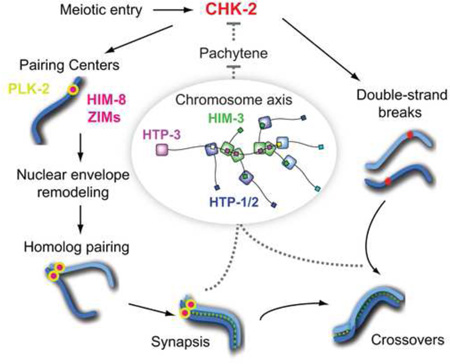
Summary
CHK-2 kinase is a master regulator of meiosis in C. elegans. Its activity is required for homolog pairing and synapsis and for double-strand break formation, but how it drives and coordinates these pathways to ensure crossover formation remains unknown. Here we show that CHK-2 promotes pairing and synapsis by phosphorylating a family of zinc finger proteins that bind to specialized regions on each chromosome known as pairing centers, priming their recruitment of the Polo-like kinase PLK-2. This knowledge enabled development of a phosphospecific antibody as a tool to monitor CHK-2 activity. When either synapsis or crossover formation is impaired, CHK-2 activity is prolonged and meiotic progression is delayed. We show that this common feedback circuit is mediated by interactions among a network of HORMA domain proteins within the chromosome axis and generates a graded signal. These findings reveal conserved regulatory mechanisms that ensure faithful meiotic chromosome segregation in diverse species.
Graphical Abstract
Introduction
Meiosis is a specialized cell division that produces haploid gametes to transmit genetic information from parent to progeny through sexual reproduction. During meiotic prophase, chromosomes are reorganized around a central axis, and then pair, synapse, and recombine with their homologs. This process generates physical linkages known as chiasmata, which enable homologs to biorient on the meiotic spindle and to disjoin during meiosis I. A fundamental mystery is how these chromosomal events are coordinated during the meiotic cell cycle. In diverse organisms, defects in synapsis (assembly of the synaptonemal complex (SC)) or meiotic recombination trigger a delay or arrest in mid-prophase (Bishop et al., 1992; Edelmann et al., 1996; Ghabrial and Schüpbach, 1999; MacQueen et al., 2002; Pittman et al., 1998), indicative of surveillance mechanisms that monitor meiotic events. This meiotic checkpoint (also referred to as the ‘pachytene checkpoint’ or the ‘meiotic recombination checkpoint’) typically prevents the meiosis I division in cells that fail to form crossovers (MacQueen and Hochwagen, 2011; Roeder and Bailis, 2000; Subramanian and Hochwagen, 2014). Because induction of programmed DNA double strand breaks (DSBs) is an integral aspect of the meiotic program, proteins involved in DNA damage response also play essential roles in meiotic checkpoints (Lydall et al., 1996).
In S. cerevisiae, Mek1, a paralog of the checkpoint kinase Chk2, mediates cell cycle arrest in response to meiotic defects (Bailis and Roeder, 2000; Xu et al., 1997). Mek1 is associated with meiotic chromosome axes, where its activation requires interactions with two other axis proteins, Hop1 and Red1 (Niu et al., 2007; Niu et al., 2005). Hop1, a founding member of the HORMA domain family (Hop1, Rev7 and Mad2) (Aravind and Koonin, 1998), is phosphorylated by ATM/ATRMec1/Tel1, and serves as a meiosis-specific adaptor for Mek1 activation (Carballo et al., 2008). Mammalian HORMA domain proteins HORMAD1 and HORMAD2 localize along unsynapsed chromosome axes and recruit ATR to activate a meiotic checkpoint (Daniel et al., 2011; Shin et al., 2010; Wojtasz et al., 2012). Thus, the chromosome axis provides a key interface for integrating meiotic chromosome dynamics and checkpoint signaling. However, how these proteins monitor chromosomal events and generate signals to regulate meiotic progression is unknown.
In the nematode C. elegans, adult germlines contain a complete progression of meiotic stages in a spatiotemporal sequence (Figure 1A). The onset of meiotic prophase is marked by a dramatic change in nuclear morphology, in which the chromosome mass becomes asymmetrically distributed within the nucleus to form a crescent shape, defining the “transition zone” region of the germline. During this stage, special regions near one end of each chromosome known as the pairing centers (PCs) establish connections to microtubules through the nuclear envelope proteins SUN-1 and ZYG-12, which play essential roles in homolog pairing and synapsis, (Penkner et al., 2007; Sato et al., 2009). Through short sequence motifs enriched over several hundred kilobases, PCs recruit a family of zinc finger proteins essential for their functions: HIM-8, ZIM-1, ZIM-2 and ZIM-3 (Phillips and Dernburg, 2006; Phillips et al., 2009; Phillips et al., 2005).
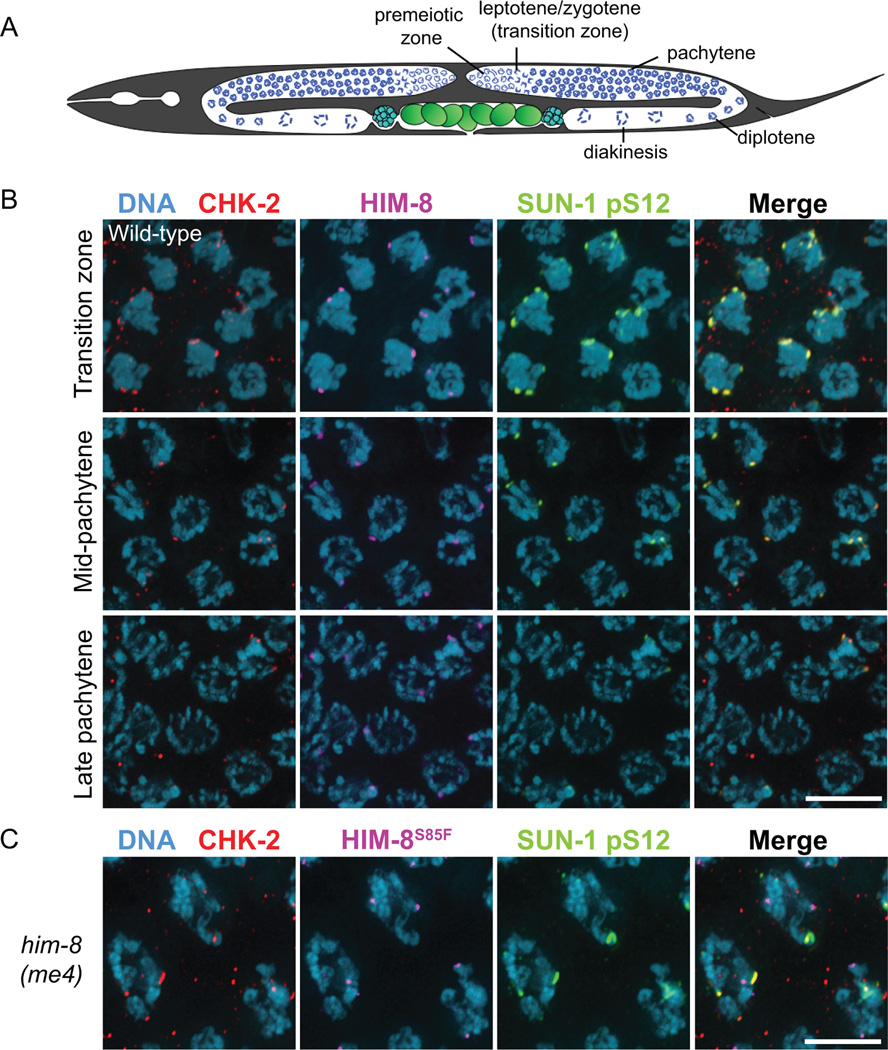
Figure 1. CHK-2 localizes to PCs in early meiotic prophase.
(A) Diagram of the C. elegans hermaphrodite germline. (B–C) Dissected gonads of wild-type (B) and him-8(me4) (C) hermaphrodites stained for CHK-2 (red), HIM-8 (purple), SUN-1 pS12 (green), and DNA (blue). Scale bars, 5 µm.
At least two kinases, PLK-2 and CHK-2, play central roles during early meiotic prophase in C. elegans. PLK-2 is one of three Polo-like kinases in C. elegans that are closely related to mammalian Plk1; PLK-1, which is essential for mitosis, can partially substitute for the function of PLK-2 in meiosis (Harper et al., 2011; Labella et al., 2011). PLK-2 localizes to PCs upon meiotic entry, which leads to aggregation of SUN-1/ZYG-12 within the nuclear envelope, initiating dynein-driven chromosome motions that promote pairing and synapsis (Harper et al., 2011; Labella et al., 2011; Rog and Dernburg, 2015; Wynne et al., 2012). CHK-2 is a meiosis-specific paralog of Chk2, and is dispensable for the canonical DNA damage response (Higashitani et al., 2000; MacQueen and Villeneuve, 2001). Instead, CHK-2 governs two major pathways essential for crossover formation: it is required for nuclear reorganization leading to homolog pairing and synapsis, and also for the programmed DSBs that initiate meiotic recombination (MacQueen and Villeneuve, 2001; Oishi et al., 2001). Upon meiotic entry, SUN-1 is phosphorylated at several sites in its nucleoplasmic domain, a subset of which requires PLK-2 and/or CHK-2 activity (Harper et al., 2011; Penkner et al., 2009). However, phosphorylation of the SUN-1 N-terminus is largely dispensable for SUN-1/ZYG-12 aggregation and homolog pairing (Woglar et al., 2013). Thus, the key targets of CHK-2 and PLK-2 that mediate pairing and synapsis remain to be identified. Recent studies have shown that CHK-2 is required for chromosomal localization of two DSB-promoting proteins, DSB-1 and DSB-2 (Rosu et al., 2013; Stamper et al., 2013). However, direct substrates of CHK-2 have not been identified, and the molecular mechanisms by which CHK-2 controls chromosome dynamics are therefore unknown.
Based on cytological observations, the existence of a mechanism linking meiotic chromosome dynamics with cell cycle progression has been inferred in C. elegans. Polarized nuclear organization is greatly extended in mutants that disrupt synapsis (Colaiacovo et al., 2003; MacQueen et al., 2002), hinting that cell cycle progression is delayed. Molecular markers of early prophase, including SUN-1 phosphorylation and chromosomal localization of DSB-1/2, also persist longer in mutants that are proficient for synapsis but fail to establish crossovers (Rosu et al., 2013; Stamper et al., 2013; Woglar et al., 2013), implying that meiotic recombination is also subject to surveillance. However, it has been unclear whether these responses occur through the same feedback circuit, since the molecular basis for this regulation has not been elucidated.
Here we identify meiotic substrates of CHK-2 through a candidate-based approach. We demonstrate that CHK-2 activity is regulated by a feedback circuit that monitors synapsis and crossover formation, establishing CHK-2 as a checkpoint target. We further elucidate how a network of meiotic HORMA domain proteins mediates this control, thereby illuminating the conserved role of chromosome axes in meiotic regulation.
Results
CHK-2 localizes to PCs in early meiotic prophase
To determine how CHK-2 promotes pairing and synapsis, we examined its localization in the C. elegans germline by immunofluorescence. In wild-type hermaphrodites, faint CHK-2 foci were detected at all PCs in transition zone nuclei, which colocalized with phosphorylated SUN-1 (pS12) patches at the nuclear envelope (Figure 1B). Overlap between CHK-2 and phospho-SUN-1 patches persisted until mid-pachytene, but was no longer detected in late pachytene except for a few “straggler” nuclei with hypercondensed chromosomes (Figure 1B) (Rosu et al., 2013; Stamper et al., 2013; Woglar et al., 2013). Localization of CHK-2 to PCs is consistent with its role in triggering nuclear reorganization and homolog pairing (MacQueen and Villeneuve, 2001). Interestingly, we found that CHK-2 did not localize to unpaired HIM-8 foci in him-8(me4) mutants, which carry a point mutation in the N-terminus (S85F) (Figures 1C and 2C). This residue is adjacent to a potential Chk2 Forkhead-associated (FHA) binding motif (pT-X-X-[I/L]) (Li et al., 2002) that is conserved in all PC proteins in Caenorhabditis species (Figure S1A). This suggests that association of CHK-2 with the PCs might be mediated through direct recruitment by HIM-8 and the ZIM proteins.
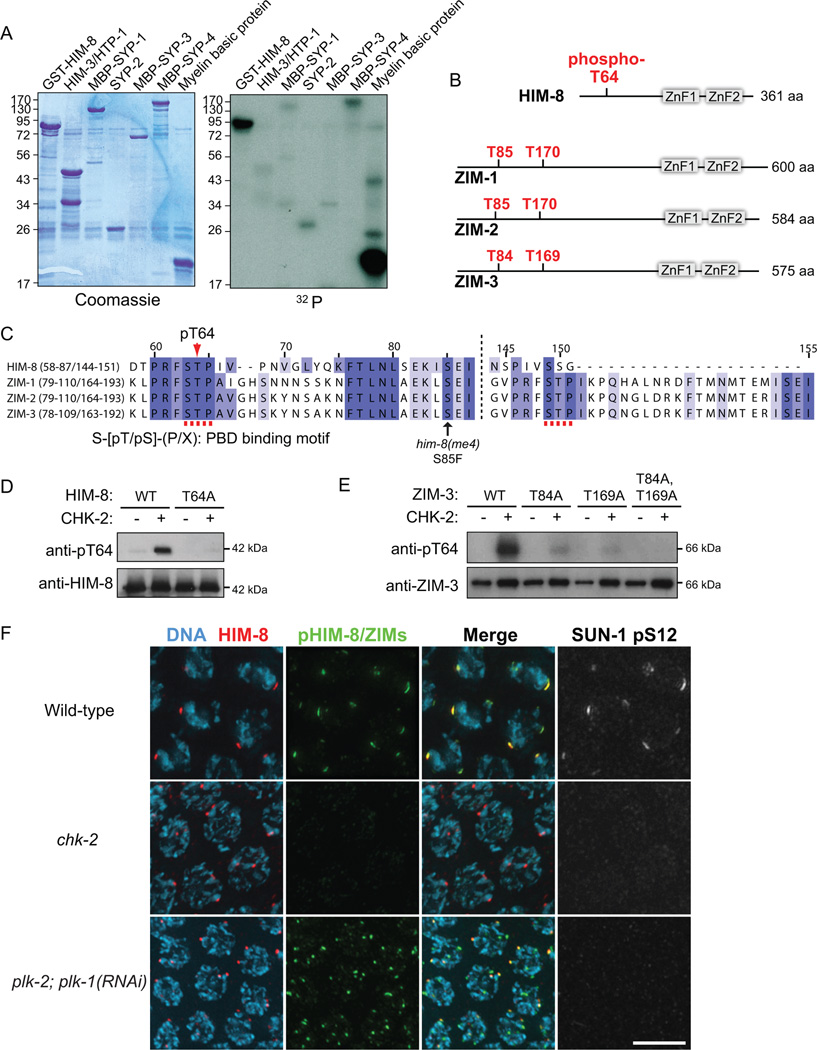
Figure 2. CHK-2 phosphorylates PRFSTP motifs within PC proteins in vitro and in vivo.
(A) In vitro kinase assays using recombinant CHK-2 and candidate substrates expressed in E. coli. GST-HIM-8, HIM-3-6His/HTP-1, MBP-SYP-1, SYP-2-6His, MBP-SYP-3, MBP-SYP-4, and myelin basic protein (positive control) were tested in reactions that included γ-32P ATP. Products were separated by SDS-PAGE. Gels were stained with Coomassie and radiolabel incorporation was analyzed by autoradiography. (B) Domain structures of HIM-8, ZIM-1, ZIM-2 and ZIM-3 and in vitro CHK-2 phosphorylation sites in HIM-8 mapped by mass spectrometry analysis. Sequence coverage was 44.9%. Corresponding threonines within PRFSTP motifs of ZIMs are also shown. (C) Sequence alignment of HIM-8 and ZIMs using the T-coffee algorithm. CHK-2 phosphorylation sites constitute the binding motif for Polo-Box Domain (PBD) proteins. (D) Immunoblot of wild-type or T64A recombinant HIM-8 phosphorylated by CHK-2 in vitro. (E) Immunoblot of recombinant ZIM-3 proteins after in vitro phosphorylation by CHK-2. (F) Projection images of transition zone nuclei from wild-type, chk-2, and plk-2; plk-1(RNAi) mutants stained for HIM-8 (red), pHIM-8/ZIMs (green), SUN-1 pS12 (white) and DNA (blue). Scale bar, 5 µm.
CHK-2 phosphorylates HIM-8 and ZIMs at conserved PRSFTP motifs in vitro and in vivo
To identify meiotic substrates of CHK-2, we purified GST-CHK-2 from insect cells, verified its kinase activity (Figure S1B), and tested its activity toward bacterially expressed proteins that play important roles in pairing and synapsis. A kinase-dead allele was included as a control. Tested candidates included the X chromosome PC protein HIM-8; two HORMA domain proteins HIM-3 and HTP-1; and four SC components, SYP-1, SYP-2, SYP-3 and SYP-4. Among the proteins tested, HIM-8 was robustly phosphorylated by CHK-2 in vitro (Figure 2A).
Using mass spectrometry, we found that the primary in vitro CHK-2 phosphorylation site on HIM-8 is T64 (Figure 2B). This residue is near the putative FHA binding motif, and lies within a sequence motif (PRFSTP) that conforms to the consensus phosphorylation motif for human Chk2 (R-X-X-S/T, where X indicates any amino acid) (O'Neill et al., 2002) and is highly conserved among all known HIM-8/ZIM family members in Caenorhabditis species (Figures 2C and S1). While HIM-8 contains a single PRFSTP motif, each ZIM protein contains 2 such motifs (Figures 2B and 2C). We generated an antibody against a phosphopeptide surrounding HIM-8 T64. The affinity-purified antibody recognized recombinant HIM-8 only when it was phosphorylated by CHK-2, and did not recognize HIM-8 containing the T64A mutation, demonstrating its phospho-specificity (Figure 2D). The antibody also recognized phosphorylated ZIM-3 in vitro (Figure 2E) due to the high conservation of this motif. Detection of ZIM-3 was diminished when the threonine in either PRFSTP motif was mutated to alanine (T84A or T169A), and was eliminated when both threonines were mutated (Figure 2E). Thus, CHK-2 phosphorylates the threonines within both PRFSTP motifs of ZIM-3 in vitro, and our antibody recognizes both motifs when phosphorylated by CHK-2.
Immunofluorescence with this antibody in C. elegans gonads revealed foci that colocalized with all PCs in transition zone nuclei (Figure 2F), indicating that all four of the zinc finger proteins are recognized. Staining was absent in chk-2 null mutants, but was detected robustly in plk-2; plk-1(RNAi) animals (Figure 2F), confirming that the phosphoepitope depends on CHK-2 but not on Polo-like kinases in vivo. Phosphorylation of HIM-8 and ZIMs was evident upon meiotic entry and persisted until mid-pachytene (Figure S2A). Staining was restricted to nuclei that had entered meiosis, even though two HIM-8 foci can be detected in pre-meiotic nuclei (Figure S2B) (Phillips et al., 2005). Phosphorylation of the PC proteins preceded homolog pairing (Figure S2B), and was temporally correlated with SUN-1 phosphorylation at S12 and chromosomal localization of DSB-1 (Figures S2A and S3A). Together, our results demonstrate that CHK-2 phosphorylates HIM-8 and ZIM proteins at the PRFSTP motifs in vivo, and that our antibody serves as a reporter for CHK-2 activity, which is detected in early meiotic prophase but not before meiotic entry.
Phosphorylation of HIM-8 by CHK-2 primes PLK-2 recruitment to the X chromosome PCs
When phosphorylated, HIM-8 T64 and the corresponding threonines in the three ZIM proteins constitute binding motifs for Polo-box domain proteins (S-[pT/pS]-(P/X)) (Figure 2C) (Elia et al., 2003). We have previously shown that this motif is important for the recruitment of PLK-2 to the X chromosome PC (Harper et al., 2011); however, a him-8T64A transgene showed variable expression, precluding systematic analysis. To better characterize the significance of HIM-8 T64 phosphorylation in vivo, we introduced the T64A mutation at the endogenous him-8 locus by Cas9/CRISPR-mediated homologous recombination (Dickinson et al., 2013). Self-progeny of hermaphrodites homozygous for the mutation were mostly viable (99%) but a large fraction of them was male (41% in him-8T64A vs. 0.2% in wild-type) (Figure S4A). This strong “high incidence of males” (Him) phenotype results from X chromosome nondisjunction during meiosis, and is quantitatively similar to that displayed by animals lacking HIM-8 (Phillips et al., 2005). We found that HIM-8T64A was expressed and localized to the X chromosome PCs, but failed to recruit PLK-2 (Figure S4C), resulting in failures in pairing and synapsis of the X chromosomes (Figure S4B). Thus, CHK-2 primes the recruitment of PLK-2 to PCs by phosphorylating the PRFSTP motifs within HIM-8 and ZIM proteins, thereby promoting homolog pairing and synapsis.
Both synapsis failures and crossover defects prolong CHK-2 activity
Using the phosphorylation status of HIM-8/ZIMs as a proxy for CHK-2 activity, we investigated how CHK-2 is regulated in response to various meiotic defects. We quantified the duration of CHK-2 activity as a fraction of the length of the germline containing nuclei in leptotene, zygotene and pachytene stages of meiosis (the LZP zone). While the phospho-staining of HIM-8/ZIMs was detected in 48% of the LZP zone of wild-type germlines (Figures S2A, 3A and 3E), mutants that fail to form crossovers due to an absence of synapsis (e.g. syp-2) (Colaiacovo et al., 2003) showed a striking extension of CHK-2 activity in the germline (up to 90% of the LZP zone) (Figures 3B and 3E). The ‘CHK-2 active zone’ was also greatly extended in plk-2; plk-1(RNAi) animals (74%) (Figures S5A and 3E), which have severe synapsis defects (Harper et al., 2011), demonstrating that Polo kinase activity is not responsible for the extension.
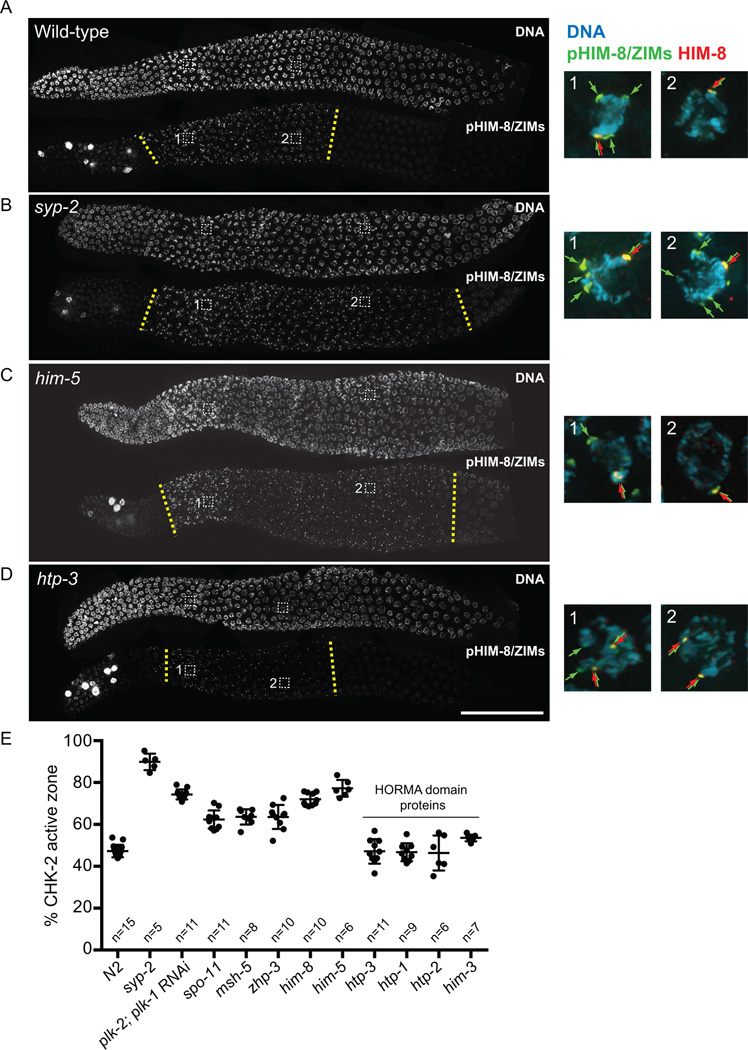
Figure 3. CHK-2 activity is prolonged in mutants that are defective in crossover formation, but not in mutants that disrupt HORMA domain proteins.
(A–D) Dissected gonads from the indicated genotypes were stained for DNA and phospho-HIM-8/ZIMs. (Note: our pHIM-8/ZIMs antibody recognizes a CHK-2–independent epitope in mitotic nuclei) Scale bar, 50 µm. Insets: representative nuclei from the transition zone and mid-prophase regions are shown for DNA (blue), pHIM-8/ZIMs (green), and HIM-8 (red). (E) Quantification of the CHK-2 active zone, expressed as a proportion by length, of the phosphoantibody-reactive region relative to the region from meiotic onset to the end of pachytene. Numbers of gonads scored are indicated below. Error bars indicate standard deviations. All genotypes show significant differences from N2 (p<0.0001), except for htp-3 (p=0.9999), htp-1 (p=0.9996), htp-2 (p=0.9993), and him-3 (p=0.0194), by ordinary one-way ANOVA.
Mutations that block processing of meiotic DSBs into crossovers without disrupting pairing or synapsis (zhp-3 and msh-5) (Bhalla et al., 2008; Kelly et al., 2000) also significantly extended the CHK-2 active zone (64% in both zhp-3 and msh-5) (Figures S5D and 3E). Importantly, spo-11 mutants, which lack DSBs (Dernburg et al., 1998), also showed extended activity of CHK-2 (62%) (Figures S5C and 3E), indicating that the response is not due to persistence of unrepaired DSBs. Mutations in him-8 or him-5, which are specifically required for X chromosome pairing/synapsis or DSB formation, respectively (Meneely et al., 2012; Phillips et al., 2005), also extended the CHK-2 active zone (72% in him-8; 77% in him-5) (Figures 3C, 3E and S5B). Thus, the absence of crossovers on a single chromosome pair is sufficient to prolong CHK-2 activity. Together, our results show that defects in synapsis and crossing-over both feed back to maintain CHK-2 activity, thereby delaying meiotic progression.
We also note both quantitative and qualitative differences in the extension of CHK-2 activity in various meiotic mutants. In general, we find that synapsis defects give rise to longer delays in CHK-2 activity than an absence of crossovers. (Figure 3E). Moreover, in synapsis-deficient mutants (e.g. syp-2, him-8 and plk-2; plk-1(RNAi)), phosphorylation persisted at all PCs (Figure 3B insets), while mutations that blocked crossover formation without impairing synapsis (e.g. zhp-3, msh-5, spo-11, and him-5) showed persistent phosphorylation only on HIM-8 (Figure 3C insets). This observation may reflect differences between HIM-8 and the autosomal ZIM proteins, as well as the stage of prophase at which the meiotic errors are generated (see Discussion).
Meiotic HORMA domain proteins mediate feedback control
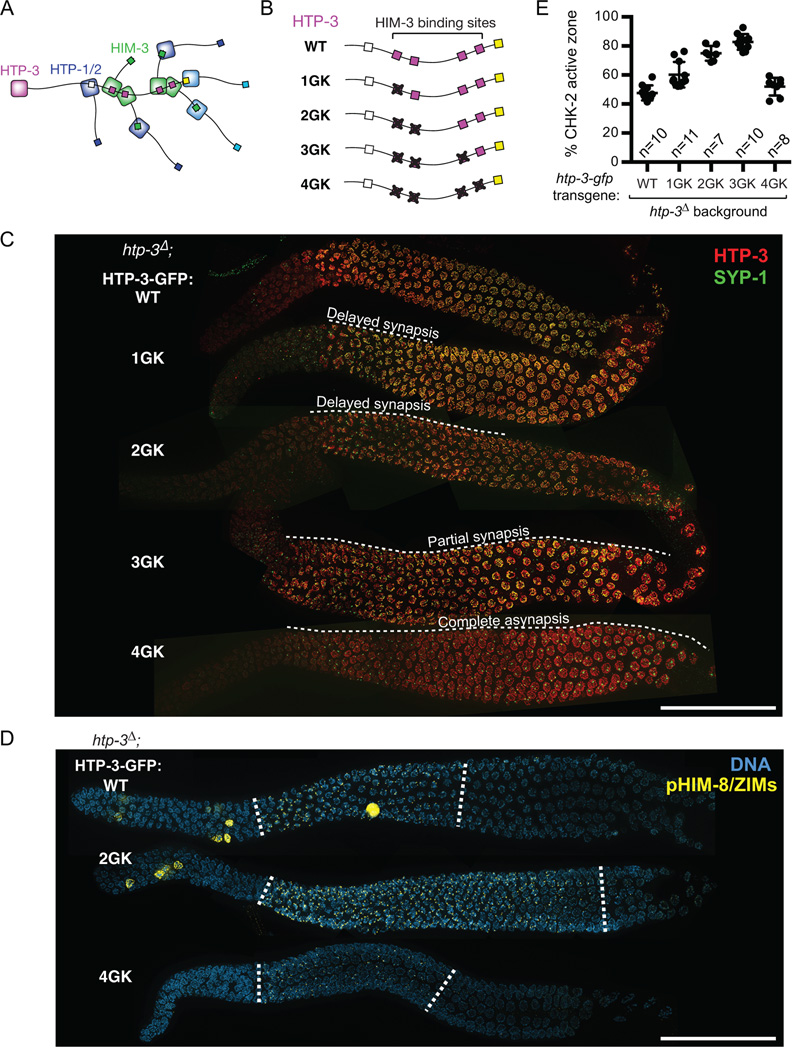
Four meiotic HORMA domain proteins in C. elegans (HIM-3, HTP-1, HTP-2, and HTP-3) associate with the chromosome axis and play essential roles in pairing, synapsis, and meiotic recombination (Couteau and Zetka, 2005; Goodyer et al., 2008; Kim et al., 2014; Martinez-Perez and Villeneuve, 2005; Zetka et al., 1999). The largest of these proteins, HTP-3, localizes to the axis in the absence of the other HORMA domain proteins, and HIM-3, HTP-1, and HTP-2 are recruited by binding of their HORMA domains to short peptide motifs (termed “closure motifs”) in the C-terminal tail of HTP-3 (Figure 5A) (Kim et al., 2014).
Figure 5. Recruitment of HIM-3 to the chromosome axis is required for feedback control of CHK-2.
(A) Schematic showing the hierarchical assembly of meiotic HORMA domain proteins in C. elegans. (B) Mutant HTP-3 transgenes containing GK mutations in four central closure motifs. (C) Dissected gonads from the indicated genotypes were stained for HTP-3 (red), and SYP-1 (green). Scale bar, 50 µm. (D) Dissected gonads from worms expressing WT, 2GK and 4GK HTP-3 were stained for DNA (blue) and pHIM-8/ZIMs (yellow). The CHK-2 active zone is marked by white dotted lines. Scale bar, 50 µm. (E) Quantification of the CHK-2 active zone. Numbers of gonads scored are indicated below. All genotypes show significant differences from WT (p<0.0001), except for HTP-34GK (p=0.4312), by ordinary one-way ANOVA.
Intriguingly, mutations that disrupt HTP-3, HTP-1, or HIM-3 showed normal timing of CHK-2 activation but no extension of HIM-8 or ZIM phosphorylation (46% CHK-2 active zone in htp-3; 47% in htp-1; 54% in him-3) (Figures 3D, 3E, S4E and S4F), despite their severe defects in synapsis and crossover formation. Mutations in HTP-2, which shares 82% sequence identity with HTP-1, do not cause obvious meiotic defects (Martinez-Perez et al., 2008) and did not extend CHK-2 activity (46%) (Figure 3E). Although polarized nuclear morphology in the transition zone was less prominent in htp-3, htp-1, and him-3 mutants, CHK-2 activity was clearly detected at the PCs upon meiotic entry (Figures 3D, S5E, and S5F).
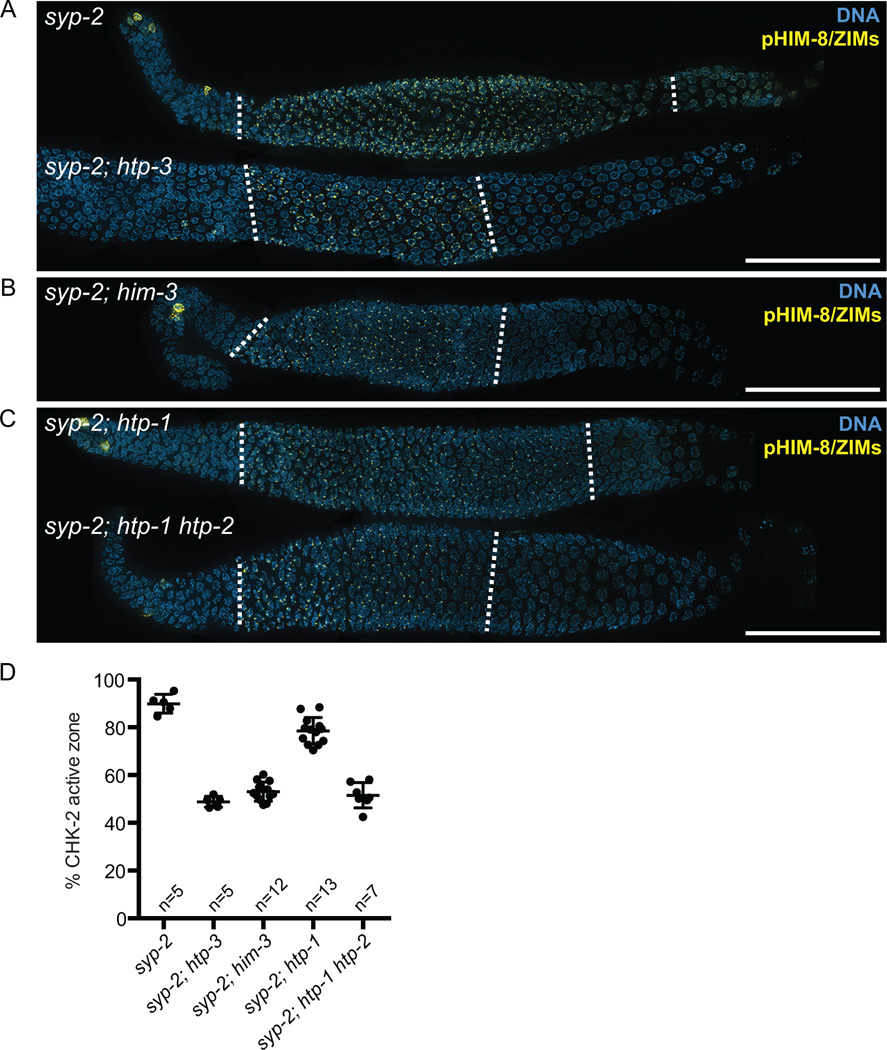
To further explore the role of the HORMA domain proteins in extending CHK-2 activity in response to meiotic defects, we crossed worms carrying mutations in these genes to animals lacking syp-2 and examined the phosphorylation of HIM-8 and ZIMs. Consistent with a role in mediating feedback control of CHK-2, deletion of HTP-3 suppressed the extension of CHK-2 activity in syp-2 mutants (49% in syp-2; htp-3 vs. 90% in syp-2) (Figures 4A and 4D). Surprisingly, deletion of HIM-3 alone fully suppressed the prolonged CHK-2 activity in syp-2 mutants (54% in syp-2; him-3) (Figures 4B and 4D), indicating that HIM-3 is essential for feedback regulation of CHK-2.
Figure 4. Deleting meiotic HORMA domain proteins suppresses the extension of CHK-2 activity in syp-2 mutants.
(A–C) Immunofluorescence images of dissected gonads from syp-2, syp-2; htp-3, syp-2; him-3, syp-2; htp-1, and syp-2; htp-1 htp-2 mutants stained with anti-pHIM-8/ZIMs (yellow) and DAPI (blue). The CHK-2 active zone is marked by dotted white lines. Scale bars, 50 µm (D) Graph showing quantification of the CHK-2 active zone in the indicated strains. Numbers of gonads scored are indicated below. syp-2 and syp-2; htp-1 mutants showed significant differences from N2 (p<0.0001), whereas syp-2; htp-3 (p=0.9926), syp-2; him-3 (p=0.0138), and syp-2; htp-1 htp- 2 (p=0.2439) did not, by ordinary one-way ANOVA.
To determine the role of HTP-1 in regulating CHK-2, we examined the phosphorylation of HIM-8/ZIMs in syp-2; htp-1 double mutants. Although deletion of HTP-1 modestly shortened the CHK-2 active zone in syp-2 mutants, it was not sufficient to fully suppress the extension of the phospho-HIM-8/ZIMs staining (79% in syp-2; htp-1) (Figures 4C and 4D). The signal from asynapsed chromosomes nonetheless appeared to be weakened in the absence of HTP-1, as persistent CHK-2 activity was found only on HIM-8, but not on ZIMs, and chromosomes did not maintain their crescent-shaped configuration, as previously described (Martinez-Perez and Villeneuve, 2005). We hypothesized that HTP-2 may play redundant roles with HTP-1 in mediating a signal from asynapsed chromosomes. Indeed, deleting both HTP-1 and HTP-2 completely suppressed the extension of CHK-2 activity in syp-2 mutants (52% in syp-2; htp-1 htp-2) (Figures 4C and 4D). Together, HTP-1 and HTP-2 play overlapping roles in signaling from asynapsed chromosomes, and both HIM-3 and HTP-1/2 are required for feedback regulation of CHK-2.
Recruitment of HIM-3 and HTP-1/2 to the chromosome axis is required for feedback control of CHK-2
HIM-3 is recruited to chromosomes through binding to four central closure motifs in HTP-3 (Figure 5A). To determine whether axis localization of HIM-3 is required for CHK-2 regulation, we engineered htp-3-gfp transgenes with a series of glycine-to-lysine (GK) mutations in these motifs (Figures 5B, S6A, and S6B). We found that the intensity of HIM-3 staining along chromosome axes correlated well with the number of intact binding sites on HTP-3 (Figure S6C), while mutating all four motifs (4GK) completely abrogated HIM-3 binding, as shown previously (Kim et al., 2014). Mutation of the HIM-3 binding sites led to commensurate reductions in progeny viability (79.2% egg viability in HTP-31GK; 57.5% in HTP-32GK; 21.2% in HTP-33GK; 3.1% in HTP-34GK), and the fraction of males among the surviving progeny also progressively increased (1.5% in HTP-31GK; 3.9% in HTP-32GK; 12.4% in HTP-33GK; 27.5% in HTP-34GK) (Figure S6B). This implies that each of the four motifs in HTP-3 can independently recruit HIM-3, and that recruiting a sufficient amount of HIM-3 to the axis is essential for accurate meiotic chromosome segregation.
Strikingly, we found that SC assembly was extremely sensitive to the amount of HIM-3 at the chromosome axis. While synapsis completely fails in the HTP-34GK mutant, completion of synapsis was significantly delayed in HTP-31GK and HTP-32GK mutants (Figure 5C). Complete synapsis was never achieved in HTP-33GK mutants, and partial synapsis was preferentially observed on X chromosomes (Figures 6C and S6D), similar to the effects of a hypomorphic allele of him-3 (Nabeshima et al., 2004). Thus, HIM-3 is required for synapsis in a dose-dependent manner, and recruitment of sufficient HIM-3 to the axis is critical to support efficient SC assembly.
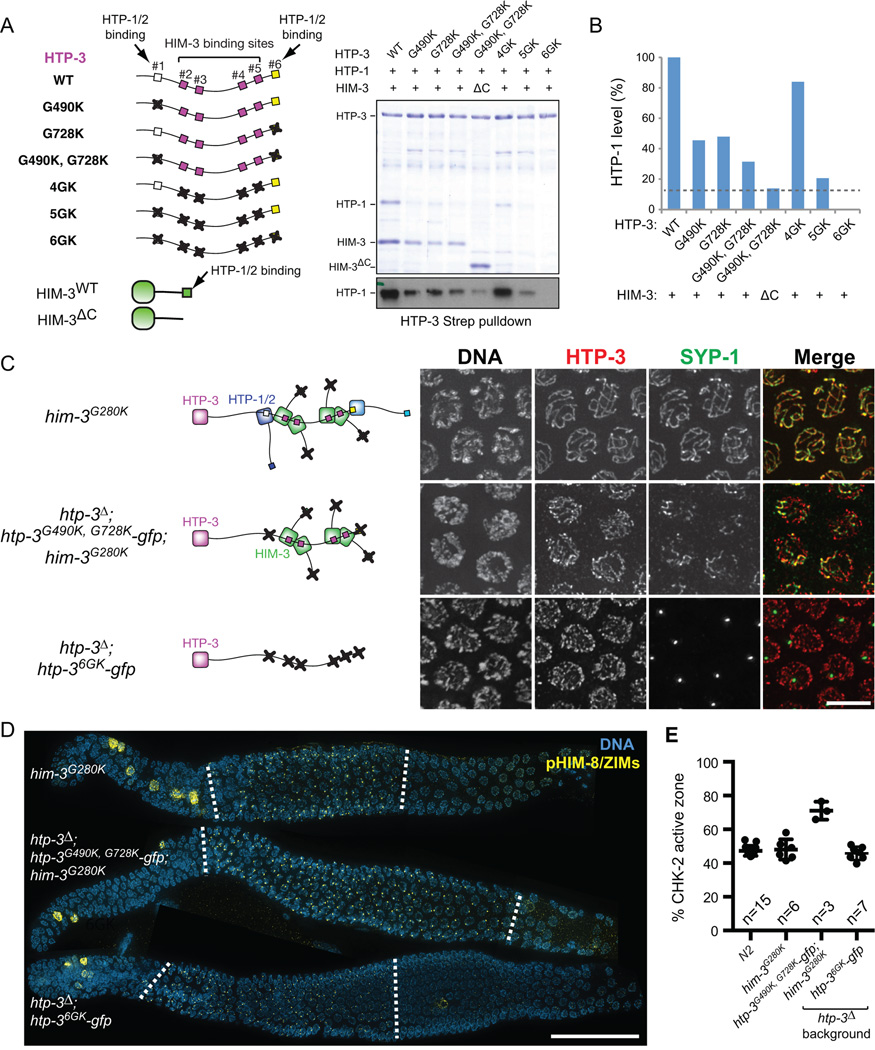
Figure 6. Axis localization of HTP-1/2 and HIM-3 is essential for feedback control of CHK-2.
(A) Strep-tagged HTP-3, either wild-type or GK mutants, was coexpressed in E. coli with untagged HTP-1 and wild-type or C-terminally truncated HIM-3 (ΔC; aa 1–245), and purified using Strep-resin to isolate proteins associated with HTP-3. Coomassie staining of SDS-PAGE shows the HTP-3-associated proteins. A Western blot for HTP-1 is shown below. (B) The amount of HTP-1 bound by HTP-3 was quantified based on the HTP-1 Western blot. (C) Schematics of HORMA domain protein assembly in the indicated genotypes are shown in the left. Mid-pachytene nuclei were stained for DNA (white), HTP-3 (red), and SYP-1 (green). Scale bar, 5 µm. (D) Immunofluorescence images of gonads dissected from him-3G280K, htp-3Δ; htp-3G490K, G728K-gfp; him-3G280K, and htp-3Δ; htp-36GK-gfp showing DNA (blue) and pHIM-8/ZIMs (yellow). White dotted lines indicate the CHK-2 active zone. Scale bar, 50 µm. (E) Quantification of the CHK-2 active zone in indicated strains. Numbers of gonads scored are indicated below. htp-3Δ; htp-3G490K, G728K-gfp; him-3G280K mutants show significant differences from N2 (p<0.0001), while others do not (him-3G280K, p=0.9608; htp-3Δ; htp-36GK-gfp, p=0.7806), analyzed by ordinary one-way ANOVA.
CHK-2 phosphorylation of HIM-8/ZIMs and crescent-shaped chromosome morphology were significantly extended in the HTP-31GK, HTP-32GK, and HTP-33GK mutants (60% CHK-2 active zone in HTP-31GK; 75% in HTP-32GK; 83% in HTP-33GK) (Figures 5D and 5E). The extent to which CHK-2 activity was prolonged correlated with the severity of synapsis defects, likely reflecting quantitative variation in the signal strength. However, in worms expressing HTP-34GK, the CHK-2 active zone was not extended (51%), despite complete failure of synapsis (Figures 5C, 5D and 5E). Thus, recruitment of HIM-3 to the meiotic chromosome axis is essential for feedback control of CHK-2, and binding of HIM-3 to a single HTP-3 closure motif is sufficient to generate a signal from asynapsed chromosomes.
HTP-1 and HTP-2 are recruited to meiotic chromosomes by redundant mechanisms, either by binding two closure motifs within HTP-3 (motifs #1 and #6) or binding to a C-terminal motif in HIM-3 (Figure 6A) (Kim et al., 2014). We created GK mutations in each of these motifs and examined complex formation by HTP-3, HTP-1, and HIM-3 in vitro using a bacterial coexpression system (Figure 6A). Mutation of HTP-3 motif #1 (G490K) or motif #6 (G728K) alone each reduced HTP-1 binding to a similar extent, and mutating both further reduced HTP-1 binding (Figure 6B). Combining the HTP-3 (G490K, G728K) mutations with a C-terminal truncation of HIM-3 (ΔC; aa 1–245) greatly reduced HTP-1 association (Figures 6A and 6B). However, a detectable amount of HTP-1 (~10% of the amount bound to the wild-type complex) remained associated in a complex with HTP-3G490K, G728K and HIM-3ΔC, whereas mutating all six motifs within HTP-3 (6GK) completely abrogated both HTP-1 and HIM-3 binding to HTP-3 (Figure 6B). This reveals previously unrecognized modes of HTP-1 association with HTP-3, which might reflect either weak binding to the four central motifs or binding to HIM-3 through an interface other than its C-terminal tail.
To determine the consequences of reducing levels of HTP-1/2 on the chromosome axis in vivo, we introduced the GK mutation in the HIM-3 closure motif (G280K) into the endogenous him-3 gene. (Figures S7A–C). Worms homozygous for the him-3G280K mutation did not extend CHK-2 activity or result in chromosome missegregation (98% egg viability; 0.5% male self-progeny; 48% CHK-2 active zone) (Figures 6D–E and S7E). We combined the htp-3G490K, G728K transgene with him-3G280K. Consistent with our in vitro results, HTP-1/2 staining was greatly reduced along the chromosome axes in htp-3G490K, G728K; him-3G280K double mutants, but faint signal was still detectable (Figure S7F). These animals exhibited a marked reduction in egg viability (17.4%) and a strong Him phenotype (15.1% males) (Figure S7E), indicating defects in meiotic chromosome segregation. Synapsis was severely defective in these animals (Figure 6C), and phosphorylation of HIM-8/ZIMs was significantly extended (70% CHK-2 active zone) (Figures 6D and 6E). Thus, ~10% of the normal amount of HTP-1/2 along the chromosomes is not sufficient to support synapsis, but nevertheless mediates robust signaling to extend CHK-2 activity. By contrast, mutation of all six closure motifs within HTP-3 (6GK), which eliminates recruitment of HTP-1/2 and HIM-3 to the axis (Figure S7F), abrogated both synapsis and feedback control (46% CHK-2 active zone) (Figures 6C–E). Importantly, HTP-36GK localized normally to chromosomes and both HIM-3 and HTP-1/2 were expressed at normal levels (not shown). Thus, we conclude that recruitment of HTP-1/2 and HIM-3 to the meiotic chromosome axis is essential for feedback regulation of CHK-2.
Feedback control of CHK-2 does not require PCs
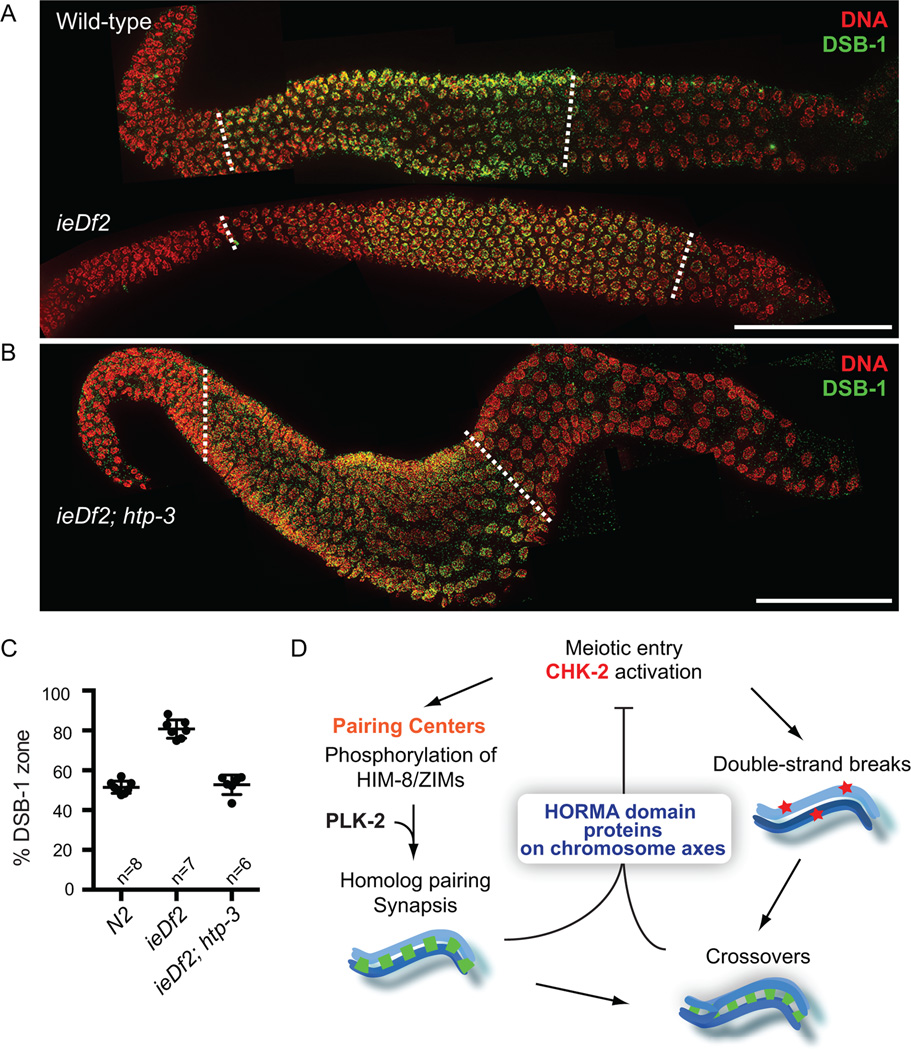
As CHK-2 was detected primarily at PCs (Figure 1), we next tested whether the PC activity is required for feedback regulation of CHK-2 using a deletion (ieDf2) that eliminates all four zinc finger proteins (Harper et al., 2011). Because the epitope recognized by our phospho-HIM-8/ZIMs antibody is absent in ieDf2 animals, we examined the chromosomal localization of DSB-1 as a reporter for CHK-2 activity. CHK-2 is required for normal chromosomal loading of DSB-1 and DSB-2 (Rosu et al., 2013; Stamper et al., 2013) and we have found that DSB-1 localization coincides with the HIM-8/ZIM phosphoepitope in wild-type and mutant animals (Figures S3A–B). We found that DSB-1 staining was greatly extended in the germline of ieDf2 animals (81% in ieDf2 vs. 52% in wild-type) (Figures 7A and 7C) in response to their synapsis and crossover defects (Harper et al., 2011). Consistent with the role of chromosome axes in mediating feedback, deletion of HTP-3 completely suppressed the extension of DSB-1 staining in ieDf2 animals (53% DSB-1 zone in ieDf2; htp-3) (Figures 7B and 7C). Thus, PCs are dispensable for both activation and the feedback control of CHK-2. Notably, CHK-2 was still activated in ieDf2; htp-3 double mutants, indicating that kinase activation occurs at meiotic entry in the absence of both functional PCs and chromosome axes.
Figure 7. Feedback control of CHK-2 does not require PCs.
(A–B) Dissected gonads from wild-type, ieDf2 and ieDf2; htp-3 hermaphrodites were stained for DAPI (red), and DSB-1 (green). Scale bars, 50 µm. The DSB-1 zone is marked by white dotted lines. (C) Quantification of the DSB-1 zone in the indicated genotypes. Numbers of gonads scored are indicated below, and error bars are standard deviations. ieDf2 mutants show significant differences from N2 (p<0.0001), while ieDf2; htp-3 mutants do not (p=0.8014), by ordinary one-way ANOVA. (D) A model for the feedback control of CHK-2. CHK-2 governs two major pathways leading to crossover formation in C. elegans. CHK-2 acts at PCs to phosphorylate HIM-8 and ZIMs and primes their recruitment of PLK-2 to promote pairing and synapsis. CHK-2 is also required for the programmed DSBs that initiate meiotic recombination, independently of PCs. Defects in synapsis or crossover formation are monitored through the meiotic HORMA domain proteins on chromosome axes, and extend CHK-2 activity, thereby delaying meiotic progression. This feedback control of CHK-2 underlies the checkpoint mechanism that ensures crossover formation in C. elegans meiosis.
Discussion
Here we establish a key kinase cascade that acts at PCs to promote homolog pairing and synapsis. CHK-2 phosphorylates the zinc finger proteins that specify PCs, which in turn primes their recruitment of PLK-2 (Figure 7D). By recruiting these two kinases, PCs serve as signaling hubs that mediate early meiotic chromosome dynamics. While most Chk2 family kinases function downstream of ATM/ATR in the DNA damage response (Matsuoka et al., 1998), C. elegans CHK-2 does not require DSBs for its initial activation and lacks clusters of SQ/TQ sites that define ATM/ATR targets. Instead, CHK-2 is essential for DSB formation and acts as a master regulator that governs pairing, synapsis, and recombination during meiotic prophase (MacQueen and Villeneuve, 2001). This rewiring of regulatory circuitry may have accompanied the emergence of homolog pairing mechanisms that function independently of meiotic recombination in C. elegans (Dernburg et al., 1998).
We demonstrate that CHK-2’s kinase activity normally declines once all chromosomes have accomplished synapsis and crossing-over, but is prolonged in mutants that disrupt synapsis or crossover formation. Adding to recent evidence for feedback regulation of crossover formation (Rosu et al., 2013; Stamper et al., 2013; Woglar et al., 2013), we now show that two distinct pathways controlled by CHK-2 (synapsis and meiotic recombination) both feed back to regulate CHK-2 activity (Figure 7D). This common circuitry delays meiotic progression and extends temporal window for active pairing, synapsis, and DSB formation. This mechanism meets the original definition of a cell cycle checkpoint (Hartwell and Weinert, 1989) in that it makes meiotic progression contingent on the formation of a crossover on each homolog pair. Our findings that mutations in the meiotic HORMA domain proteins fail to extend CHK-2 activity despite severe defects in pairing, synapsis, and crossover formation highlights the central and conserved role of these proteins in checkpoint control.
Although CHK-2 is primarily detected at PCs, it clearly acts elsewhere within the nucleus. Our evidence demonstrates that the meiotic checkpoint is fully functional even in the absence of PC activity and that CHK-2 feedback regulation is rather a nucleus-wide response. This directly refutes the recent proposal that HIM-8 is required for feedback in early meiotic prophase (Silva et al., 2014). This confusion arose because the previous study did not directly monitor CHK-2 activity, but instead the recruitment of PLK-2 to PCs, which requires not only CHK-2 activity but also the zinc finger proteins, specifically HIM-8 under conditions where the ZIMs do not remain phosphorylated.
Our work explains the previously enigmatic observation that the polarized nuclear organization that defines the transition zone is greatly extended in synapsis-defective mutants (Colaiacovo et al., 2003; MacQueen et al., 2002; Phillips et al., 2005). Synapsis is initiated in the transition zone, and thus a nucleus-wide signal generated from asynapsed chromosomes (e.g. in syp-2, him-8, and plk-2; plk-1(RNAi)) can maintain the phosphorylation of all PC proteins by CHK-2. On the other hand, a signal from chromosomes that have synapsed but lack crossovers (e.g. in zhp-3, msh-5, spo-11, and him-5) would be generated in early pachytene, when ZIMs are no longer detected at the autosomal PCs and transition zone morphology is no longer observed (Phillips and Dernburg, 2006). At this stage of meiosis, only the phosphoepitope on HIM-8 normally remains evident, and thus only HIM-8 phosphorylation appears to be prolonged by crossover failures. Why HIM-8 phosphorylation and binding to the X chromosome PC persists longer than for the ZIMs is unknown; nevertheless, feedback regulation maintains the status of the meiotic cell cycle at which errors are first detected.
Different meiotic lesions extend the zone of CHK-2 activity to different degrees, indicating that feedback is graded rather than binary. Clear evidence for this comes from our analysis of a series of mutations in HTP-3 that disrupt individual HIM-3 recruitment: as the synapsis defects become progressively more severe, the duration of CHK-2 activation increases (Figure 5). This graded meiotic checkpoint resembles the rheostat-like behavior of the spindle assembly checkpoint (SAC) that monitors kinetochore-microtubule attachment during mitosis (Collin et al., 2013; Dick and Gerlich, 2013). The strength of the SAC correlates with the amount of Mad2 recruited to kinetochores, which in turn depends on a hierarchical assembly of other checkpoint proteins at the kinetochore (London and Biggins, 2014). We have now shown that both HTP-1 and HTP-2 are required to generate a maximal signal from asynapsed chromosomes, and that both HIM-3 and HTP-1/2 are required for the feedback regulation. HTP-3 contains two binding sites for HTP-1/2 and four for HIM-3, which can recruit additional HTP-1/2 through its C-terminal motif (Kim et al., 2014). Thus, HTP-3 acts as a scaffold for checkpoint activation, and the hierarchical network of HIM-3 and HTP-1/2 may allow flexibility in responding to diverse meiotic defects.
A recent report showed that C. elegans homozygous for a point mutation within the HORMA domain of HTP-1 (M127K), which disrupts its association with chromosome axes, displayed an extended zone of PLK-2 localization to PCs in response to asynapsis (Silva et al., 2014), which we show here is a direct consequence of prolonged CHK-2 activity. This apparent feedback activity was attributed to a signal from the soluble nuclear pool of mutant HTP-1 protein. We note that this conclusion is inconsistent with our findings that mutating the six closure motifs in HTP-3, which prevents recruitment of HTP-1/2 (and HIM-3) to the chromosome axes, fully abrogates feedback regulation of CHK-2 (Figures 5 and 6). We also report that recruitment of a small amount of HTP-1 and/or HTP-2 to chromosomes is sufficient to support feedback signaling. We therefore think it likely that feedback regulation in htp-1M127K mutants is mediated by HTP-2 along the chromosome axis, rather than the mutant version of HTP-1 in the nucleoplasm.
As the formation of chromosome axes is prerequisite for pairing, synapsis, and meiotic recombination, the HORMA domain proteins are ideally situated to monitor chromosomal events during meiotic prophase. This implies that the axis must undergo structural or biochemical changes during meiotic progression to sense and/or signal the status of meiotic events. Indeed, meiotic HORMA domain proteins in S. cerevisiae (Hop1) and mice (HORMAD1/2) are removed from synapsed axes by the AAA+ ATPase Pch2/Trip13 (Börner et al., 2008; Joshi et al., 2009; Roig et al., 2010; Wojtasz et al., 2009), suggesting a mechanism by which the checkpoint may be silenced in response to synapsis. However, the C. elegans ortholog of Pch2/Trip13, PCH-2, is largely dispensable for meiotic processes, and is not required for axis remodeling (not shown). Therefore, how the assembly of meiotic HORMA domain proteins is dynamically regulated to monitor chromosomal events and coordinate with meiotic progression remains to be determined.
We recently confirmed that HIM-3 and HTP-1/2 are structurally very similar to Mad2 (Kim et al., 2014), raising the intriguing possibility that the meiotic HORMA domain proteins might undergo a Mad2-like dimerization and/or conformational change to mediate checkpoint signaling (De Antoni et al., 2005; Luo et al., 2004). However, mutating the αC helix of HTP-1 (F180A, K184A), corresponding to the dimerization interface of Mad2 (Mapelli et al., 2007), did not abrogate the feedback regulation (not shown), indicating that this interface is not essential for signal transduction by HTP-1. Moreover, it is not yet known whether the meiotic HORMA proteins bind to downstream effectors through their HORMA domains in the same way that Mad2 transduces checkpoint signaling to the anaphase-promoting complex. Future identification of signaling effectors that directly regulate CHK-2 (e.g., phosphatases) will help to reveal how axis proteins collaborate to mediate the meiotic checkpoint.
Experimental Procedures
For full experimental details, see Supplementary Material.
Antibody production
Polyclonal CHK-2 antibodies were raised in rabbits against a KLH-conjugated synthetic peptide containing the N-terminal 14 amino acids of CHK-2 (Pocono Rabbit Farm), and the immune serum was directly used for immunofluorescence.
A synthetic phospho-peptide (DTPRFSpTPIVPNVC) corresponding to the region of HIM-8 flanking T64 (Biomatik) was coupled to maleimide-activated KLH according to the manufacturer’s instructions (Thermo Scientific) and injected into rabbits (Covance). Polyclonal pT64 antibodies were affinity-purified by passing the immune serum through SulfoLink Coupling Resins (Thermo Scientific) coupled to a nonphospho-peptide (DTPRFSTPIVPNVC), followed by binding to and elution from phosphopeptide-coupled resins.
Protein expression and purification
The full-length open reading frame (ORF) of CHK-2 was amplified from a C. elegans cDNA library and cloned into pFastBac1 (Life Technologies). For protein purification, CHK-2 was tagged at its N-terminus with GST. GST-CHK-2 was expressed in Sf9 cells using the Bac-to-Bac system (Life Technologies) and purified over Glutathione Sepharose (GE Life Sciences) using standard protocols.
Full-length ORFs of HIM-8, ZIM-3, HIM-3, HTP-1, SYP-1, SYP-2, SYP-3, and SYP-4 were amplified from a C. elegans cDNA library and cloned into either pGEX6P-1 (GE Life Sciences) or pET23d (EMD Millipore). HIM-8 and ZIM-3 were tagged at their N-termini with GST, and SYP-2 was tagged at its C-terminus with 6His. HTP-1 and HIM-3 fused with 6His at their C-termini were co-expressed as previously described (Kim et al., 2014). SYP-1, SYP-3, and SYP-4 were tagged at their N-termini with 6His-Maltose Binding Protein (MBP) to improve protein solubility. All protein expression was induced at 20°C for 16 hours with 0.1 mM IPTG in Rosetta(DE3)pLysS, and proteins were purified using either Glutathione Sepharose (GE Life Sciences) or HisTrap HP (GE Life Sciences), followed by ion-exchange chromatography.
For HTP-3 Strep pulldown assays, full-length ORFs of HTP-3, HTP-1, and HIM-3 were cloned into a polycistronic vector for expression. HTP-3 was tagged N-terminally with Strep-tag II, and the triple-protein complexes of HTP-3:HTP-1:HIM-3 were expressed and purified using StrepTrap HP (GE Healthcare) as previously described (Kim et al., 2014). For HTP-3 GK mutants, site-directed mutagenesis and isothermal assembly with synthetic gene blocks were used to generate GK mutations in the noted positions (G490K, G532K, G560K, G652K, G683K, or G728K).
In vitro kinase assays
In vitro kinase assays were performed in 20 mM HEPES pH 7.5, 25 mM KCl, 1 mM MgCl2, 1 mM DTT in the presence of 0.2 mM Mg-ATP and 50 µCi/ml γ -32P ATP. 2 µM of GST-HIM-8, HTP-1:HIM-3-6His, 6His-MBP-SYP-1, SYP-2-6His, 6His-MBP-SYP-3 and 6His-MBP-SYP-4 were incubated with 0.2 µM of GST-CHK-2 at room temperature for 1 hour. Kinase reactions were terminated by addition of sample buffer and analyzed by SDS-PAGE.
C. elegans strains and immunofluorescence
him-8T64A and him-3G280K strains were generated by Cas9/CRISPR-mediated homologous recombination. Strains carrying wild-type and mutant htp-3-gfp transgenes were generated by Mos1-mediated single copy insertion (MosSCI) (Frøkjaer-Jensen et al., 2008), as previously described (Kim et al., 2014). Immunofluorescence of dissected gonads was performed as previously described (Phillips et al., 2009).
Supplementary Material
Acknowledgements
We thank Ann Fischer for assistance with Sf9 cell culture. Mass spectrometry was performed by Lori Kohlstaedt in the Proteomics/Mass Spectrometry Laboratory at UC Berkeley, supported in part by NIH S10 Instrumentation Grant S10RR025622. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by funding from the Damon Runyon Cancer Research Foundation (DRG 2084-11) to YK, and the National Institute of Health (GM065591) and the Howard Hughes Medical Institute to AFD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
YK and AFD conceived and designed the study. YK conducted the experiments, assisted by NK. YK and AFD wrote the manuscript.
References
- Aravind L, Koonin EV. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends in Biochemical Sciences. 1998;23:284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Pachytene Exit Controlled by Reversal of Mek1-Dependent Phosphorylation. Cell. 2000;101:211–221. doi: 10.1016/S0092-8674(00)80831-4. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS genetics. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Börner GV, Barot A, Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proceedings of the National Academy of Sciences. 2008;105:3327–3332. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo JsA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the Axial Element Protein Hop1 by Mec1/Tel1 Ensures Meiotic Interhomolog Recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal Complex Assembly in C. elegans Is Dispensable for Loading Strand-Exchange Proteins but Critical for Proper Completion of Recombination. Developmental Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nature Cell Biology. 2013;15:1378–1385. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes & Development. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nature Cell Biology. 2011;13:599–610. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Current Biology. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic Recombination in C. elegans Initiates by a Conserved Mechanism and Is Dispensable for Homologous Chromosome Synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Dick AE, Gerlich DW. Kinetic framework of spindle assembly checkpoint signalling. Nature Cell Biology. 2013;15:1370–1377. doi: 10.1038/ncb2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. Meiotic Pachytene Arrest in MLH1-Deficient Mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Elia AEH, Cantley LC, Yaffe MB. Proteomic Screen Finds pSer/pThr-Binding Domain Localizing Plk1 to Mitotic Substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Schüpbach T. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nature cell biology. 1999;1:354–357. doi: 10.1038/14046. [DOI] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. HTP-3 Links DSB Formation with Homolog Pairing and Crossing Over during C. elegans Meiosis. Developmental Cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Harper NC, Rillo R, Jover-Gil S, Assaf Z, Bhalla N, Dernburg AF. Pairing Centers Recruit a Polo-like Kinase to Orchestrate Meiotic Chromosome Dynamics in C. elegans. Developmental Cell. 2011;21:934–947. doi: 10.1016/j.devcel.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Aoki H, Mori A, Sasagawa Y, Takanami T, Takahashi H. Caenorhabditis elegans Chk2-like gene is essential for meiosis but dispensable for DNA repair. FEBS letters. 2000;485:35. doi: 10.1016/s0014-5793(00)02178-5. [DOI] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, Börner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS genetics. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 Is Required for Both Normal and Radiation-Induced Meiotic Crossing Over but Not for Completion of Meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Rosenberg SC, Kugel CL, Kostow N, Rog O, Davydov V, Su TY, Dernburg AF, Corbett KD. The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins. Developmental Cell. 2014;31:487–502. doi: 10.1016/j.devcel.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S, Woglar A, Jantsch V, Zetka M. Polo Kinases Establish Links between Meiotic Chromosomes and Cytoskeletal Forces Essential for Homolog Pairing. Developmental Cell. 2011;21:948–958. doi: 10.1016/j.devcel.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and Functional Versatility of the FHA Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2. Molecular Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nature Reviews Molecular Cell Biology. 2014;15:736–748. doi: 10.1038/nrm3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nature Structural & Molecular Biology. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. 1996 doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MnP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Development. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Hochwagen A. Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends in cell biology. 2011;21:393–400. doi: 10.1016/j.tcb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes & Development. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Schvarzstein M, Barroso C, Lightfoot J, Dernburg AF, Villeneuve AM. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes & Development. 2008;22:2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes & Development. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to Cell Cycle Regulation by the Chk2 Protein Kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Meneely PM, McGovern OL, Heinis FI, Yanowitz JL. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics. 2012;190:1251–1266. doi: 10.1534/genetics.111.137463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Villeneuve A, Hillers K. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Molecular and cellular biology. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Molecular biology of the cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill T, Giarratani L, Chen P, Iyer L, Lee C-H, Bobiak M, Kanai F, Zhou B-B, Chung JH, Rathbun GA. Determination of Substrate Motifs for Human Chk1 and hCds1/Chk2 by the Oriented Peptide Library Approach. Journal of Biological Chemistry. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- Oishi I, Iwai K, Kagohashi Y, Fujimoto H, Kariya K-I, Kataoka T, Sawa H, Okano H, Otani H, Yamamura H, et al. Critical Role of Caenorhabditis elegans Homologs of Cds1 (Chk2)-Related Kinases in Meiotic Recombination. Mol Cell Biol. 2001;21:1329–1335. doi: 10.1128/MCB.21.4.1329-1335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The Nuclear Envelope Protein Matefin/SUN-1 Is Required for Homologous Pairing in C. elegans Meiosis. Developmental Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y, et al. Meiotic Chromosome Homology Search Involves Modifications of the Nuclear Envelope Protein Matefin/SUN-1. Cell. 2009;139:920–933. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Dernburg AF. A Family of Zinc-Finger Proteins Is Required for Chromosome-Specific Pairing and Synapsis during Meiosis in C. elegans. Developmental Cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol. 2009;11:934–942. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF. HIM-8 Binds to the X Chromosome Pairing Center and Mediates Chromosome-Specific Meiotic Synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Molecular cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM. The pachytene checkpoint. Trends in Genetics. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- Rog O, Dernburg AF. Direct Visualization Reveals Kinetics of Meiotic Chromosome Synapsis. Cell reports. 2015;10:1639–1645. doi: 10.1016/j.celrep.2015.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, Keeney S. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS genetics. 2010;6:e1001062. doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM. The C. elegans DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance. PLoS genetics. 2013;9:e1003674. doi: 10.1371/journal.pgen.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal Forces Span the Nuclear Envelope to Coordinate Meiotic Chromosome Pairing and Synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y-H, Choi Y, Erdin SU, Yatsenko SA, Kloc M, Yang F, Wang PJ, Meistrich ML, Rajkovic A. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS genetics. 2010;6:e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N, Ferrandiz N, Barroso C, Tognetti S, Lightfoot J, Telecan O, Encheva V, Faull P, Hanni S, Furger A, et al. The Fidelity of Synaptonemal Complex Assembly Is Regulated by a Signaling Mechanism that Controls Early Meiotic Progression. Developmental Cell. 2014;31:503–511. doi: 10.1016/j.devcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in Caenorhabditis elegans, Illuminates a Crossover Assurance Checkpoint. PLoS genetics. 2013;9:e1003679. doi: 10.1371/journal.pgen.1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, Hochwagen A. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harbor perspectives in biology. 2014;6:a016675. doi: 10.1101/cshperspect.a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, La Volpe A, Jantsch V. Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement. PLoS genetics. 2013;9:e1003335. doi: 10.1371/journal.pgen.1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Remenyi A, Turner JMA, et al. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and - dependent mechanisms. Genes & Development. 2012;26:958–973. doi: 10.1101/gad.187559.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS genetics. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. The Journal of Cell Biology. 2012;196:47–64. doi: 10.1083/jcb.201106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes & Development. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Development. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.