SUMMARY
In metazoans, microRNAs play a critical role in the post-transcriptional regulation of genes required for cell proliferation and differentiation. microRNAs themselves are regulated by a multitude of mechanisms influencing their transcription and post-transcriptional maturation. However, there is only sparse knowledge on pathways regulating the mature, functional form of a microRNA. Here, we identify a new player in the control of microRNA turnover, the decapping scavenger protein DCS-1. In Caenorhabditis elegans, mutations in dcs-1 increase the levels of functional microRNAs. We demonstrate that DCS-1 interacts with the exonuclease XRN-1 in vivo and in vitro, that this interaction promotes microRNA degradation and that this new function does not require its decapping scavenger activity, establishing independence of these two processes. Our findings thus indicate that DCS-1 is part of a degradation complex that performs microRNA turnover in animals.
INTRODUCTION
MicroRNAs (miRNAs) are ~22 nucleotide long non-coding RNAs that regulate gene expression at the post-transcriptional level by binding to partially complementary sequences of target mRNAs (Ebert and Sharp, 2012). miRNA genes are mostly transcribed by RNA polymerase II to yield a primary miRNA transcript (pri-miRNA). The pri-miRNA undergoes processing by a multiprotein complex known as the microprocessor, to produce an intermediate called the precursor miRNA (pre-miRNA). After nuclear export, the pre-miRNA is cleaved by Dicer resulting in the mature miRNA. Subsequently, mature miRNAs are bound by an Argonaute protein to form the core of a multisubunit effector complex termed miRISC (miRNA-induced silencing complex) (reviewed in Krol et al., 2010; Kim et al., 2009). miRISC binds to partially complementary sequences found typically in the 3′ untranslated region (3′UTR) of mRNAs, leading to their translational repression and/or degradation (reviewed in Huntzinger and Izaurralde, 2011; Pasquinelli, 2012).
miRNA-mediated gene regulation is involved in diverse biological functions, including the control of development in metazoans (reviewed in Ebert and Sharp, 2012) as well as cellular pathways such as DNA damage and stress responses (reviewed in Hu and Gatti, 2011; Leung and Sharp, 2010). Notably, miRNAs are dysregulated in many diseases, such as cancer (reviewed in Esteller, 2011; Mendell and Olson, 2012). Thus, it is essential for a cell to tightly control miRNA biogenesis and turnover.
Like other RNA Polymerase II-transcribed genes, miRNA loci are subject to widespread transcriptional regulation. Additionally, the different biogenesis steps are controlled by cellular factors that either modulate the activity of processing factors or bind miRNA precursor molecules to interfere with the processing of a subset of miRNAs (reviewed in Bajan and Hutvagner, 2011; Krol et al., 2010; Newman et al., 2011). More recently, evidence has emerged that miRNA decay pathways contribute to the control of miRNA levels (Kai and Pasquinelli, 2010; Ruegger and Grosshans, 2012). In particular, the small RNA degrading nucleases (SDNs) mediate 3′-to-5′ turnover of miRNAs in plants (Ramachandran and Chen, 2008) and the exoribonucleases XRN-1 and XRN-2 function in 5′-to-3′ miRNA degradation in C. elegans (Chatterjee et al., 2011; Chatterjee and Grosshans, 2009).
In this study, we identify the DeCapping Scavenger enzyme 1 (DCS-1, also known as DcpS) as a new player in the C. elegans miRNA turnover pathway. Our data reveal that DCS-1 stimulates XRN-1-mediated miRNA degradation through a physical interaction and that this process is independent of the decapping scavenger activity previously assigned to DCS-1.
RESULTS
Mutations in dcs-1 lead to precocious adult fate and enhanced repression of miRNA targets
In order to discover new factors implicated in the miRNA pathway, we performed a genetic screen for genes that interact synthetically with alg-2, one of the two C. elegans Argonautes that functions in the miRNA pathway (Grishok et al., 2001). The rationale of the screen was based on the observation that an alg-2 mutant (alg-2(ok304)) is viable whereas the combined loss of alg-2 and its paralog alg-1 results in lethality (Grishok et al., 2001; Vasquez-Rifo et al., 2012). We generated an alg-2(ok304) strain that carries an extrachromosomal array expressing wild-type alg-2 and a GFP marker. Since alg-2 is not required for viability, the array is lost stochastically from this strain. Following chemical mutagenesis, we screened for animals that remained GFP positive, indicating a potential requirement for the array, and thus alg-2, for survival. Among the six complementation groups isolated from this screen, we characterize here one group of two mutant alleles, qbc2 and qbc3, that were lesions in the dcs-1 gene (Figure S1).
dcs-1 encodes the decapping scavenger enzyme, which has been shown to hydrolyze the residual cap structure that results from 3′-to-5′ decay of mRNAs by the exosome (Cohen et al., 2004; Liu et al., 2002). Whereas alg-2(ok304); dcs-1(qbc3) double mutant animals arrested in development as early as embryos post-fertilization (data not shown), dcs-1(qbc3) single mutant embryos developed normally. However, in contrast to wild-type animals, animals carrying lesions in the dcs-1 gene displayed alae structures at the larval L4 stage, one developmental stage earlier than wild-type (Figure 1A; the developmental stage was confirmed by examining the vulval and gonad morphology (Figure S2A)). Re-establishing the expression of wild-type dcs-1 using a transgenic array rescued the precocious formation of the alae (Figure 1A), confirming that this developmental phenotype is caused by the mutation in the dcs-1 gene.
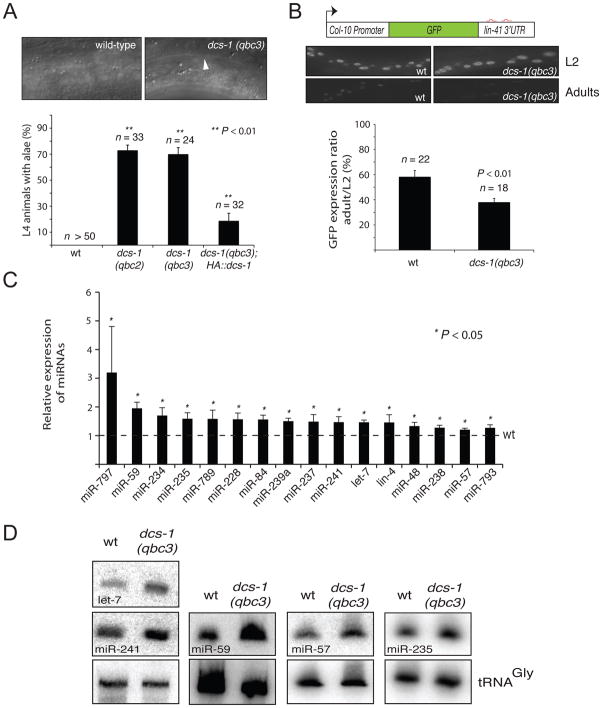
Figure 1. The loss of dcs-1 affects miRNA-mediated gene regulation and miRNA levels.
(A) Alae are produced precociously in dcs-1 animals. L4 animals were precisely staged by observing vulval and gonad formation by Nomarski optics (Figure S2A) and the percentage (%) of animals with precocious alae (white arrow; bottom picture) was scored. Magnification is x 1,000. (B) To study lin-41 regulation in vivo, we monitored the expression of Green Fluorescent Protein (GFP) under the control of a hypodermis specific col-10 promoter and the lin-41 3′ UTR containing the let-7 miRNA binding sites (red; diagram). GFP is expressed during early larvae stages (L2) and downregulated when animals reach young adulthood (Adults). Young adult dcs-1(qbc3) animals repressed the miRNA-sensitive reporter more strongly than wild-type animals (Middle). Quantification of GFP in adults relative to L2 stage animals. The quantification of the GFP signal was performed by measuring the mean of the GFP detected in five different cells for each animal (Below). The magnification of all pictures is x 1,000. The number of animals (n) scored are indicated in parenthesis (A–B). (C–D) miRNAs levels of are increased in the dcs-1 mutant. (C) The level of miRNAs found in dcs-1(qbc3) young adult animals were measured by quantitative reverse-transcription-PCR (TaqMan Assay) and compared with the levels found in wild-type animals (wt; 1). TaqMan assays for the small nucleolar RNAs sn2841 was used as the normalization control. The error bars represent the 95% confidence interval of three independent experiments. P values were obtained using normalized delta delta Ct values. Normalized delta delta Ct values were obtained by subtracting the mean of delta delta Ct from all experiments. (D) The levels of significantly increased miRNAs (let-7, miR-57, miR-59, miR-235 and miR-241) in dcs-1 mutant were detected by Northern blot hybridization of RNA samples purified from young adult animals. tRNAGly was used as loading control.
LIN-41 is an important regulator of the L4-to-adult developmental transition in C. elegans. During the L4-stage, lin-41 mRNA levels are repressed by the let-7 miRNA allowing developmental progression (Reinhart et al., 2000; Slack et al., 2000). Similar to what we observe for dcs-1, lin-41 loss-of-function alleles cause precocious formation of alae (Slack et al., 2000). To test whether dcs-1(qbc3) leads to misregulation of lin-41, a GFP reporter under the control of the lin-41 3′UTR was used. Consistent with the fact that the let-7 miRNA is only expressed during late larval stages (Reinhart et al., 2000; Slack et al., 2000), we found, that in wild-type worms the GFP signal was present in L2-stage but decreased in adult animals. Although the signal was comparable for wild-type and dcs-1(qbc3) animals at the L2 stage, repression appeared enhanced in adults dcs-1 mutant animals relative to wild-type (Figure 1B). This effect was accompanied by significantly decreased endogenous lin-41 mRNA levels in dcs-1(qbc3) relative to wild-type adult animals (Figure S2B). Notably, the effect on the lin-41 mRNA level also suggest that the role dcs-1 in this process does not involve the decapping scavenger activity, since its loss lead to an increase of overall mRNA levels (Liu and Kiledjian, 2005). Thus, our data indicate that the precocious alae observed in dcs-1 mutant animals are a consequence of precocious down regulation of lin-41.
Mutations in dcs-1 lead to an increase of functional miRNA levels
A possible explanation for the robust repression of a miRNA-targeted gene such as lin-41 in dcs-1 mutants is that the loss of dcs-1 increases the levels of functional miRISC. To test this hypothesis, we monitored the level of two core constituents of miRISC, namely miRNAs and the Argonaute protein ALG-1 (Hutvágner et al., 2004). Whereas alg-1 mRNA and protein levels were unaltered in dcs-1(qbc3) (Figure S2C–D), the levels of 16 out of 97 tested miRNAs, including let-7, were significantly increased relative to wild-type (Figure 1C–D). It remains to be shown what the common feature is that qualifies the miRNAs that change as DCS-1 targets since many miRNAs are not affected by the loss of dcs-1 (Table S1).
To determine whether the elevated miRNA levels are due to enhanced miRNA transcription or processing, we examined the levels of let-7 miRNA precursors and did not detect any significant change in the level of pre-let-7 and pri-let-7 molecules between wild-type and dcs-1(qbc3) animals (Figure S3A–B). We conclude that the increase in mature let-7 is not a consequence of increased transcription, enhanced stability of the pri- or pre-miRNA molecules or enhanced processing which is consistent with a function for dcs-1 that is not related to the decapping scavenger activity, but instead reflects an effect on the stability of mature miRNA molecules.
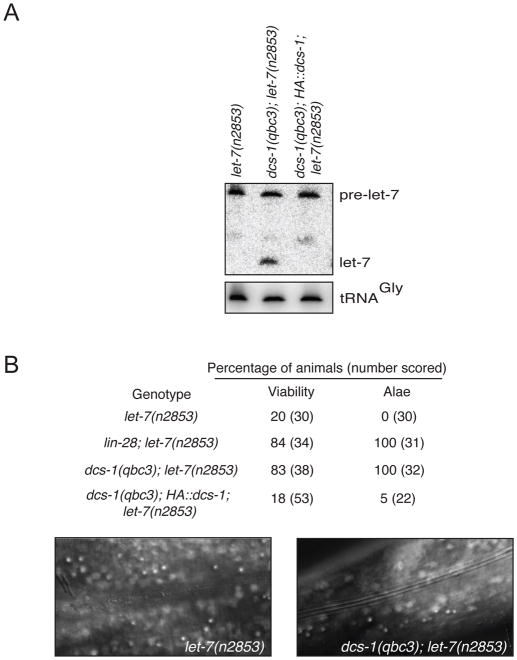
To obtain further evidence that the miRNAs that accumulate in dcs-1 animals are functional, we investigated let-7(n2853) mutant animals. This point mutation in the mature let-7 miRNA causes reduced let-7 levels, as well as temperature-sensitive alae defects and lethality due to vulval bursting (Reinhart et al., 2000). Similar to wild-type, the let-7 RNA was significantly increased during the development of dcs-1(qbc3); let-7(n2853) animals (Figure S3C), with no significant changes in the pre-let-7 RNA levels (Figure 2A). Moreover, this increase in let-7 levels coincided with suppression of both the lethality and alae defects of let-7(n2853) animals to an extend comparable to that seen with lin-28 knock-out, a conserved negative regulator of let-7 production (Figure 2B; Lehrbach et al., 2009). We conclude that the loss of dcs-1 leads to accumulation of functional miRNAs in animals.
Figure 2. The loss of dcs-1 function rescues let-7 mutant developmental defects by increasing mature let-7 miRNA levels.
(A) Detection by Northern blotting of let-7 molecules in various genetic backgrounds of young adult animals. The detection of tRNAGly by Northern hybridization acted as loading control. (B) Viability and complete adult alae were scored at 20°C on young adult animals in the genetic background listed. The number of animals scored are indicated in parenthesis.
DCS-1 is required for the degradation of miRNAs that are released from miRISC
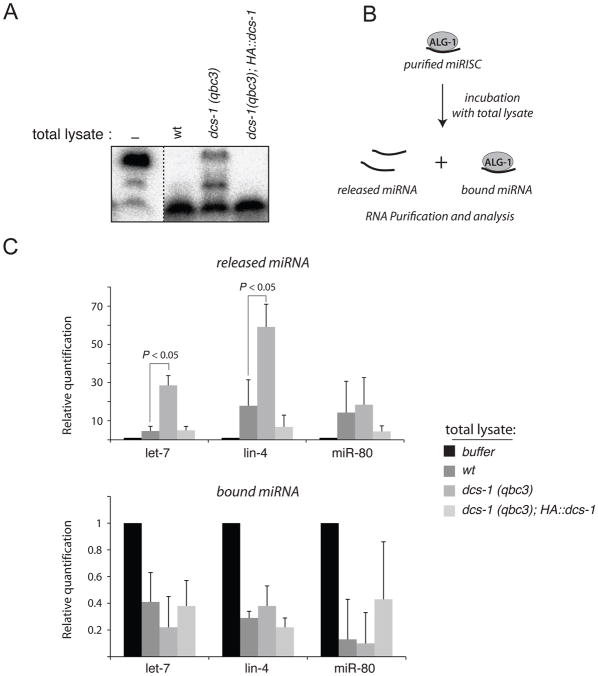
Because our observations argue against a role of DCS-1 in miRNA transcription or biogenesis, we next sought to evaluate a potential role of DCS-1 in miRNA degradation. To this end, we performed miRNA degradation assays by incubating radiolabeled synthetic miRNA molecules with total worm lysates, as described previously (Chatterjee and Grosshans, 2009). While we observed complete degradation of synthetic let-7 after incubation with lysates prepared from wild-type animals, degradation was severely impaired in lysates prepared from dcs-1(qbc3) mutant animals (Figures 3A and S3D). This effect was specific because lysate from dcs-1(qbc3) animals that expressed a dcs-1 transgene restored decay (Figure 3A). We conclude that DCS-1 is required for efficient miRNA degradation in vitro.
Figure 3. DCS-1 is required for miRNA degradation.
(A) Degradation assays of a 5′-P32 21nt long RNA incubated for 15 minutes with total worm lysates produced from wild-type (wt), dcs-1(qbc3) mutant and dcs-1(qbc3) rescued with a HA::dcs-1 transgene (HA::dcs-1) animals. Dashed lines indicate that unrelated lanes have been removed between samples. (B) Schematic of the miRNA release assay. (C) The miRISC purified by immunoprecipitation of endogenous ALG-1 was incubated with micrococcal nuclease-treated worm lysate and the let-7, lin-4 and miR-80 miRNAs that are released into the supernatant (top panel) or remains associated with ALG-1 complexes (bottom panel) were quantified by quantitative reverse-transcription-PCR (TaqMan Assay). The error bars represent the 95% confidence interval of three independent experiments. P values were obtained using a two-sided Student’s test with the normalized Ct values.
To further characterize the function of DCS-1 in miRNA degradation, we performed a miRNA release assay. In this assay, miRISC is first purified by immunoprecipitation of endogenous ALG-1, and then incubated with a miRNA-depleted worm lysate (by treating the extract with micrococcal nuclease). Subsequently, the miRNA molecules that remain associated with, or are released from, miRISC are quantified (Figure 3B; Chatterjee and Grosshans, 2009). Incubation of miRISC with wild-type lysate led to efficient release of miRNAs, and this was unchanged when dcs-1 mutant lysate was used (Figure 3C). However whereas wild-type lysate caused substantial degradation of the released miRNAs, which thus was detectable in only small amounts in the supernatant, dcs-1(qbc3) lysate caused a significant accumulation in the supernatant of let-7 and lin-4 (Figure 3C). By contrast, miR-80, a miRNA that is not affected by the loss of dcs-1 did not accumulate in the supernatant either (Figure 3C; Table S1). These data suggest that DCS-1 is important for the degradation of some miRNAs released from the miRISC.
DCS-1 interacts with XRN-1 to form a miRNA degradation complex
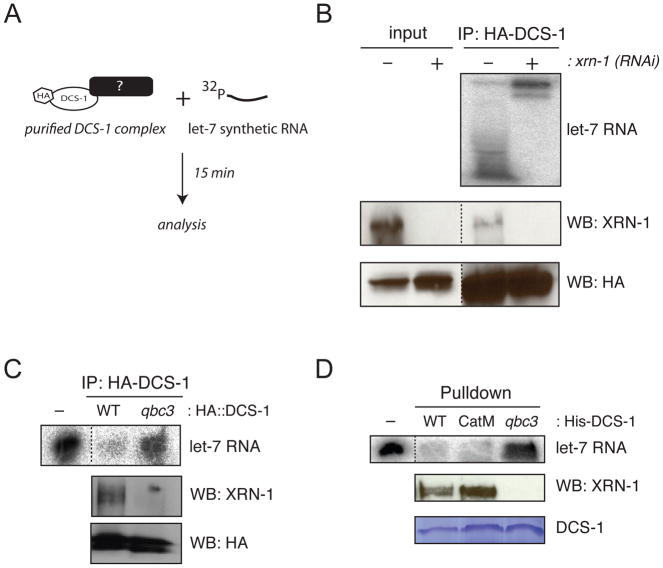
Since our previous results did not rule out the possibility that DCS-1 affected miRNA turnover indirectly, we immunoprecipitated HA-tagged DCS-1 complexes from rescued dcs-1(qbc3) animals and performed a miRNA degradation assay (Figure 4A). As previously observed with total lysates, the purified DCS-1 complex efficiently degraded exogenously supplied radiolabeled miRNA (Figure 4B). This confirms that miRNA degrading activity is associated with DCS-1.
Figure 4. DCS-1 and XRN-1 form a miRNA degradation complex.
(A) Schematic of the immunoprecipitation assay. (B) Transgenic animals are first exposed to either control (−) or xrn-1 (+) dsRNA-expressing bacteria for 38 hrs followed by total protein extraction and HA::DCS-1 complex purification with beads coupled with anti-HA monoclonal antibody. Inputs represent 10% of the total protein lysate used for the immunoprecipitation (bottom panels). Dashed lines indicate that unrelated lanes have been removed between samples. (C) Same as (B) with transgenic lines expressing either wild-type (WT) HA-tagged DCS-1 or HA-DCS-1 with the point mutation found in dcs-1(qbc3) allele (qbc3). In both cases, the detection of HA::DCS-1 and XRN-1 was achieved by Western blotting. The minus (−) lane represents the amount of 5′-P32 let-7 RNA used for the assay. (D) The point mutation in DCS-1 abrogates the XRN-1 interaction and the exonuclease activity. Beads coupled with His tagged recombinant wild-type (WT), mutated in the catalytic Histidine triad (CatM) or mutated at the leucine residue 32 to proline (qbc3) DCS-1 protein were incubated with worm total protein extracts. Each DCS-1 purified complex were then incubated with the 5′-P32 21nt long let-7 RNA incubated for 15 minutes. The minus (−) lane represents the amount of radiolabeled let-7 RNA used for the assay. Coomassie staining monitored His-DCS-1 proteins and Western blotting was used to detect XRN-1 association.
DCS-1 has dinucleoside triphosphate hydrolase activity, which allows it to remove the m7G cap from capped oligonucleotides, but is not known to degrade oligoribonucleotides themselves (Cohen et al., 2004). Hence, we hypothesized that a co-purifying RNase could endow DCS-1 immunoprecipitates with miRNA degrading activity. XRN-1 and XRN-2 appeared suitable candidates as these two 5′-to-3′ exonucleases are associated with miRNA degradation in C. elegans (Chatterjee et al., 2011; Chatterjee and Grosshans, 2009). Because DCS-1 (Lall et al., 2005), and GFP-tagged XRN-1 but not XRN-2 are localized in the cytoplasm (Figure S4A–B), we investigated whether the DCS-1 purified complex contained the XRN-1 protein. Using an antibody specific to endogenous XRN-1 (SR & HG, unpublished data), we found that the functional HA::DCS-1 fusion protein did contain endogenous XRN-1 (Figure 4B).
To confirm that this interaction was functionally relevant for decay, we purified DCS-1 complexes from animals depleted for XRN-1 by RNAi and observed that miRNA degradation was lost (Figure 4B). Moreover, when we introduced a HA::dcs-1 transgene carrying the L32P missense mutation found in dcs-1(qbc3) animals (Figure S1), both, the interaction with XRN-1 as well as the miRNA degrading activity of the immunopurified DCS-1 was lost (Figure 4C). We conclude that XRN-1 is the catalytic engine driving miRNA turnover in the DCS-1 complex.
Finally, to determine whether the decapping scavenger activity of DCS-1 participates in miRNA degradation and to verify that the qbc3 mutation affects binding to XRN-1, we performed pulldown experiments with recombinant wild-type, catalytically inactive (CatM) and qbc3 mutant DCS-1 proteins (Figure S1). Whereas the wild-type and catalytically inactive DCS-1 interacted with XRN-1 and retain the degradation activity in the complex, the interaction as well as the degradation activity was lost with DCS-1 qbc3 protein (Figure 4D). We therefore conclude that the implication of DCS-1 in miRNA turnover is uncoupled from its decapping scavenger activity. Because we did not observe any changes in xrn-1 mRNA and protein levels in dcs-1(qbc3) animals (Figure S4C–D), our data support the notion that an interaction with DCS-1 promotes the enzymatic activity of XRN-1, rather than DCS-1 stabilizing XRN-1. Altogether, we conclude that DCS-1 and XRN-1 form an enzymatic complex that performs miRNA degradation in C. elegans.
DISCUSSION
Overall, our observations demonstrate that the interaction between DCS-1 and XRN-1 promotes the degradation of miRNAs. Consistent with this notion, the orthologous S. cerevisiae proteins Dcs1p and Xrn1p have recently been shown to interact, and Dcs1p is essential for Xrn1p enzymatic activity in vitro and, at least in the presence of a nonfermentable carbon source, in vivo (Sinturel et al., 2012). Strikingly, Sinturel and colleagues further observed that the stimulation of Xrn1p by Dcs1p did not require Dcs1p catalytic activity. Similarly, our in vitro results indicate that the catalytic activity of C. elegans DCS-1 is dispensable for XRN-1 interaction and stimulation. Therefore our data demonstrate that in addition to its previously characterized role in the degradation of the cap structure of mRNAs (Cohen et al., 2004; Liu et al., 2002; Wang and Kiledjian, 2001), DCS-1 contributes to miRNA turnover in animals by promoting the exonuclease activity of XRN-1 on miRNAs.
Currently, it is unknown how miRNAs are released from the miRISC for degradation by the DCS-1/XRN-1 complex. Recent crystal structures of yeast and human Argonautes showed that the 5′ end of the small RNA is embedded within the Mid domain of the protein (Elkayam et al., 2012; Nakanishi et al., 2012; Schirle and MacRae, 2012) and thus most likely not accessible to the 5′-to-3′ exonuclease complex. Our observation that DCS-1 does not promote miRNA release in vitro, but facilitates degradation of the released miRNA, is indeed consistent with the notion that DCS-1 and XRN-1 act after the release step, once the miRNA 5′ end has become available. In vivo, our analysis clearly indicates that not only miRNA levels, but also miRNA activity are increased in dcs-1 mutant animals, which suggests that miRNA will, at least in part, be retained on Argonaute. It therefore seems possible that the release of the miRNA from the Argonaute protein is driven by a dedicated factor that itself needs to unload the miRNA onto the DCS-1/XRN-1 complex to promote further rounds of release.
Our finding that dcs-1 loss-of-function mutations are embryonic lethal when combined with loss of the Argonaute alg-2 in embryos, but promote miRNA accumulation in the presence of alg-2 is puzzling, since this finding suggests that DCS-1 can have both positive and negative effects on miRNA activity. However, it seems possible that impaired miRNA decay alone might account for both phenotypes. In our recent effort to characterize alg-1 and alg-2, we observed that while nearly all C. elegans miRNAs are associated with both Argonaute proteins, a small subset of miRNAs remains specifically interacting with ALG-1 or ALG-2 (Vasquez-Rifo et al., 2012). Therefore, if loss of ALG-2 leads to increased competition among miRNAs for access to ALG-1, overaccumulation of selected miRNAs by loss of DCS-1 could further compromise Argonaute loading of a subset of miRNAs. If these include miRNAs essential for embryonic viability, synthetic lethality might result. Future work on the function of DCS-1 during embryogenesis may help us to confirm or refute this idea.
Our data, along with previous observations, support that the level of miRNAs in C. elegans must be carefully controlled to enable precisely timed developmental transitions in animal development. Loss or excess of miRNAs can severely impair developmental timing, leading to heterochronic phenotypes, e.g., animals that adopt adult cell fates prematurely or not at all (Resnick et al., 2010). The fact that DCS-1 expression is developmentally regulated in C. elegans (Kwasnicka et al., 2003; Figure S4E), suggested that modulating DCS-1 levels may represent an efficient way to regulate XRN-1 activity and thus, rapidly turning over miRNAs at a specific point during development. This timely controlled expression of DCS-1 may also explain why some miRNAs are less sensitive to the loss of dcs-1 function in animals. We speculate that the role of DCS-1 in miRNAs turnover would be more prominent during important developmental switches where animals undergo significant changes in genes expression, such as the initial steps of embryogenesis and at the larvae-adult transition (Kaufman and Miska, 2010).
EXPERIMENTAL PROCEDURES
Nematode methods
C. elegans strains were grown under standard conditions (Brenner, 1974). Animal transgenic lines were produced by microinjections, as described in (Mello and Fire, 1995). All worm culture was performed at 20°C unless otherwise noted.
Synthetic lethal forward screen
A population of 500,000 alg-2(ok304)Ex[alg-2;sur-5::GFP] animals was mutagenized with 50 mM of ethylmethanesulfonate (EMS) for 4 hours. A total of four GFP positive F2 animals per F1 were isolated from 1,000 GFP positive F1. We screened the F3 population from 1,556 F2 to obtain six unlinked mutations.
Total worm lysate preparation
Populations of synchronized animals were harvested at the adult stage. Harvested animals were homogenized in ice-cold lysis buffer (100 mM Potassium Acetate, 30 mM Hepes-KOH [pH 7.4], 2mM Magnesium Acetate, 1 mM DDT, 0.5% [v/v] Triton X-100, 2% [v/v] SUPERaseIn (Ambion) and Mini Complete EDTA-free Protease Inhibitor Cocktail (1 tablet/5 ml solution) (Roche)). The homogenized extract was clarified by centrifugation at 13,817 × g for 10 min at 4°C and the protein concentrations of the different samples were normalized using Dc protein Assay (Bio-Rad).
DCS-1 protein purification
HA-tagged DCS-1 wild-type and mutant proteins were expressed and purified as reported in (Cohen et al., 2004).
Pulldown assays
Purified recombinant DCS-1 proteins were first incubated with total worm protein lysate for 1h at 4°C followed by purification on a Talon affinity column (Clontech). Purified DCS-1 complexes were washed three times with lysis buffer and proteins associated with DCS-1 were identified by Western blotting.
MicroRNA degradation assays
Assays were performed as described previously in (Chatterjee and Grosshans, 2009).
Supplementary Material
Acknowledgments
We would like to thank Dr. Julie Claycomb, Dr. Gyorgy Hutvagner and members of our laboratories for comments on the manuscript. We also like to thank Eric Paquet for statistical support. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). G.D.B. was a recipient of a Fonds de Recherche en Santé du Québec Scholarship and now is a Natural Sciences and Engineering Research Council of Canada Graham-Bell Scholar. S.R. gratefully acknowledges his Boehringer Ingelheim Fonds PhD fellowship. Work in the lab of M.J.S has been funded by the Canadian Institutes of Health Research (CIHR). Work in the lab of H.G. is funded by grants from the European Research Council (miRTurn; ERC 2419845), the Swiss National Science Foundation (SNF 31003A_127052), and Friedrich Miescher Institute, which is supported by the Novartis Research Foundation. Contributions of VA and MO were supported by funding from the US NIH grant (R01 GM24028). M.J.S. is a Junior 2 scholar from Fonds de Recherche du Québec en Santé.
GDB and MJS conceived and designed nearly all experiments. SR and HG conceived and designed the XRN-1/-2 localization experiments and contributed tools. MCO and VRA conceived and designed the TaqMan miRNA profiling. ELR performed the screen. AVR contributed to the identification of mutant alleles from the screen. GDB, SR and MCO performed the experiments. GDB, SR, MCO, VRA, HG and MJS analyzed the data and wrote the paper.
Footnotes
Supplementary experimental procedures, a list of oligonucleotide sequences, four figures and one table can be found online.
References
- Bajan S, Hutvagner G. Another “loophole” in miRNA processing. Mol Cell. 2011;44:345–347. doi: 10.1016/j.molcel.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Fasler M, Bussing I, Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell. 2011;20:388–396. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- Cohen LS, Mikhli C, Friedman C, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE. Nematode m7GpppG and m3(2,2,7)GpppG decapping: activities in Ascaris embryos and characterization of C. elegans scavenger DcpS. RNA. 2004;10:1609–1624. doi: 10.1261/rna.7690504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The Structure of Human Argonaute-2 in Complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Fay DS, Keenan S, Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–517. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3:151–158. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman EJ, Miska EA. The microRNAs of Caenorhabditis elegans. Semin Cell Dev Biol. 2010;21:728–737. doi: 10.1016/j.semcdb.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kwasnicka DA, Krakowiak A, Thacker C, Brenner C, Vincent SR. Coordinate expression of NADPH-dependent flavin reductase, Fre-1, and Hint-related 7meGMP-directed hydrolase, DCS-1. J Biol Chem. 2003;278:39051–39058. doi: 10.1074/jbc.M306355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Piano F, Davis RE. Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol Biol Cell. 2005;16:5880–5890. doi: 10.1091/mbc.E05-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kiledjian M. Scavenger decapping activity facilitates 5′ to 3′ mRNA decay. Mol Cell Biol. 2005;25:9764–9772. doi: 10.1128/MCB.25.22.9764-9772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods in Cell Biology. 1995;48:451–482. [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–1803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Resnick TD, McCulloch KA, Rougvie AE. miRNAs give worms the time of their lives: small RNAs and temporal control in Caenorhabditis elegans. Dev Dyn. 2010;239:1477–1489. doi: 10.1002/dvdy.22260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger S, Grosshans H. MicroRNA turnover: when, how, and why. Trends Biochem Sci. 2012;37:436–446. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F, Brechemier-Baey D, Kiledjian M, Condon C, Benard L. Activation of 5′-3′ exoribonuclease Xrn1 by cofactor Dcs1 is essential for mitochondrial function in yeast. Proc Natl Acad Sci U S A. 2012;109:8264–8269. doi: 10.1073/pnas.1120090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Vasquez-Rifo A, Jannot G, Armisen J, Labouesse M, Bukhari SI, Rondeau EL, Miska EA, Simard MJ. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One. 2012;7:e33750. doi: 10.1371/journal.pone.0033750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.