13749–13754. doi: 10.1073/pnas.0506346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., Friesner R.A., Berne B.J. Replica exchange with solute scaling: a more efficient version of replica exchange with solute tempering (REST2) J. Phys. Chem. B. 2011;115:9431–9438. doi: 10.1021/jp204407d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terakawa T., Kameda T., Takada S. On easy implementation of a variant of the replica exchange with solute tempering in GROMACS. J. Comput. Chem. 2011;32:1228–1234. doi: 10.1002/jcc.21703. [DOI] [PubMed] [Google Scholar]
- 22.Bussi G. Hamiltonian replica exchange in GROMACS: a flexible implementation. Mol. Phys. 2014;112:379–384. [Google Scholar]
- 23.Miller C.M., Brown A.C., Mittal J. Disorder in cholesterol-binding functionality of CRAC peptides: a molecular dynamics study. J. Phys. Chem. B. 2014;118:13169–13174. doi: 10.1021/jp5106423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang K., García A.E. Acceleration of lateral equilibration in mixed lipid bilayers using replica exchange with solute tempering. J. Chem. Theory Comput. 2014;10:4264–4272. doi: 10.1021/ct500305u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole D.J., Tirado-Rives J., Jorgensen W.L. Molecular dynamics and Monte Carlo simulations for protein-ligand binding and inhibitor design. Biochim. Biophys. Acta. 2015;1850:966–971. doi: 10.1016/j.bbagen.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornak V., Abel R., Simmerling C. Comparison of multiple AMBER force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darden T., York D., Pedersen L. Particle mesh Ewald—an NLog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 29.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 30.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Race P.R., Bentley M.L., Banfield M.J. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J. Biol. Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J., Zhou H.X. Dynamically driven protein allostery exhibits disparate responses for fast and slow motions. Biophys. J. 2015;108:2771–2774. doi: 10.1016/j.bpj.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patriksson A., van der Spoel D. A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 34.Cascioferro S., Totsika M., Schillaci D. Sortase A: an ideal target for anti-virulence drug development. Microb. Pathog. 2014;77:105–112. doi: 10.1016/j.micpath.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Chan A.H., Wereszczynski J., Clubb R.T. Discovery of Staphylococcus aureus sortase A inhibitors using virtual screening and the relaxed complex scheme. Chem. Biol. Drug Des. 2013;82:418–428. doi: 10.1111/cbdd.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krebs W.G., Gerstein M. The Morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.